Summary
The RD1 genomic region is present in virulent strains of Mycobacterium tuberculosis (MTB), missing from the vaccine strain M. bovis BCG, and its importance to virulence has been established experimentally. Based on in silico analysis, it has been suggested that RD1 may encode a novel secretion system, but the mechanism by which this region affects virulence is unknown. Here we examined mutants disrupted in five individual RD1 genes. Both in vitro and in vivo, each mutant displayed an attenuated phenotype very similar to a mutant missing the entire RD1 region. Genetic complementation of individual genes restored virulence. Attenuated mutants could multiply within THP-1 cells, but they were unable to spread to uninfected macrophages. We also examined export of two immunodominant RD1 proteins, CFP-10 and ESAT-6. Export of these proteins was greatly reduced or abolished in each attenuated mutant. Again, genetic complementation restored a wild-type phenotype. Our results indicate that RD1 genes work together to form a single virulence determinant, and argue that RD1 encodes a novel specialized secretion system that is required for pathogenesis of MTB.
Introduction
Though Mycobacterium tuberculosis (MTB) has infected humans for thousands of years, its virulence mechanisms remain largely undefined. The availability of genomic sequence (Cole et al., 1998) has facilitated development of new tools to probe for genes involved in pathogenesis (Sassetti and Rubin, 2002). Comparisons between the avirulent M. bovis BCG (the MTB vaccine strain) and virulent MTB led to the identification of several genetic regions of difference (Mahairas et al., 1996; Behr et al., 1999; Gordon et al., 1999). Only one of these, region of difference 1 (RD1), is absent from every strain of BCG and present in every virulent strain tested (Mahairas et al., 1996; Talbot et al., 1997; Behr et al., 1999; Kearns et al., 1999). RD1-encoded proteins have been investigated for utility in diagnosing tuberculosis (Harboe et al., 1996; Pollock and Andersen, 1997) and for differentiating latent infection from BCG vaccination (Lalvani et al., 2001). In addition, the link between RD1 and MTB virulence has been demonstrated experimentally. Deletion of the 9.5 kb RD1 region from MTB results in attenuation similar to BCG in cultured macrophages and mice (Lewis et al., 2003). In another study, disruption of the RD1-encoded esat-6 of M. bovis produced an attenuated phenotype in guinea pigs (Wards et al., 2000). Finally, on introduction of RD1 into BCG, the RD1-restored strain persisted longer in immunocompetent mice and grew to higher numbers in SCID mice than the BCG Pasteur parent (Pym et al., 2002), though virulence was still much reduced relative to MTB.
Though none of the nine open reading frames (ORFs) that comprise RD1 have a biochemically assigned function, this region has been carefully scrutinized in silico (Tekaia et al., 1999; Gey Van Pittius et al., 2001; Pallen, 2002). Rv3870 (just upstream of the 5′ RD1 border) and Rv3871 are predicted to be part of a larger transcriptional unit that includes an ATP-dependent chaperone and a membrane-bound protein of unknown function (Neuwald et al., 1999; Gey Van Pittius et al., 2001; Pym et al., 2003). The products of Rv3870 and Rv3871 may work together to form a membrane-bound ATPase. Rv3872 is a PE and Rv3873 is a PPE, two superfamilies of repetitive proteins with no known function (Cole et al., 1998), though some family members may play roles in mycobacterial pathogenesis (Camacho et al., 1999; Ramakrishnan et al., 2000). Rv3874 (also known as cfp-10, 10 kDa culture filtrate protein) and Rv3875 (also known as esat-6, 6 kDa early secretory antigenic target) are co-transcribed, exported proteins (Berthet et al., 1998) of the ESAT-6/WXG-100 superfamily (Pallen, 2002) that form a tight 1:1 complex with each other in vitro (Renshaw et al., 2002). CFP-10 and ESAT-6 are potent T-cell antigens that are recognized by TB patient sera (Ulrichs et al., 1998; Lein et al., 1999; Skjot et al., 2000; Brusasca et al., 2001). Rv3876 encodes a protein with a proline-rich N-terminal region and homology in its C-terminal portion to a Pseudomonas aeruginosa protein involved in flagellar biosynthesis, including a deviant Walker A motif (Koonin, 1993). Rv3877 is an integral membrane protein of unknown function. The last ORFs in RD1 (Rv3878 and 3879c) are not essential for MTB virulence as they are interrupted in at least one virulent clinical MTB isolate (Pym et al., 2003).
The predicted functions of several RD1 region genes suggest they may have roles in protein translocation. Further, as CFP-10 and ESAT-6 lack clear secretion signals, they may require a novel secretion machinery for export (Braunstein and Belisle, 2000), and components of RD1 may form that machinery (Cole et al., 1998; Tekaia et al., 1999; Gey Van Pittius et al., 2001; Pallen, 2002; Lewis et al., 2003). Consistent with this idea, recombinant BCG expressed but did not secrete ESAT-6 unless a heterologous signal sequence was added (Bao et al., 2003), and complementation of BCG with a 32 kb segment including the entire RD1 region resulted in export of CFP-10 and ESAT-6 into the culture supernatant (Pym et al., 2003).
We envision three broad possibilities by which RD1 genes might influence the pathogenesis of MTB: (i) a single gene may be responsible; (ii) two or more genes may contribute to virulence independently of one another; (iii) multiple RD1-encoded proteins may work together to form a single virulence machine. By making mutations in individual genes within RD1 we can test each of these possibilities. Here we describe studies with mutants in five relevant genes: Rv3870, Rv3871, Rv3874 (cfp-10), Rv3875 (esat-6) and Rv3876. We have tested these mutants in the human macrophage-like THP-1 cell line and in a murine infection model. To assess more directly a possible role for RD1 in protein export, we examined effects of individual mutations on secretion of CFP-10 and ESAT-6. Our results indicate that components of RD1 are required both for translocation of CFP-10 and ESAT-6 to the culture supernatant and for virulence of MTB.
Results
Mutation of genes within RD1
Mutants in individual genes within RD1 should be useful in defining the mechanism by which RD1 affects MTB virulence. We assembled a series of relevant mutants (Table 1). C. Sassetti and E. Rubin (Harvard University) generously provided transposon insertion mutants. We defined the precise site of transposon insertion in each case: H37Rv:tn3870, H37Rv:tn3871, H37Rv:tn3874 and H37Rv:tn3876 (data not shown). We also analysed mutants H37Rv:tn3864, and H37Rv:tn0982. Rv3864 is only ∼7 kb upstream of the RD1 deletion of BCG, and it is part of the related RD1 deletion of M. microti (Brodin et al., 2002). The mutant in Rv0982 (mprB, a probable two-component sensor kinase), a physically distant and unlinked gene, was included as a potential negative control.
Table 1.
Strains and plasmids used in this study.
| Strain name | Marker | Features | Phenotype in vitro (* and in vivo) |
|---|---|---|---|
| H37Rv | none | wild-type | virulent* |
| H37Rv:tn0982 | kan | tn Rv0982 | virulent |
| H37Rv:tn3864 | kan | tn Rv3864 | virulent |
| H37Rv:ΔRD1 | kan | deletion of RD1 | attenuated* |
| H37Rv:tn3870 | kan | tn Rv3870 | attenuated* |
| H37Rv:tn3871 | kan | tn Rv3871 | attenuated |
| H37Rv:tn3874 | kan | tn Rv3874 | attenuated |
| H37Rv:Δ3875 | hyg | deletion of Rv3875 | attenuated* |
| H37Rv:tn3876 | kan | tn Rv3876 | attenuated* |
| Plasmid | |||
| pKG18 | hyg | Rv3870, Rv3871, Rv3872 | |
| pMH406 | kan | Rv3874, Rv3875 | |
| pMH408 | hyg | Rv3874, Rv3875 | |
| pMH429 | hyg | Rv3876 |
kan = kanamycin, hyg = hygromycin, tn = transposon insertion.
ESAT-6 (Rv3875) is the best-characterized protein within RD1. It has been recognized as an important stimulator of T-cells in vitro and in vivo (Ulrichs et al., 1998; Lein et al., 1999; Skjot et al., 2000; Brusasca et al., 2001), and we wanted to examine its possible role in MTB virulence. We created a precise deletion of Rv3875 from H37Rv using the conditionally replicating phage transduction system (Bardarov et al., 2002; Braunstein et al., 2002). We confirmed the deletion (H37Rv:Δ3875) by Southern blot (data not shown).
Whether produced by precise deletion or transposon insertion, mutations sometimes have unexpected effects on neighbouring genes. To assess whether such effects occurred here, we generated RNA from log-phase cultures and measured gene expression by real-time PCR. We analysed mutants (genes assayed in parentheses): H37Rv:tn3870 (Rv3871, Rv3872, Rv3875), H37Rv:tn3874 (Rv3871, Rv3875, Rv3877), H37Rv:Δ3875 (Rv3877) and H37Rv:tn3876 (Rv3871, Rv3875, Rv3877). In each of three biological samples from each mutant, all ORFs tested were expressed at wild-type levels (data not shown). Also, depending on the site of insertion, transposon mutagenesis may leave residual gene activity. Because none of the RD1 region genes have defined biochemical functions, we cannot completely exclude this possibility at this time.
Individual mutants are attenuated for growth and cytotoxicity in THP-1 cells
Macrophages are among the first cells encountered by MTB in vivo and growth in macrophages has been associated with virulence in vivo (Fenton and Vermeulen, 1996; Silver et al., 1998; Zhang et al., 1998). We have previously used growth in cultured macrophages to assess virulence of the ΔRD1 mutant (Lewis et al., 2003). In both peripheral blood mononuclear cells and the human macrophage-like THP-1 cell line, H37Rv multiplied and rapidly destroyed the cell monolayer, whereas H37Rv:ΔRD1 grew minimally and produced no discernable cytotoxicity. To determine the effect of disrupting individual genes within RD1, we studied the ability of mutant strains to grow and cause cytotoxicity in THP-1 cells (Fig. 1A and B). Each RD1-related mutant (H37Rv:tn3870, H37Rv:tn3871, H37Rv:tn3874, H37Rv:Δ3875 and H37Rv:tn3876) was attenuated for growth and cytotoxicity to THP-1 cells. Results were indistinguishable from those obtained with H37Rv:ΔRD1. In contrast, mutants H37Rv:tn3864 and H37Rv:tn0982 exhibited growth and cytotoxicity very similar to wild-type H37Rv.
Fig. 1.
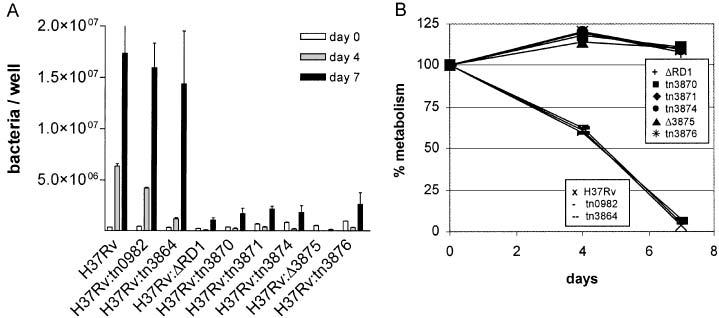
Bacterial growth (A) and cytotoxicity (B) by individual mutants in the human macrophage-like THP-1 cell line. Bacteria were quantified by luciferase assay or by plating for colony forming units. Macrophage metabolism was measured by a redox dye, and a decrease in metabolism is taken as an indication of cytotoxicity. Means and standard deviation (SD) for each time-point are from one infection done in triplicate, representative of (A) two or (B) three infections.
Complementation of mutants restores virulence
If attenuation results from mutations in individual RD1 region genes, introducing a functional copy of the ORF into each mutated strain should restore virulence. To test our ability to complement mutations in this manner, we produced a series of integrating plasmids with a strong, constitutive promoter driving expression of the relevant genes (Table 1). To complement H37Rv:tn3870 and H37Rv:tn3871 we made pKG18, which expresses Rv3870-Rv3871-Rv3872. For H37Rv:tn3874 and H37Rv:Δ3875 we made pMH408 and pMH406, respectively, both of which express Rv3874-Rv3875. For H37Rv:tn3876 we created pMH429, expressing Rv3876. These constructs were introduced by electroporation (Wards and Collins, 1996) and inserted into the chromosome of the appropriate mutants at the phage L5 attachment site (Stover et al., 1991); the presence of the transforming DNA was confirmed by PCR (data not shown).
We assessed the effects of genetic complementation in the THP-1 cell assay. For each of five mutants tested, introduction of functional copies of the mutated genes resulted in reversion to a virulent phenotype. In every case, growth (data not shown) and cytotoxicity (Fig. 2) were very similar to wild-type H37Rv. Collectively these THP-1 assay results are consistent with the idea that the RD1 genes under study work together to affect virulence, since individual mutations produce the same effect as deleting the entire RD1 region.
Fig. 2.
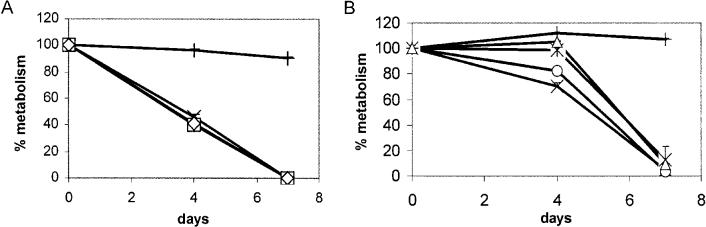
Complementation of mutants in THP-1 cells. Cells were infected with control strains H37Rv (X), H37Rv:ΔRD1 (+), and complements (A) H37Rv:tn3870::KG18 (□) and H37Rv:tn3871::KG18 (◇) or (B) H37Rv:tn3874::MH408 (○), H37Rv: Δ3875::MH406 (△) and H37Rv:tn3876::MH429 ([unk]). Data for each time point are means and SD from one infection done in triplicate, representative of two infections.
Mutant bacilli can grow within THP-1 cells, but are deficient at spreading to new cells
To examine the conduct of bacilli within THP-1 cells in more detail, we followed the infection by microscopy. Bacilli within cells on coverslips were stained with TB Quick and the percentage of infected cells was determined (Fig. 3A). With H37Rv, the percentage of infected cells increased steadily throughout the experiment, reaching a maximum value of 59% by day 7. By this time, the number of viable THP-1 cells remaining was substantially diminished (Fig. 1B). In contrast, after day 0 the ability of H37Rv:ΔRD1 to infect new cells was greatly reduced. As an indication of intracellular growth, we also enumerated the number of bacilli per infected cell (Fig. 3B). Throughout the assay, the great majority of H37Rv-infected cells contained ≤ 10 bacilli, even as the number of H37Rv-infected cells increased. MTB is toxic to macrophages (Molloy et al., 1994) and cells lysed as the bacilli grew. However, while the overall number of H37Rv:ΔRD1-infected cells remained almost constant, the proportion of cells containing > 10 bacilli steadily increased, indicating that H37Rv:ΔRD1 is capable of intracellular growth. Examination of cells infected with mutants H37Rv:tn3870 and H37Rv:Δ3875 revealed kinetics strikingly similar to H37Rv:ΔRD1 (data not shown). We conclude that RD1 region mutants are capable of intracellular growth but are deficient at spreading to new cells.
Fig. 3.
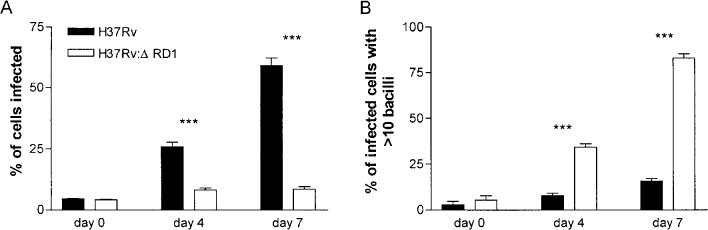
Enumeration of bacilli within THP-1 cells. Infected cells were stained with TB Quick and bacilli visualized by microscopy. Data are from one infection, representative of two. ***P < 0.0001.
Mutant bacilli are attenuated in mice
To characterize further the effects of single gene mutations within RD1 on growth and virulence of MTB, we examined the phenotype of selected mutants in vivo. We used a murine aerosol infection model, which closely mimics the normal route of human infection. We introduced strains H37Rv:tn3870, H37Rv:Δ3875 and H37Rv:tn3876 into C57BL/6 mice and measured bacterial burden in lung at 4 weeks post infection. Consistent with their phenotype in THP-1 cells, these three mutants were attenuated for growth in the lung (Fig. 4). Dissemination of mutants to the spleen was also impeded. Spleens of mice infected with H37Rv harboured ∼105 bacilli at 4 weeks, whereas spleens of mice infected with the mutants carried 100- to 1000-fold fewer bacilli (data not shown). Growth and dissemination of all three mutants was clearly distinct from H37Rv and generally similar to H37Rv:ΔRD1.
Fig. 4.
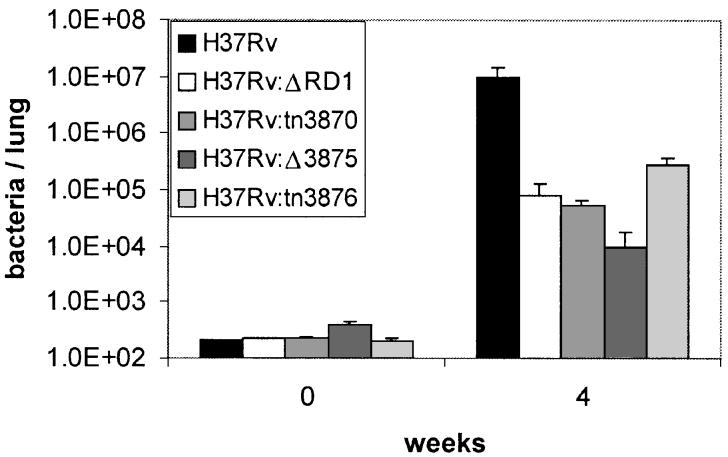
Bacterial growth in lungs after aerosol infection of 6–8 week-old C57BL/6 mice. Data at each time-point are the mean and SD of five mice per strain from one infection, representative of two.
Lung histopathology at 4 weeks post infection also revealed that single gene RD1 mutants are dramatically attenuated relative the H37Rv, similar to H37Rv:ΔRD1. Lungs of H37Rv-infected mice displayed early granulomas with inflammation involving ∼50% of the lung tissue (Fig. 5A). In contrast, lungs from mice infected with H37Rv:ΔRD1, H37Rv:tn3870 or H37Rv:Δ3875 were normal, with no inflammation detectable (Fig. 5B–D). Lungs from mice infected with H37Rv:tn3876 were predominantly normal, with very mild lymphocytic infiltration and inflammation restricted to airways, involving <5% of the lung tissue (Fig. 5E).
Fig. 5.
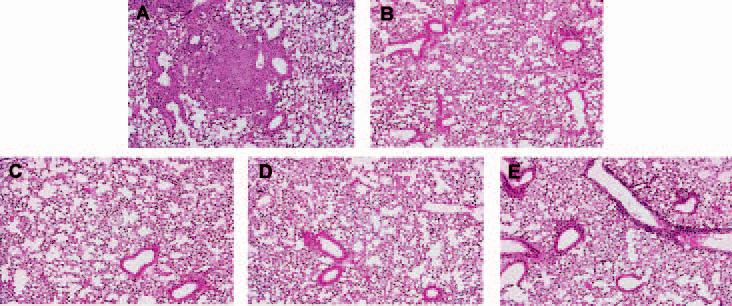
Lung histopathology of infected C57/BL6 mice at 4 weeks post infection. Mice were infected with (A) H37Rv (B) H37Rv:ΔRD1 (C) H37Rv:tn3870 (D) H37Rv:Δ3875 (E) H37Rv:tn3876.
Mutants are defective in export of ESAT-6
Based on the bioinformatics analysis of RD1 region genes and the link between RD1 and MTB virulence, several groups have proposed that RD1 encodes a novel secretion system that could export CFP-10 and ESAT-6 (Cole et al., 1998; Tekaia et al., 1999; Gey Van Pittius et al., 2001; Pallen, 2002; Lewis et al., 2003). Recent experiments with recombinant BCG expressing ESAT-6 have lent some experimental support to this idea (Bao et al., 2003; Pym et al., 2003).
To investigate this question in more detail and determine whether individual RD1 region genes play a role in protein translocation, we assayed several mutants for their ability to express and appropriately export ESAT-6 protein. Cells were grown from an OD600 = 0.1 for 6 days, harvested, and filtered culture supernatants were subjected to Western immunoblotting (Fig. 6). As expected, ESAT-6 was not detectable in H37Rv:ΔRD1, but it was abundant in culture filtrates of H37Rv. ESAT-6 was not detected in H37Rv:Δ3875. When complemented, however, this strain produced ESAT-6 and exported it to the culture filtrate. In the mutants H37Rv:tn3870 and H37Rv:tn3874 no secretion of ESAT-6 was observed (Fig. 6) even though the protein was detectable within cells (data not shown). This result is consistent with recent work showing that transformation of BCG with esat-6 yielded expression but not export of ESAT-6 protein (Bao et al., 2003). Complementation of these mutants resulted in detection of ESAT-6 in the culture filtrates.
Fig. 6.
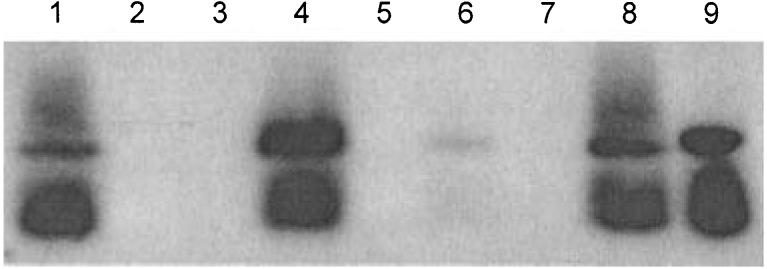
Detection of ESAT-6 in culture filtrates by Western immunoblot. Samples are (1) H37Rv (2) H37Rv: ΔRD1 (3) H37Rv:tn3870 (4) H37Rv:tn3870::KG18 (5) H37Rv:tn3874 (6) H37Rv:tn3874::MH408 (7) H37Rv:Δ3875 (8) H37Rv: Δ3875::MH406 (9) purified ESAT-6 protein.
None of the attenuated mutant strains exported any detectable ESAT-6 into the culture filtrates. Even those mutants that contained the gene and produced ESAT-6 (H37Rv:tn3870, H37Rv:tn3874) were unable to secrete it. However, in all cases examined, complementation restored both ESAT-6 export and virulence. It seemed unlikely that ESAT-6 secretion by the complemented strains could be due to a non-specific increase in cell permeability. However, to address this possibility we assayed supernatant fractions with antibodies raised against a culture filtrate protein preparation. The amount of reactive material in the supernatants was roughly equivalent from all strains (data not shown). In addition, H37Rv and H37Rv:ΔRD1 were identical in their sensitivity to the detergent sodium dodecyl sulphate (data not shown). These results argue that the export of ESAT-6 from complemented strains was not the result of some global permeability change.
Mutants are defective in export of CFP-10
To examine secretion of another RD1 protein CFP-10, an ELISPOT assay using the CD4+ T cell clone 38.1–1 (Lewinsohn et al., 2003) was employed. In these assays, the T cell clone was co-incubated with an autologous lymphoblastoid cell line (LCL) in the presence of either recombinant CFP-10 protein or mycobacterial cell pellet or supernatant. The release of IFN-γ was assessed by ELISPOT, where the maximum achievable spot number was 234 ± 13 spots using recombinant CFP-10 protein. At the minimum CFP-10 concentration tested (2 ng ml−1) 27 ± 4 spots were detected, indicating a minimum sensitivity for the assay. As shown in Fig. 7, incubation with either H37Rv cell pellet or culture filtrate resulted in high levels of T cell stimulation, corresponding to high levels of CFP-10 protein (Fig. 7B). As expected, mutation of Rv3874 (cfp-10) abolished T cell stimulation, as no IFN-γ was elicited by either fraction. Interestingly, deletion of Rv3875 (esat-6) resulted in no detectable CFP-10 in the culture filtrate, and a markedly diminished signal in the cell pellet. ESAT-6 and CFP-10 form a tight 1 : 1 dimer in vitro (Renshaw et al., 2002), and dimer formation may be important for CFP-10 stability in vivo. Levels of T cell stimulation from pellets of mutants H37Rv:tn3870, H37Rv:tn3871 and H37Rv:tn3876 were very similar to levels from H37Rv. However, the levels of T cell stimulation was markedly reduced from the culture filtrates of these attenuated mutants. Culture filtrates from complemented strains H37Rv:tn3870::KG18, H37Rv:tn3871::KG18, H37Rv:tn3874::MH408, H37Rv:Δ3875::MH406 and H37Rv:tn3876::MH429 with restored virulence all produced high levels of T cell stimulation.
Fig. 7.
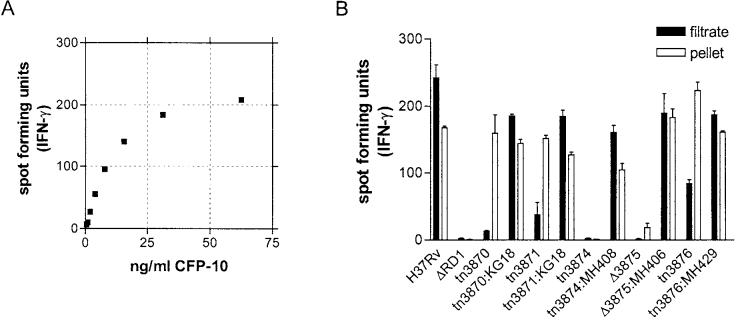
IFN-γ ELISPOT assay to determine the presence of CFP-10 protein. A. Standard curve with purified CFP-10 protein. Fewer than 10 spots were detected in the absence of antigen. Using recombinant protein, maximum achievable spot number was 234 ± 13. At a CFP-10 concentration of 2 ng ml−1, 27 ± 4 spots were detected indicating a minimum sensitivity for the assay.B. ELISPOT results from cultured filtrate and cell pellet samples. Data are mean and standard error of duplicate determinations, representative of two independent experiments.
Discussion
The attenuation of M. bovis to create BCG 80 years ago was an experiment in pathogenesis that is still not fully understood. We recently showed that deletion of RD1 from M. tuberculosis produced attenuation strikingly similar to BCG (Lewis et al., 2003). In addition, knock-in of RD1 into BCG resulted in a modest increase in virulence (Pym et al., 2002), and an earlier report showed that deletion of esat-6 from M. bovis yielded reduced pathology in guinea pigs (Wards et al., 2000). The mechanism by which RD1 affects MTB pathogenesis has remained unclear. To investigate this matter, we characterized MTB mutants disrupted at individual genes in and around the RD1-region. In the human macrophage-like THP-1 cell line, the single-gene RD1 mutants that we studied all showed attenuation equivalent to H37Rv:ΔRD1, which is missing the entire RD1 region (Fig. 1). In each case tested, re-introduction of the disrupted gene restored ability both to multiply within THP-1 cells at levels similar to wild-type (data not shown) and to destroy the macrophage monolayer (Fig. 2). These data argue that RD1-region genes work in concert to form a single virulence entity, since individual gene mutants are phenotypically identical to deletion of the entire RD1 region.
Examination of intracellular bacteria by microscopy revealed that infection with either H37Rv or H37Rv:ΔRD1 results in very similar uptake by THP-1 cells. However, over the course of a 7-day infection significant differences emerged. Whereas H37Rv continuously grows, lyses cells, and spreads to other cells, H37Rv:ΔRD1 is able to grow within cells but is deficient at causing cell lysis and subsequent spread (Fig. 3). At the same time, we observe only a small increase in the total numbers of bacilli in H37Rv:ΔRD1-infected cells (Fig. 1A), possibly because of some inadequacy in our measurement (perhaps from clumping of bacilli tightly packed within cells) and/or some killing of bacilli by cells. These results indicate that RD1 genes act not at initial bacterial uptake or even intracellular growth, but at a later step crucial to virulence, perhaps in lysis of the cell.
Animal infections with selected mycobacterial strains produced comparable results. Four weeks following low dose aerosol infection of C57BL/6 mice, bacterial numbers of H37Rv:tn3870, H37Rv:tn3876 and H37Rv:Δ3875 were clearly reduced relative to H37Rv, similar to H37Rv:ΔRD1 (Fig. 4). By histopathology, individual RD1 region mutants were also attenuated compared with H37Rv and similar to H37Rv:ΔRD1. Interestingly however, the histopathology produced by H37Rv:tn3876 was subtly different (Fig. 4E). This difference may indicate that the function of this gene product is somewhat less critical to the virulence entity encoded by RD1. Alternatively, the transposon insertion within Rv3876 may have left some portion of its biological function intact. In this light it is interesting that both pellets and filtrates of H37Rv:tn3876 contained more CFP-10 than any other attenuated mutant (Fig. 7B).
It has been argued that RD1 encodes a novel multicomponent secretion machine for translocation of CFP-10 and ESAT-6 (Cole et al., 1998; Tekaia et al., 1999; Gey Van Pittius et al., 2001; Pallen, 2002; Lewis et al., 2003), but the function of this region and individual genes within it has remained unresolved. By Western blot, strains mutated in Rv3870, Rv3871 and Rv3874 (cfp-10) were negative for ESAT-6 in the culture supernatant (Fig. 6), although the protein was produced as it was weakly detectable in cell pellets (data not shown). The more sensitive ELISPOT assay for CFP-10 yielded very similar results (Fig. 7B). Mutants in the RD1-region genes Rv3870, Rv3871, Rv3875 (esat-6) and Rv3876 all produced CFP-10 but the level of this protein in supernatants was significantly reduced relative to wild-type H37Rv or complemented mutants. Thus individual genes in the RD1 region are required for secretion of ESAT-6 and CFP-10. A picture is starting to emerge as to how the RD1 region contributes to MTB virulence. Mutants lacking individual genes within RD1 cannot export ESAT-6 or CFP-10, and are avirulent. These mutants are capable of growth within macrophages but are deficient at spreading to new cells. In other bacteria, attenuating mutations with similar phenotypes have mapped to specialized secretion systems (Sansonetti, 2001; Stevens et al., 2002).
It could still be argued that the influence of Rv3870, Rv3871 and Rv3876 on protein export may be indirect. If these proteins affect the stability of ESAT-6, CFP-10 or the heterodimer, then reduced levels of exported protein may simply reflect less available substrate for export. We consider this possibility unlikely for three reasons. First, esat-6 and cfp-10 are co-transcribed (Berthet et al., 1998) and can form a tight heterodimer in the absence of any other MTB proteins (Renshaw et al., 2002). Second, bioinformatics clearly places Rv3870/Rv3871 and Rv3876 in the cell membrane, where they are well-positioned to act on secretion as opposed to protein stability. Finally, in all three mutants the level of CFP-10 in pellets was roughly the same or greater than in H37Rv (Fig. 7), yet the amount secreted was dramatically reduced. Likewise, the presence of some CFP-10 in the filtrate of mutants H37Rv:tn3870, H37Rv:tn3871 and H37Rv:tn3876 could suggest that secretion still operates, but more likely reflects protein released from lysed cells. However we cannot at present exclude the possible existence of an alternate means of protein export operating at a low level.
Altogether our experiments reveal that individual genes within RD1 work together to export important antigens ESAT-6 and CFP-10, and that export is linked with MTB virulence. All the MTB strains and complements that were positive for export of ESAT-6 were virulent in cell culture and animal assays, whereas all strains negative for ESAT-6 export were avirulent. Similarly, all strains shown to export CFP-10 at high levels (> 150 spot forming units) were virulent in cell culture and animals, whereas strains with less CFP-10 in supernatants were avirulent. These data argue that the RD1 region encodes a specialized protein secretion system that is tightly linked to the virulence of MTB. Specialized secretion systems are key virulence determinants of many other bacterial pathogens (Lee and Schneewind, 2001).
The genetic boundaries of the RD1 secretion system remain to be defined. Genes upstream of Rv3865 are apparently not essential, as we have shown that a mutant of Rv3864 has a wild-type phenotype (Fig. 1). Bioinformatics suggests that at least Rv3868 (a putative ATP-dependent chaperone) and Rv3869 (a putative membrane protein) could possibly play roles in protein secretion. At the 3′ boundary, Rv3876 is clearly required for secretion and virulence but our data do not address Rv3877, a putative integral membrane protein of unknown function that may be involved as well. The remaining RD1 ORFs (Rv3878 and 3879c) are presumably not part of a virulence-linked secretion machine as they are interrupted in at least one virulent clinical MTB isolate (Pym et al., 2003).
Our data are consistent with the possibility that ESAT-6 and/or CFP-10 are virulence factors with toxic effects on mammalian cells. However, THP-1 cells incubated with purified ESAT-6 either alone or in combination with CFP-10 at concentrations up to 5 μg/well showed no toxic effects (data not shown). Alternatively, it has been suggested that ESAT-6 may be primarily associated with the cell wall rather than secreted (Pym et al., 2002). Perhaps ESAT-6 and CFP-10 are part of the secretion machinery located on the cell wall, and their presence in cell supernatants merely a result of protein sloughing (Pallen, 2002). In either case, we cannot yet state whether other MTB proteins may also prove to be components of or substrates for this secretion system. It should be possible to address this question with careful comparison of proteins in the cell wall and supernatants of H37Rv and the RD1 region mutants.
Experimental procedures
Bacterial cultures
H37Rv (ATCC 27294), H37Rv:ΔRD1, transposon mutants and H37Rv:Δ3875 were grown in roller bottles with Middlebrook 7H9 media (Becton Dickinson) supplemented with 0.05% Tween-80 and albumin, dextrose, catalase (ADC) supplement (BBL) or on Middlebrook 7H10 plates at 37°C, as described elsewhere (Sherman et al., 1995). Bacterial strains were stored as 1 ml aliquots in 15% glycerol at −80°C.
Transposon mutants
Transposon mutants were a generous gift from Chris Sassetti and Eric Rubin (Sassetti et al., 2003). To confirm position of transposon insertion, arbitrary primer PCR was used (O'Toole and Kolter, 1998). A primary PCR reaction was done with a primer of known transposon sequence and an arbitrarily chosen primer with a 3′ tail of ‘Ns’ (to allow for promiscuous binding), followed by a nested PCR reaction with a distal transposon primer and the arbitrary primer without the ‘N’ tail. This resulted in the amplification of genomic DNA adjacent to the transposon, which was sequenced using the distal transposon primer. Resultant sequence was then compared to the H37Rv genome, and insertion site determined.
Deletion of Rv3875
Two primer pairs were used to amplify 831 bp upstream and 858 bp downstream of the esat-6 coding sequence. The products were confirmed by DNA sequencing, cloned into cosmid pYUB854 flanking the hygromycin-resistance marker and packaged into phasmid phAE87 to create the specialized transducing phage phMH421. Transduction of H37Rv to delete the esat-6 gene was performed as previously described (Bardarov et al., 2002).
Complementation vectors
To generate pKG18, genes Rv3870, Rv3871 and Rv3872 were subcloned from a BAC library (clone 414) (Brosch et al., 1998) with restriction enzymes BamHI and NotI. The 5.1 kb fragment was ligated upstream of the mop promoter (George et al., 1995) in an integrating plasmid carrying a hygromycin resistance cassette. For complementation of H37Rv:tn3874 and H37Rv:Δ3875, the Rv3874 and Rv3875 open reading frames were amplified from genomic DNA using the following primers: (A) 5′-GTCCGAATTCAGGAGTCCAGCATGGCAGAGATG-3′ and (B) 5′-ACCCATCGATGTAGTCGGCCGCCATGACAAC-3′. Primer A introduces a single base change (T > G) generating a consensus ribosome binding site 9 bp upstream of the initiation codon. The product was digested with restriction enzymes EcoRI and ClaI, then subcloned downstream of the mop promoter into vectors containing a kanamycin (pMH406) or hygromycin (pMH408) resistance cassette. To complement H37Rv:tn3876, the open reading frame of Rv3876 was amplified using the following primers: (C) 5′-CAACAAGCTTTGAGCGCACTCTGAGAGGTTG-3′ and (D) 5′-CATAGGTACCGCAGGTGCGCTCAACGACGTC-3′, digested with HindIII and Asp718, and subcloned into a vector containing the mop promoter and hygromycin resistance cassette as described above.
Infection of THP-1 cells
THP-1 cells were differentiated as described previously (Lewis et al., 2003). For each infection, bacterial stocks were grown to mid-log phase in 7H9 media, diluted in RPMI 1640 with 10% FCS and added to each well at an MOI of 1–2.5. After 4 h of uptake, cells were washed three times and then reincubated with fresh RPMI 1640 with 10% FCS. Pilot experiments indicated that uptake of bacteria by THP-1 cells was variable (ranging from 18% to 90%; average = 70%). However no differences were noted in the uptake of different bacterial strains (data not shown). At each time-point, triplicate wells for each infecting strain were processed. For measurement of cytotoxicity, supernatant was removed and RPMI 1640 with 10% FCS containing 1× alamarBlue (Biosource International) was added. After 6 h incubation, results were measured at OD570 and OD600. Per cent reduction of alamarBlue was calculated according to manufacturer's instructions. Reduction by uninfected cells at day 0 was set as 100% metabolism. For enumeration of bacilli, supernatant was removed, adherent cells were lysed with 1% Triton X-100, and both samples were plated for colony forming units and/or tested by luciferase assay (for strains containing pMH109, with the mop promoter driving luciferase expression). Luciferase assay reagent (Promega) was added to an equal volume of sample in an opaque 96-well plate, incubated for 10 min and relative light units (rlus) were read in a Lumi/96 luminometer (Bioscan). Rlus were converted to number of bacilli by applying a strain-specific conversion factor derived from several growth curve experiments: H37Rv, 0.0138 rlu/bacillus; H37Rv:ΔRD1, 0.0147 rlu/bacillus; H37Rv:tn0982, 0.0129 rlu/bacillus; H37Rv:tn3864, 0.0206 rlu/bacillus; H37Rv:tn3870, 0.0121 rlu/bacillus; H37Rv:tn3871, 0.0116 rlu/bacillus; H37Rv:tn3874, 0.0126 rlu/bacillus; H37Rv:tn3876, 0.0117 rlu/bacillus. For microscopic examination, cells were differentiated with coverslips in wells and at each time point were fixed for 15 min with 4% paraformaldehyde, permeabilized for 4 min with 0.1% Triton-X 100, and stained with TB Quick Stain (Becton Dickinson) per manufacturer's instructions. Pilot experiments with in vitro-grown bacilli indicated that most but not all bacteria could be detected microscopically, and that staining of both wild-type and RD1 mutant strains was equivalent (data not shown).
Aerosol infection of mice
Aerosol infections of mice were performed as described previously (Smith et al., 2002; Lewis et al., 2003). C57BL/6 mice aged 6–8 weeks (Jackson Laboratories) were maintained in a biosafety-level 3 animal facility. For each infection, frozen bacterial stocks were thawed, sonicated, diluted to ∼106 bacteria ml−1 and nebulized in an aerosol infection chamber (Salter Laboratories) containing the mice. Infectious dose was determined by plating whole lung homogenates on day 1. At each time-point, five mice per group were killed by cervical dislocation. The right lung was homogenized in PBS/0.05% Nonidet P-40 and plated as serial dilutions on 7H10 plates. Colonies were counted after 2–3 weeks incubation at 37°C. To examine histopathology, the left lung was removed and inflated, fixed in 10% buffered formalin, embedded in paraffin, sectioned and stained with hemotoxylin-eosin.
Western immunoblot assay
Cell pellets were lysed as previously described (Sherman et al., 2001). Culture filtrates were prepared and concentrated by established methods (Sorensen et al., 1995). Ten μg of protein were separated by SDS/PAGE, transferred to PVDF membrane (Millipore), probed with α-ESAT-6 monoclonal antibody HYB 76–8 (supplied by T. Mark Doherty, Staten Serum Institute) and detected by SuperSignal West Pico (Pierce).
LCL
EBV-transformed B-cell lines (lymphoblastoid cell lines, LCL) capable of processing and presenting exogenous antigen were generated using supernatants from the cell line 9B5-8 (Lewinsohn et al., 1998). LCL were cultured in RPMI supplemented with HEPES (25 mM), gentamicin (5 μg ml−1), and 10% fetal calf serum.
T cell clones
Clone 38.1–1 is a CD4+ T cell clone specific for CFP-10 and recognizes the epitope STNIRQAGVQYSRAD (CFP1073–87) (Lewinsohn et al., 2003). To expand the T cell clones, a rapid expansion protocol utilizing anti-CD3 monoclonal antibody stimulation was used (Riddell et al., 1992). T cell clones were cultured in the presence of irradiated allogeneic PBMC (2.5 × 107), irradiated allogeneic LCL (5 × 106) and anti-CD3 mAb (30 ng ml−1; Chiron) in RPMI media with 10% human serum in a T-25 upright flask in a total volume of 30 ml. The cultures were supplemented with IL-2 (1 ng ml−1; Chiron) on Days +1, +4, +7 and +10 of culture. The cell cultures were washed on day 4 to remove remaining soluble anti-CD3 mAb.
IFN-γ ELISPOT assay
MTB-specific effectors were detected from purified CD8+ T cells by ELISPOT, as described with minor modifications (Lalvani et al., 1997). Briefly, 96-well nitrocellulose-backed plates (MAHA S4510; Millipore) were coated as recommended by the manufacturer with 10 μg ml−1 capture mouse anti-IFN-γ (1-D1K; Mabtech AB) overnight at 4°C. Plates were then washed six times with PBS/0.05% Tween 20 (Sigma) and blocked with RPMI/10% HS for 1 h at room temperature. T cell clones (n = 750) and LCL (n = 20 000) were added, and the plate incubated overnight at 37°C. PHA (10 μg ml−1) was used as a positive control. After washing with PBS/0.05% Tween 20, 100 μl of 1 μg ml−1 biotinylated secondary anti-IFN-γ mAb (7B6-1; Mabtech AB) was added. After 2 h of incubation at room temperature, plates were washed six times and 100 μl avidin/biotinylated enzyme (HRP) complex (Vectastain ABC Elite Kit; Vector Laboratories) was added to wells, and the plates were incubated for an additional 1 h. Plates were then washed six times, and 100 μl AEC substrate (Vectastain AEC substrate kit, Vector Laboratories) was added. After 4–7 min, the colorimetric reaction was stopped by washing with distilled water. Spots were enumerated using a Zeiss Axioplan 2 microscope with 3200 K incident illumination equipped with a Epiplan Neofluar 5x/0.15 objective, Sony DXC 950 CCD camera, Märzhäuser scanning stage, MCP4 control unit and KS ELISPOT software (Carl Zeiss Vision).
Other procedures
Escherichia coli was used for routine DNA manipulations. Real-time PCR was done on a Bio-Rad iCycler and products were quantified using iQ SYBR Green Supermix (Bio-Rad), per the manufacturer's instructions.
Acknowledgements
Anti-ESAT-6 antibodies (mAb HYB 76–8) were kindly provided by T. Mark Doherty (Staten Serum Institute), and antibodies against an MTB culture filtrate protein preparation (C-193) were provided by Colorado State University as part of the NIH NAIAD TB Material Contract #N01 AI-75320. We thank Chris Sassetti and Eric Rubin (Harvard University) for generously supplying transposon mutants, Stoyan Bardarov and Bill Jacobs (Albert Einstein College of Medicine) for supplying reagents and advice for the phage transduction, and Marcel Behr, Lalita Ramakrishnan, Wes Van Voorhis and members of the Sherman laboratory for helpful discussions. We gratefully acknowledge funding from NIH (HL64550 and HL68533) and from the Sequella Global Tuberculosis Foundation, of which D.R.S. is a Core Scientist.
Footnotes
Note added in press
Between acceptance and publication of this manuscript, complementary manuscripts were published by the laboratories of William R. Jacobs and Jeffrey S. Cox.
References
- Bao L, Chen W, Zhang H, Wang X. Virulence, immunogenicity, and protective efficacy of two recombinant Mycobacterium bovis bacillus Calmette-Guerin strains expressing the antigen ESAT-6 from Mycobacterium tuberculosis. Infect Immun. 2003;71:1656–1661. doi: 10.1128/IAI.71.4.1656-1661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov S, Bardarov S, Jr, Pavelka MS, Sambandamurthy V, Larsen M, Tufariello J, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144:3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Belisle JT. Genetics of protein secretion. In: Hatfull GF, Jacobs WR Jr, editors. Molecular Genetics of Mycobacteria. American Society for Microbiology Press; Washington, D.C.: 2000. pp. 203–220. [Google Scholar]
- Braunstein M, Bardarov SS, Jacobs WR., Jr Genetic methods for deciphering virulence determinants of Mycobacterium tuberculosis. Methods Enzymol. 2002;358:67–99. doi: 10.1016/s0076-6879(02)58081-2. [DOI] [PubMed] [Google Scholar]
- Brodin P, Eiglmeier K, Marmiesse M, Billault A, Garnier T, Niemann S, et al. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect Immun. 2002;70:5568–5578. doi: 10.1128/IAI.70.10.5568-5578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Billault A, Garnier T, Eiglmeier K, Soravito C, et al. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusasca PN, Colangeli R, Lyashchenko KP, Zhao X, Vogelstein M, Spencer JS, et al. Immunological characterization of antigens encoded by the RD1 region of the Mycobacterium tuberculosis genome. Scand J Immunol. 2001;54:448–452. doi: 10.1046/j.1365-3083.2001.00975.x. [DOI] [PubMed] [Google Scholar]
- Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Fenton MJ, Vermeulen MW. Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect Immun. 1996;64:683–690. doi: 10.1128/iai.64.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George KM, Yuan Y, Sherman DR, Barry CE., III The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Identification and functional analysis of CMAS-2. J Biol Chem. 1995;270:27292–27298. doi: 10.1074/jbc.270.45.27292. [DOI] [PubMed] [Google Scholar]
- Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD.The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria Genome Biol 20012research0044.0041–0044.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- Harboe M, Oettinger T, Wiker HG, Rosenkrands I, Andersen P. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect Immun. 1996;64:16–22. doi: 10.1128/iai.64.1.16-22.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns AM, Magee JG, Gennery A, Steward M, Graham C, Seiders PR, Freeman R. Rapid identification of Mycobacterium bovis BCG by the detection of the RD1 deletion using a multiplex PCR technique. Int J Tuberc Lung Dis. 1999;3:635–638. [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalvani A, Nagvenkar P, Udwadia Z, Pathan AA, Wilkinson KA, Shastri JS, et al. Enumeration of T cells specific for RD1-encoded antigens suggests a high prevalence of latent Mycobacterium tuberculosis infection in healthy urban Indians. J Infect Dis. 2001;183:469–477. doi: 10.1086/318081. [DOI] [PubMed] [Google Scholar]
- Lee VT, Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15:1725–1752. doi: 10.1101/gad.896801. [DOI] [PubMed] [Google Scholar]
- Lein AD, von Reyn CF, Ravn P, Horsburgh CR, Jr, Alexander LN, Andersen P. Cellular immune responses to ESAT-6 discriminate between patients with pulmonary disease due to Mycobacterium avium complex and those with pulmonary disease due to Mycobacterium tuberculosis. Clin Diagn Laboratory Immunol. 1999;6:606–609. doi: 10.1128/cdli.6.4.606-609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn DM, Alderson MR, Briden AL, Riddell SR, Reed SG, Grabstein KH. Characterization of human CD8+ T cells reactive with Mycobacterium tuberculosis-infected antigen-presenting cells. J Exp Med. 1998;187:1633–1640. doi: 10.1084/jem.187.10.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn DA, Heinzel AS, Gardner JM, Zhu L, Alderson MR, Lewinsohn DM. Mycobacterium tuberculosis-specific CD8+ T cells preferentially recognize heavily infected cells. Am J Respir Crit Care Medical. 2003 doi: 10.1164/rccm.200306-837OC. in press. [DOI] [PubMed] [Google Scholar]
- Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy A, Laochumroonvorapong P, Kaplan G. Apoptosis, but not necrosis, of infected monocytes is coupled with killing of intracellular bacillus Calmette-Guerin. J Exp Med. 1994;180:1499–1509. doi: 10.1084/jem.180.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Pallen MJ. The ESAT-6/WXG100 superfamily – and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- Pollock JM, Andersen P. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J Infectious Dis. 1997;175:1251–1254. doi: 10.1086/593686. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002;46:709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, et al. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan L, Federspiel NA, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Hewinson RG, Williamson RA, Carr MD. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1: 1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J Biol Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Micro-biol Rev. 2001;25:3–14. doi: 10.1111/j.1574-6976.2001.tb00569.x. [DOI] [PubMed] [Google Scholar]
- Sassetti C, Rubin EJ. Genomic analyses of microbial virulence. Curr Opin Microbiol. 2002;5:27–32. doi: 10.1016/s1369-5274(02)00281-3. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Sherman DR, Sabo PJ, Hickey MJ, Arain TM, Mahairas GG, Yuan Y, et al. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc Nat Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci USA. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RF, Li Q, Ellner JJ. Expression of virulence of Mycobacterium tuberculosis within human monocytes: virulence correlates with intracellular growth and induction of tumor necrosis factor alpha but not with evasion of lymphocyte-dependent monocyte effector functions. Infect Immun. 1998;66:1190–1199. doi: 10.1128/iai.66.3.1190-1199.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjot RL, Oettinger T, Rosenkrands I, Ravn P, Brock I, Jacobsen S, Andersen P. Comparative evaluation of low-molecular-mass proteins from Mycobacterium tuberculosis identifies members of the ESAT-6 family as immunodominant T-cell antigens. Infect Immun. 2000;68:214–220. doi: 10.1128/iai.68.1.214-220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Liggitt D, Jeromsky E, Tan X, Skerrett SJ, Wilson CB. Local role for tumor necrosis factor alpha in the pulmonary inflammatory response to Mycobacterium tuberculosis infection. Infect Immun. 2002;70:2082–2089. doi: 10.1128/IAI.70.4.2082-2089.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AL, Nagai S, Houen G, Andersen P, Andersen AB. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, et al. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol. 2002;46:649–659. doi: 10.1046/j.1365-2958.2002.03190.x. [DOI] [PubMed] [Google Scholar]
- Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Talbot EA, Williams DL, Frothingham R. PCR identification of Mycobacterium bovis BCG. J Clin Microbiol. 1997;35:566–569. doi: 10.1128/jcm.35.3.566-569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F, Gordon SV, Garnier T, Brosch R, Barrell BG, Cole ST. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- Ulrichs T, Munk ME, Mollenkopf H, Behr-Perst S, Colangeli R, Gennaro ML, Kaufmann SH. Differential T cell responses to Mycobacterium tuberculosis ESAT6 in tuberculosis patients and healthy donors. Eur J Immunol. 1998;28:3949–3958. doi: 10.1002/(SICI)1521-4141(199812)28:12<3949::AID-IMMU3949>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Wards BJ, Collins DM. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol Lett. 1996;145:101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x. [DOI] [PubMed] [Google Scholar]
- Wards BJ, de Lisle GW, Collins DM. An esat6 knockout mutant of Mycobacterium bovis produced by homologous recombination will contribute to the development of a live tuberculosis vaccine. Tuber Lung Dis. 2000;80:185–189. doi: 10.1054/tuld.2000.0244. [DOI] [PubMed] [Google Scholar]
- Zhang M, Gong J, Lin Y, Barnes PF. Growth of virulent and avirulent Mycobacterium tuberculosis strains in human macrophages. Infect Immun. 1998;66:794–799. doi: 10.1128/iai.66.2.794-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


