Abstract
Single-stranded mRNA molecules form secondary structures through complementary self-interactions. Several hypotheses have been proposed on the relationship between the nucleotide sequence, encoded amino acid sequence and mRNA secondary structure. We performed the first transcriptome-wide in silico analysis of the human and mouse mRNA foldings and found a pronounced periodic pattern of nucleotide involvement in mRNA secondary structure. We show that this pattern is created by the structure of the genetic code, and the dinucleotide relative abundances are important for the maintenance of mRNA secondary structure. Although synonymous codon usage contributes to this pattern, it is intrinsic to the structure of the genetic code and manifests itself even in the absence of synonymous codon usage bias at the 4-fold degenerate sites. While all codon sites are important for the maintenance of mRNA secondary structure, degeneracy of the code allows regulation of stability and periodicity of mRNA secondary structure. We demonstrate that the third degenerate codon sites contribute most strongly to mRNA stability. These results convincingly support the hypothesis that redundancies in the genetic code allow transcripts to satisfy requirements for both protein structure and RNA structure. Our data show that selection may be operating on synonymous codons to maintain a more stable and ordered mRNA secondary structure, which is likely to be important for transcript stability and translation. We also demonstrate that functional domains of the mRNA [5′-untranslated region (5′-UTR), CDS and 3′-UTR] preferentially fold onto themselves, while the start codon and stop codon regions are characterized by relaxed secondary structures, which may facilitate initiation and termination of translation.
INTRODUCTION
In 1972 White et al. (1) suggested that the redundancy in the genetic code permits extensive variation of the nucleotide sequence and allows the satisfaction of requirements for both protein structure and RNA structure. Ball (2) proposed three alternative hypotheses on the relationship between amino acid sequences and mRNA secondary structure. First, the choice of codons and their sequence in the message could be independent of the resulting secondary structure of the mRNA. Second, optimization of mRNA secondary structure may occur only within the limits of encoded amino acid sequence. Third, selection pressure for specific RNA secondary structure could affect the choice of nucleotide at both synonymous and non-synonymous positions. Fitch (3) examined these hypotheses and found evidence of the use of the degeneracy of the genetic code to optimize base pairing in mRNA molecules. He discussed the third hypothesis as being biologically plausible. However, he did not find evidence for (or against) the notion that the needs of RNA structure and function must compete with the needs of protein structure and function.
Since then, the idea that the redundancy of the genetic code allows preservation of mRNA folding has been supported by several lines of evidence. Periodical patterns complementary to the proof-reading site in the ribosome and presumably involved in the translation frame monitoring mechanism have been found in many transcripts (4). It was shown that synonymous substitutions affect mRNA translation in different organisms (5–7). Strong mRNA secondary structures formed due to gene-specific codon usage have been implicated in discontinuous translation and pauses in synthesis of insect silk fibroin, chicken collagen and other proteins (8,9). These and similar works [for a review see (10)] gave rise to the expectations that secondary structures can interfere with translation and therefore should be avoided in mRNA coding regions. Contrarily to this opinion, significant biases in favor of local RNA structures have been found in several bacterial species and the yeast (11). Although evolutionarily conserved local secondary structures were described in eukaryotic and mammalian mRNAs and pre-mRNAs (12), no conclusive evidence has been found to confirm or disprove the hypothesis that selection for RNA structure can lead to non-optimal amino acid usage. Seffens and Digby (13) reported that native mRNAs have a lower calculated folding free energy than random sequences. Correlations between mRNA and protein secondary structures have been noted (14). Following Jia et al. (14), Chamary and Hurst (15) suggested that base composition at the third synonymous site is driven by the nucleotide usage of amino acids, and the requirement for elevated C at 4-fold degenerate sites is related to usage of encoded amino acids in alpha helices and beta sheets.
Non-random use of synonymous triplets coding for same amino acids has been observed in genomes from different life forms. The biological basis of unequal codon choice is not completely clear. Positive correlation between synonymous codon usage bias and gene expression level was established in bacteria (5,16), yeast (5), nematode (17) and insect (18). In many cases, preferred codons correspond to the most abundant iso-accepting tRNAs, which was explained by evolutionary selection for efficient translation (5,16–18). In mammals, however, evidence supporting translational selection of codon choice is arguable (19–21). No correspondence between the usage of a codon in human protein coding sequences and the abundance of iso-accepting tRNA has been found in several studies (22–26). Recently, Lavner and Kotlar (27) reported a weak positive correlation between expression level and frequency of optimal codons for human genes.
Important observations suggesting the functional role for degenerate sites in the maintenance of mRNA secondary structure were derived from mutational studies. Analysis of synonymous nucleotide polymorphism in enteric bacteria and compensatory nucleotide substitutions in Drosophila suggested selective constraint on mRNA secondary structures (28,29). Conservation of secondary structure features was demonstrated for retroviral mRNA. It was shown that folding in RNA stem regions disrupted by silent mutations on one strand of retroviral RNA is restored by compensatory mutations on the other strand (30). Mutations in GC-rich secondary structures in complex 5′-untranslated regions (5′-UTRs) that provide skaffold for interactions with trans-acting proteins can have implications in disease and tumorogenesis (31). It was shown that the location of synonymous mutations in the mouse lineages is non-random with respect to mRNA stability (15), and substitutions at the third synonymous positions affect mRNA decay rates (32) and translation (5–7). Moreover, synonymous mutations affecting mRNA structure and decay rate can be highly deleterious and have implications in disease in humans (33,34).
Here we report results of the transcriptome-scale in silico analysis of the human and mouse mRNAs. We describe general structural properties of mammalian protein coding transcripts and demonstrate that the structure of the genetic code creates specific periodic pattern of nucleotide base pairing in mRNA coding regions. We show that degenerate codon sites are important for maintaining a more ordered and stable mRNA secondary structure in the protein coding regions. We also demonstrate that mRNA functional domains (5′-UTR, CDS, 3′-UTR) preferentially self-fold, and regions involved in translation initiation and termination are characterized with reduced levels of secondary structures.
MATERIALS AND METHODS
Non-redundant datasets of the human (19 317 sequences) and mouse (20 892 sequences) mRNAs were compiled from the RefSeq database for the human and mouse genomes (ftp://ftp.ncbi.nlm.nih.gov/genomes). Only annotated mRNA sequences with complete CDSs (400 nt or longer) possessing the 5′- and 3′-UTRs (50 nt or longer) were analyzed in this study. A dataset of 6919 orthologous human–mouse mRNA pairs with annotated 5′-UTRs and 3′-UTRs and with properly aligned start and stop codons used in this study was described previously (35). Vista computational tool was used for alignment visualization (36,37). Symmetrical best hits between proteins from the respective genomes were identified using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence alignments for identified orthologous pairs of human–mouse 5′-UTRs and 3′-UTRs were produced with the OWEN program (38). For the CDS, the alignment of the nucleotide sequences was guided by the amino acid sequence alignments. The positions of 5′- and 3′-UTRs were taken from the feature tables of the GenBank entries. The degree of conservation at each nucleotide position was calculated as the number of matches over the number of pairwise alignments. The start codon and the stop codon provided natural reference points for this analysis, the position number was always determined as a distance from one of these codons. Relative abundance of human mRNAs was estimated from the numbers of the corresponding expressed sequence tags (ESTs) from normal tissues in GenBank.
To investigate the role of the genetic code and codon usage in mRNA secondary structure formation, we compared the folding of native mRNAs with foldings computed for sequences randomized with different methods. For each mRNA sequence, we constructed several randomized sequences using procedures similar to those described by Seffens and Digby (13). The first randomization procedure shuffled nucleotides at the third 4-fold synonymous codon sites, retaining nucleotide composition and amino acid sequence of the native mRNAs. The second randomization procedure preserved the amino acid sequence, but eliminated codon usage bias at the third codon positions by randomly choosing synonymous codons from the genetic code table with equal probability. The resulting shuffled sequences were ∼80% identical to the corresponding native mRNA sequences at the nucleotide level (data not shown). The third randomization procedure preserved the amino acid sequence and created ‘codon flat’ coding sequence by randomly choosing all synonymous codons from the genetic code table. The forth procedure randomly shuffled all nucleotides in CDSs of the native mRNAs, preserving only the original nucleotide content. Nucleotides in the 5′-UTRs and 3′-UTRs were shuffled in all randomization procedures retaining nucleotide composition. Additionally, we employed two dinucleotide randomization procedures described by Katz and Burge (11). The first dinucleotide randomization preserved dinucleotide frequencies, codon frequencies, codon usage and nucleotide composition of native mRNAs. The second dinucleotide randomization randomly shuffled all dinucleotides, retaining nucleotide composition of native mRNAs.
Native mRNAs and randomly generated sequences were computationally ‘folded’ and the predicted minimum free secondary structure energy was calculated, using our implementation of the dynamic programming algorithm described by Zuker (39) that employs nearest neighbor parameters for evaluation of free energy. Energy minimization was performed by dynamic programming method that finds the secondary structure with the minimum free energy with sums contributing from stacking, loop length and the like using a new algorithm for evaluation of internal loops (40). Our program ‘folds’ sequences up to the 28 000 nucleotide long. The sequence fold variant with the lowest secondary structure energy was used in our analysis. P-values for randomizations were determined by paired t-tests. To test the program performance, a part of the sequence dataset was folded with the mfold v.3.2 server (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/) and RNAalifold program from Vienna server (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi), which uses comparative sequence information. All programs produced similar results. As an additional control, we folded the complete set of human tRNAs from an RNA database (http://lowelab.ucsc.edu/GtRNAdb/). Distributions of tRNA preferred base pairing are presented in Supplementary Figure 2.
RESULTS AND DISCUSSION
Sequence conservation and mRNA stability
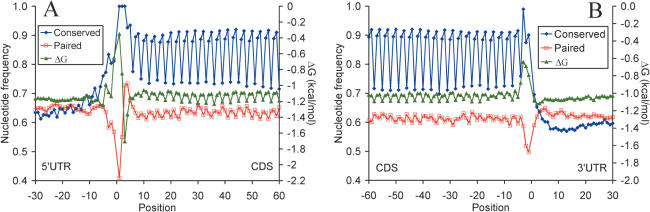
To study the relationship between the genetic code and mRNA secondary structure, we evaluated sequence conservation, free Gibbs energy of secondary structure formation and nucleotide involvement in secondary structure elements. A total of 19 317 human and 20 892 mouse mRNA sequences were folded in silico. Profiles of nucleotide base pairing and secondary structure stability in the 5′-UTR, CDS and 3′-UTR in the human mRNAs, and profiles of sequence conservation in the human and mouse mRNAs are shown on Figure 1. A well-defined periodic pattern of conservation is observed in the coding region. Nucleotides in the first two codon sites are the most conserved, while those in the third positions are the least conserved. In the CDSs, we also found a pronounced periodic pattern of nucleotide base pairing and mRNA secondary structure stability. Frequencies of paired nucleotides at the first, second and third codon sites differed significantly (0.62, 0.608 and 0.631, respectively, P < 10−5). Notably, base pairing at the third GC-rich codon sites and their contribution to mRNA secondary structure stability are significantly higher than contributions of the first G-rich sites or the second AU-rich sites. The same periodic pattern of nucleotide base pairing was observed in the mouse mRNAs (Supplementary Figure 1).
Figure 1.
Profiles of nucleotide involvement in secondary structures, free energy of secondary structure formation and sequence conservation around the start codon (A) and the stop codon (B) in human mRNAs. Positions from −30 to −1 correspond to 5′-UTRs and positions from 1 to 60 correspond to CDSs (A). Positions from −60 to −1 correspond to CDSs and positions from 1 to 30 correspond to 3′-UTRs (B). Blue, sequence conservation in 6919 orthologous human and mouse mRNAs. Red, base paired nucleotides in 19 317 human mRNAs. Green, free Gibbs energy of base pairing in 19 317 human mRNAs.
Periodic patterns of nucleotide base pairing and secondary structure stability in the CDSs follow the triplet pattern of nucleotide conservation created by the genetic code. Correlation coefficients between sequence conservation and nucleotide base pairing at different codon sites were as follows: −0.373 for site 1 (P < 0.005), −0.516 for site 2 (P < 10−5) and 0.733 for site 3 (P < 10−11). Correlation coefficients between sequence conservation and free energy of base pairing at different codon sites were as follows: 0.438 for site 1(P < 0.0003), 0.662 for site 2 (P < 10−8) and −0.836 for site 3 (P < 10−17). Importantly, a significant negative correlation was observed between sequence conservation and the free energy of base pairing at the third codon sites, as opposite to the first and the second codon sites (Figure 1), indicating that the third sites make the greatest contribution to mRNA secondary structure stability.
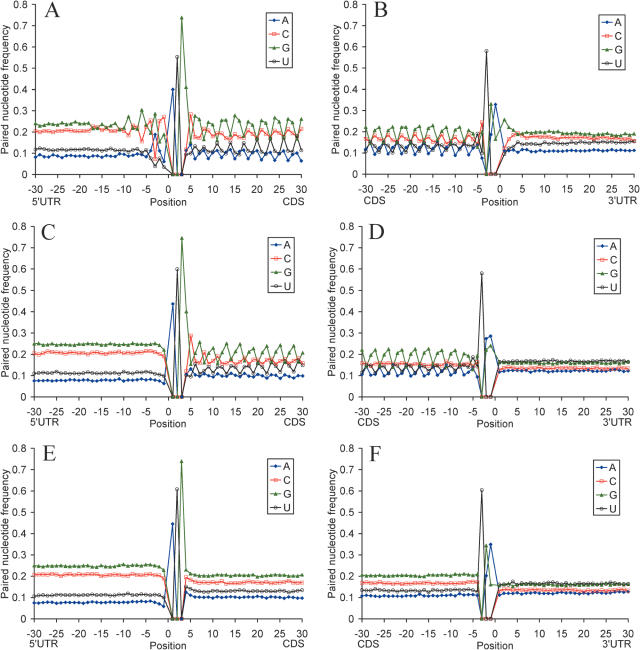
The periodic pattern of nucleotide base pairing becomes even more apparent when involvement of individual nucleotides in the secondary structure formation is considered (Figure 2). This pattern emerges at nt −9 upstream from the start codon and terminates at the stop codon. Nucleotide G preferentially participates in the secondary structures at codon site 1, nucleotides U and A at codon site 2, G and C at codon site 3. These preferences are in agreement with the relative nucleotide abundance (Supplementary Table 1) and codon position biases in the human genes (41).
Figure 2.
Profiles of nucleotide base pairing around the start codon (A, C and E) and the stop codon (B, D and F) for 19 317 human mRNAs (A and B), for sequences with randomly chosen synonymous codons (C and D), and sequences with randomly shuffled nucleotides and the same nucleotide composition as native mRNAs (E and F). Blue, guanine; red, cytosine; green, adenosine; orange, uridine.
Pattern of nucleotide base pairing created by the genetic code
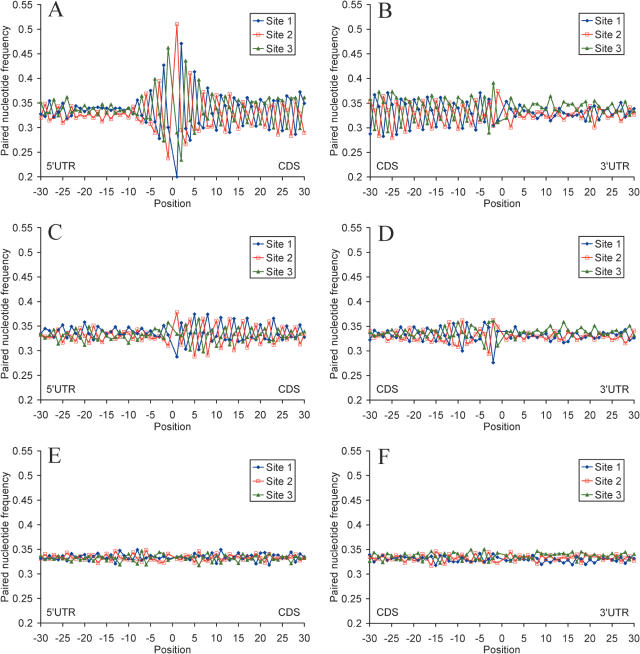
Messenger RNA self-folding allows three types of nucleotide base pairing in the CDS in relation to codon sites (Figure 3), which we denote as base pairing phase 1 (sites [123] paired with sites [132]), phase 2 (sites [123] paired with sites [321]), and phase 3 (sites [123] paired with sites [213]). As seen from Figure 3, phases 1, 2 and 3 are supported by nucleotide base pairing at the first, second and third codon sites, correspondingly. A pronounced bias for preferential base pairing in phase 3 is observed in the CDSs of human mRNAs (Figure 4 and Table 1). Obvious reasons for preferential realization of pairing phase 3 are the elevated GC content and the near equivalent frequencies of base pairing nucleotides (A ≈ U, C ≈ G) at the third codon site, relative to other sites (Supplementary Table 1). Another reason is that frequencies of trinucleotides at codon sites [2,3,1] are close to frequencies of the corresponding complementary trinucleotides at the same sites, which results in a similarly high frequencies of paired nucleotides (Supplementary Table 4). Furthermore, as seen from Table 1, phase 3 allows a more dense nucleotide pairing, as compared with phases 1 and 2. All this promotes preferential mRNA self-folding in pairing phase 3, relative to other pairing phases. This type of mRNA folding observed in the human and mouse transcripts may also be preferentially realized in transcripts from other taxa. For example, Carlini et al. (42) found that base pairing between synonymous sites 3–3 and 3–1 are most frequent in the drosophilid alcohol dehydrogenase genes.
Figure 3.
The three phases of nucleotide base pairing in the mRNA CDS. The numbers denote codon sites.
Figure 4.
Profiles of base pairing for nucleotides around the start codon (A, C and E) and the stop codon (B, D and F) with different codon sites for 19 317 human mRNAs (A and B), for sequences with randomly chosen synonymous codons (C and D), and sequences with randomly shuffled nucleotides and the same nucleotide composition as native mRNAs (E and F). Nucleotides paired with codon sites 1, 2 and 3 are shown in blue, red and green, respectively.
Table 1.
Frequencies of base paired nucleotides at different codon sites in the human mRNAs and randomized sequences
| Sequences | Codon site 1 | Codon site 2 | Codon site 3 | |||
|---|---|---|---|---|---|---|
| Frequency | P-value | Frequency | P-value | Frequency | P-value | |
| Base paired G | ||||||
| Real mRNAs | 0.242 | 0.157 | 0.228 | |||
| Random CC | 0.239 | 0.097 | 0.151 | 10−8 | 0.202 | 10−38 |
| Random NS | 0.204 | 10−47 | 0.205 | 10−124 | 0.205 | 10−32 |
| Base paired C | ||||||
| Real mRNAs | 0.169 | 0.160 | 0.195 | |||
| Random CC | 0.157 | 10−22 | 0.163 | 0.12 | 0.156 | 10−93 |
| Random NS | 0.169 | 0.42 | 0.168 | 10−8 | 0.169 | 10−66 |
| Base paired U | ||||||
| Real mRNAs | 0.103 | 0.159 | 0.127 | |||
| Random CC | 0.114 | 10−51 | 0.155 | 10−3 | 0.148 | 10−54 |
| Random NS | 0.131 | 10−131 | 0.131 | 10−67 | 0.131 | 10−5 |
| Base paired A | ||||||
| Real mRNAs | 0.108 | 0.133 | 0.080 | |||
| Random CC | 0.100 | 10−19 | 0.129 | 0.0011 | 0.097 | 10−60 |
| Random NS | 0.103 | 10−10 | 0.103 | 10−65 | 0.102 | 10−77 |
| Nucleotides paired with codon site 1 | ||||||
| Real mRNAs | 0.290 | 0.354 | 0.358 | |||
| Random CSx4 | 0.297 | 10−8 | 0.350 | 10−6 | 0.353 | 10−3 |
| Random CCx4 | 0.315 | 10−119 | 0.335 | 10−71 | 0.352 | 10−5 |
| Random CC | 0.322 | 10−177 | 0.345 | 10−44 | 0.338 | 10−160 |
| Random NS | 0.328 | 10−210 | 0.336 | 10−162 | 0.335 | 10−202 |
| Random DCS | 0.287 | 10−5 | 0.352 | 10−2 | 0.361 | 10−10 |
| Random DNS | 0.328 | 10−103 | 0.335 | 10−95 | 0.337 | 10−80 |
| Nucleotides paired with codon site 2 | ||||||
| Real mRNAs | 0.344 | 0.293 | ||||
| Random CS×4 | 0.341 | 10−3 | 0.309 | 10−20 | ||
| Random CC×4 | 0.339 | 10−8 | 0.319 | 10−113 | ||
| Random CC | 0.327 | 10−63 | 0.322 | 10−250 | ||
| Random NS | 0.330 | 10−57 | 0.334 | 0 | ||
| Random DCS | 0.347 | 10−7 | 0.291 | 10−4 | ||
| Random DNS | 0.331 | 10−16 | 0.334 | 10−159 | ||
| Nucleotides paired with codon site 3 | ||||||
| Real mRNAs | 0.358 | |||||
| Random CS×4 | 0.352 | 10−12 | ||||
| Random CC×4 | 0.333 | 10−80 | ||||
| Random CC | 0.339 | 10−104 | ||||
| Random NS | 0.331 | 10−165 | ||||
| Random DCS | 0.356 | 10−3 | ||||
| Random DNS | 0.329 | 10−93 | ||||
Abbreviations: Real mRNAs, coding sequences of 19 317 native human mRNA; Random CC, mRNA sequences with randomly chosen synonymous codons; Random NS, sequences with randomly shuffled nucleotides and the same nucleotide composition as native mRNAs; Random CS×4, sequences with shuffled 4-fold degenerate synonymous codons; Random CC×4, sequences with randomly chosen 4-fold degenerate synonymous codons; Random DCS, dicodone shuffling that preserved dinucleotide frequencies, encoded amino acid sequence and codon usage of native mRNAs; Random DNS, random shuffling of all dinucleotides that retained nucleotide composition of native mRNAs. Start and stop codons were excluded from this analysis.
To investigate the role of the genetic code and codon usage in the formation of mRNA secondary structure, we compared the folding computed for native mRNAs with the folding computed for sequences randomized with different methods. First, we eliminated codon usage bias by randomly choosing synonymous codons from the genetic code table. This significantly decreased base pairing for C and G at the third codon sites (Figure 2 and Table 1), pattern of base pairing relative to codon sites (Figure 4 and Table 1) and markedly reduced free Gibbs energy of secondary structure formation at all codon sites (Table 2). Shuffling of nucleotides at the third synonymous codon sites which retained nucleotide composition and amino acid sequence of the native mRNAs affected nucleotide base pairing to some extend due to changes in codon context (Table 1). Random shuffling of all nucleotides in the CDS while preserving the original nucleotide content of the native mRNAs completely eliminated all the secondary structure patterns and differences in Gibbs free energy between all codon sites (Figures 2 and 4 and Tables 1 and 2).
Table 2.
Free Gibbs energy (ΔG) for dinucleotide interaction in secondary structure formation at different codon sites in 19 317 human mRNAs, randomized sequences and in subsets of abundant and rare transcripts
| Sequences | Codon sites [1,2] | P-value | Codon sites [2,3] | P-value | Codon sites [3,1] | P-value | Average |
|---|---|---|---|---|---|---|---|
| ΔG (kcal/mol) | ΔG (kcal/mol) | ΔG (kcal/mol) | ΔG (kcal/mol) | ||||
| Real mRNAs | −2.204 | −2.200 | −2.332 | −2.245 | |||
| Random CC | −2.173 | 0.0148 | −2.080 | 10−47 | −2.193 | 10−38 | −2.149 |
| Random NS | −2.196 | 0.270 | −2.206 | 0.189 | −2.210 | 10−31 | −2.204 |
| Abundant | −2.216 | −2.224 | −2.332 | −2.257 | |||
| Rare | −2.198 | 0.03 | −2.185 | 10−7 | −2.323 | 0.05 | −2.235 |
Abbreviations are the same as in Table 1. Subsets of abundant and rare mRNAs (3227 and 3639 sequences, correspondingly) were compiled based on the numbers of corresponding EST sequences in GenBank. Selection limits were ≤15 and ≥108 EST sequences for rare and abundant mRNAs, correspondingly.
Since it was argued that dinucleotide content is important when assessing the predicted free energy of RNA secondary structure (43), we additionally employed dinucleotide randomization procedures suggested by Katz and Burge (11). Results of these randomization experiments are presented in Table 1. Dinucleotide shuffling that preserved dinucleotide and codon frequencies, nucleotide composition, and codon usage of native mRNAs had little effect on RNA base pairing pattern. Random shuffling of all dinucleotides while preserving the original nucleotide content of the native mRNAs completely eliminated all the secondary structure patterns, similar to random nucleotide shuffling.
We also evaluated thermodynamic stability of native human mRNAs and randomized sequences. As Table 3 shows, all shuffling procedures, with the exception of dicodon shuffling, led to statistically significant increase in calculated free energy of mRNA secondary structure formation. Analysis of base pairing frequencies and differences in base pairing levels between codon sites (Table 1 and Supplementary Table 5) demonstrates that all randomization procedures, with the exception of dicodon shuffle, changed base pairing pattern of native human mRNAs. Thus, both synonymous codon usage at degenerate sites and the dinucleotide relative abundances at codon positions [1,2], [2,3] and [3,1] [defined as genome specific codon signature by Karlin and Mrazek (41)] are important for maintaining secondary structures in human transcripts. Specifically, base pairing at the first and the second codon positions is determined by the structure of the genetic code and dinucleotide frequencies, while base pairing at the third codon positions is largely determined by the usage of 4-fold degenerate codons. Taken together, results of our shuffling experiments indicate that periodic pattern of secondary structure in mRNA coding regions is largely determined by the structure of the genetic code, with contribution from synonymous codon usage bias at degenerate codon sites. Our data suggest that synonymous sites are under selection for a more ordered and more stable mRNA secondary structure.
Table 3.
Stability of the human mRNA secondary structures and randomized sequences
| Sequences | ΔG (kcal/mol) | P-value |
|---|---|---|
| Real mRNAs | −1032.0 ± 4.39 | |
| Random CC | −952.67 ± 3.98 | 10−40 |
| Random NN | −986.38 ± 4.19 | 10−13 |
| Random CSx4 | −993.01 ± 4.24 | 10−10 |
| Random CCx4 | −976.42 ± 4.39 | 10−20 |
| Random DCS | −1025.6 ± 4.37 | 0.143 |
| Random DNS | −984.54 ± 4.39 | 10−15 |
Means ± SEM are shown. N = 19 317. Abbreviations are the same as in Table 1.
A characteristic feature of mRNA folding in the CDS is periodic alternation of AT and GC base pairing, which may be important for reduction of local strong secondary structures in the protein coding regions. This alternation is largely due to relatively high frequencies of AT at the second codon sites and GC at the third codon sites in the human genome (41). Our results on relative dinucleotide abundancy and codon position bias for the human mRNAs (Supplementary Tables 2 and 3) are consistent with data reported for the human genes (22,41). The observed high frequency of dinucleotide GA at sites [1,2] reflects high proportion of glutamate and aspartate in human proteins. The relative abundance of AG and AC at codon sites [2,3] may be explained with high usage of specific codons for glutamine (CAG), lysine (AAG), histidine (CAC), glutamate (GAG) and aspartate (GAC) in the human protein coding genes. Two dinucleotides, UA and CG, were underrepresented at all codon sites. The deficiency of CG has been explained by the high mutability of this dinucleotide in the nuclear DNA (44). The deficiency of UA is considered adaptive. This dinucleotide is most successible for RNase activity, and underrepresentation of UA in mRNAs may reflect a requirement for transcript stability (45).
mRNA secondary structure and transcript abundance
To study the relationship between mRNA secondary structure and transcript abundance, we calculated mRNA folding, codon frequencies and dinucleotide frequencies for different subsets of the human mRNA dataset. The average ΔG of dinucleotide interaction was significantly lower for abundant messages (Table 2). We found no major difference in the pattern and frequencies of base paired nucleotides between the groups of abundant and rare transcripts, and between the groups of long and short transcripts (data not shown). At the same time, we observed notable differences in the dinucleotide frequencies and codon frequencies between abundant and rare mRNAs (Supplementary Tables 2 and 3). Frequencies of codons for histidine, proline, cysteine and tryptophan are significantly enhanced, and codon frequencies for lysine, asparagine, aspartate and glutamate are significantly reduced in abundant transcripts, relatively to rare transcripts. However, this appear to reflect differences in the amino acid content between the groups of proteins encoded by abundant and rare transcripts, and have little effect on the pattern and frequencies of nucleotide base pairing.
Folding and conservation in mRNA functional domains
We studied distribution of secondary structures in different domains of human mRNAs (i.e. within 5′-UTR, CDS or 3′-UTR). Overall, 5′-UTRs are enriched with the secondary structures, as compared with 3′-UTRs, which can be explained by higher GC content of 5′-UTRs. Frequencies of paired nucleotides in the 5′-UTRs and 3′-UTRs were 0.64 and 0.60, correspondingly. In the 5′-UTRs, the most pronounced conservation (over 75% identity) is seen in the nine positions immediately upstream of the start codon (Figure 1A). The nucleotide conservation level in the 30 nt region upstream of the start codon is comparable with that of the synonymous third positions in the CDSs. Further upstream into the 5′-UTR, conservation level decreases to a plateau of ∼50% identity (data not shown), which is close to the neutral conservation level in human–mouse alignments (46,47). In the 3′-UTR, the level of conservation is the lowest within the 30 nt region immediately downstream from the stop codon and steadily rises further in the 3′-UTR (Figure 1B). The frequency of paired nucleotides within this 30 nt region is highest, and steadily declines in the downstream 3′-UTR region (data not shown). Similar results were obtained for the mouse mRNAs (Supplementary Figure 1). These data are in agreement with the idea that the 30 nt GC-rich region downstream of the stop codon could be involved in the post-termination scanning and dissociation of the ribosome from the mRNA (35). The requirement for the 3′-UTR to bind miRNAs and proteins which are responsible for transcript stability often results in local instability around the binding site, which provides another reason to suggest that thermodynamic stability of mRNA molecule is optimized, and not minimized (48–50).
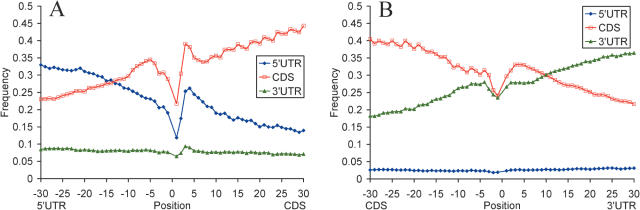
We evaluated levels of secondary structure formation within and between mRNA functional domains. As Figure 5 shows, levels of base pairing within same domains are the highest, while levels of base pairing between non-adjacent domains (5′-UTR–3′-UTR) are the lowest. Moreover, levels of base pairing with a neighboring domain rapidly decrease, as the distance to the neighboring domain increases. As seen from the figure, these levels drop 2-fold within 60 nt regions around the start codon (5′-UTR–CDS border) and the stop codon (CDS–3′-UTR border). These observations underscore major role of local secondary structures in mRNA folding. Our results indicate that secondary structures are predominantly formed within the same functional domain, and the three functional domains preferentially fold onto themselves.
Figure 5.
Profiles of base pairing for nucleotides around the start codon (A) and the stop codon (B) with different mRNA structural domains. Blue, nucleotides paired with the 5′-UTRs; red, nucleotides paired with the CDSs; green, nucleotides paired with the 3′-UTRs. Data for 19 317 human mRNAs.
Secondary structure is avoided at translation initiation and termination sites
The 5′-UTR–CDS and CDS–3′-UTR boundaries are characterized with conserved secondary structures. Sequence conservation and nucleotide base pairing profiles around the start codon show a degree of symmetry around the AUG (Figures 1, 2 and 4). Apparently, the first 9 nt upstream of the start codon, and the synonymous positions of codons directly following the AUG are subject to a stronger purifying selection than those in the rest of the CDS and the 5′-UTR. Thus, there seems to be a distinct functional signal around the start codon that extends into the 5′-UTR and CDS beyond the Kozak sequence. mRNA secondary structures tend to be less stable at the first two positions of the start codon and at the position −1 upstream of the start codon, and more stable at the last position of the start codon and the first position of the second codon. The first and second nucleotides of the start codon and three upstream nucleotides tend to be unpaired, while the third nucleotide of the start codon and several downstream nucleotides preferentially base pair with the 5′-UTR. Similarly, stop codons are less frequently base paired than the rest of mRNA sequence, and ∼10 first nucleotides of the 3′-UTR preferentially pair with the CDS (Figures 1 and 5).
These data indicate that start codons and stop codons of a large proportion of the human and mouse mRNAs reside in local loop structures. This is consistent with the observation that the start codon of the CAV1 and WNT2 transcripts are contained in evolutionarily conserved loop regions of hairpin-like secondary structure elements (12). Such secondary structures around the start and stop codon may be common in mammals and may be important for efficient initiation and termination of translation. Functional importance of the nucleotide context flanking the AUG was demonstrated in several studies that traced hereditary diseases to point mutations around the start codon [for a review see (10)]. Each of these mutations was shown to cause a decrease in translation.
CONCLUSIONS
In this work, we computationally folded the human and mouse mRNA sequences on the transcriptome scale and found a pronounced periodic pattern of mRNA secondary structure created by the structure of the genetic code. The dinucleotide relative abundances at codon positions [1,2] and [2,3] that induce periodic alteration of GC and AU base pairings, and [3,1] that define the usage of 4-fold degenerate synonymous codons are important for the maintenance of this pattern. Although the third GC-rich codon positions support mRNA folding in pairing phase 3 and contribute significantly to the transcript thermodynamic stability, periodic pattern of mRNA folding is also well pronounced in the absence of synonymous codon usage bias. Our results convincingly support the hypothesis that the structure of the genetic code contains provisions for the optimal secondary structure of mRNA (1,2). While all codon sites are important for the maintenance of mRNA secondary structure, degeneracy of the code allows regulation of mRNA secondary structure stability and periodicity.
Our results support the idea that selection is operating on synonymous codon sites to maintain structural features of mRNA. The distribution of base paired nucleotides in pairing phase 3 is mostly determined by the relative abundance of C and G nucleotides at the third codon sites and the avoidance of CpG and UpA context (Supplementary Figure 2 and Table 4). This is in agreement with published data on weak selection in favor of G and C at third 4-fold degenerate sites (51,52), on an elevated rate of evolution of synonymous sites in Drosophila and in hominids where selection favors GC pairs (53), and with observation that the location of synonymous mutations in the mouse lineages is non-random with respect to mRNA stability (15). It is known that synonymous sites are occupied by C and G more often than intron sites, especially in potential CpG sites despite their enhanced mutability (54). A plausible cause is synergistic epistasis due to the involvement of synonymous sites in maintaining the structure of mRNA (51,55).
Periodicity in mRNA secondary structure facilitates formation of intramolecular helices and a more compact transcript folding which may enhance resistance of the genetic message to degradation and modification. We found that the average ΔG of dinucleotide interaction was lower for abundant messages, as compared with rare messages, although this difference was not dramatic. Thus, selection seems to operate not for the most stable, but for optimally stable and ordered mRNA secondary structure. At the same time, periodic alteration of GC and AU base pairings prevents the formation of strong local secondary structures that may stall ribosome translocation and impede translation. Another possibility, suggested by Lagunez-Otero and Trifonov (4), is a potential role of local periodicities in mRNA structure in the control of reading frame during translation. In this work, we analyzed mRNA secondary structures with the lowest predicted free energy, and did not take into account dynamic behavior of mRNA molecules. In the cell, the observed periodic properties may be even more important for highly dynamic mRNA secondary structure. Our results demonstrate that the genetic code allows preservation of both protein and RNA structure, and underscore the importance of the structure of the genetic code and degenerate codon sites for the maintenance of mRNA folding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Eugene Koonin, John Atkins, Luda Diatchenko and Alexey Spiridonov for critical reading of the manuscript; Michael Roytberg for helpful discussions. This research was supported by the Intramural Research Program of the NIH, NLM. Funding to pay the Open Access publication charges for this article was provided by National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.White H.B., III, Laux B.E., Dennis D. Messenger RNA structure: compatibility of hairpin loops with protein sequence. Science. 1972;175:1264–1266. doi: 10.1126/science.175.4027.1264. [DOI] [PubMed] [Google Scholar]
- 2.Ball L.A. Secondary structure and coding potential of the coat protein gene of bacteriophage MS2. Nature New Biol. 1973;242:44–45. doi: 10.1038/newbio242044a0. [DOI] [PubMed] [Google Scholar]
- 3.Fitch W.M. The large extent of putative secondary nucleic acid structure in random nucleotide sequences or amino acid derived messenger-RNA. J. Mol. Evol. 1974;3:279–291. doi: 10.1007/BF01796043. [DOI] [PubMed] [Google Scholar]
- 4.Lagunez-Otero J., Trifonov E.N. mRNA periodical infrastructure complementary to the proof-reading site in the ribosome. J. Biomol. Struct. Dyn. 1992;10:455–464. doi: 10.1080/07391102.1992.10508662. [DOI] [PubMed] [Google Scholar]
- 5.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 6.Sorensen M.A., Kurland C.G., Pedersen S. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 7.Sharp P.M., Averof M., Lloyd A.T., Matassi G., Peden J.F. DNA sequence evolution: the sounds of silence. Philos. Trans. R Soc. Lond. B Biol. Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- 8.Mita K., Ichimura S., Zama M., James T.C. Specific codon usage pattern and its implications on the secondary structure of silk fibroin mRNA. J. Mol. Biol. 1988;203:917–925. doi: 10.1016/0022-2836(88)90117-9. [DOI] [PubMed] [Google Scholar]
- 9.Zama M. Correlation between mRNA structure of the coding region and translational pauses. Nucleic Acids Symp. Ser. 1999;42:81–82. doi: 10.1093/nass/42.1.81. [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz L., Burge C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer I.M., Miklos I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seffens W., Digby D. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 1999;27:1578–1584. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia M., Luo L., Liu C. Statistical correlation between protein secondary structure and messenger RNA stem–loop structure. Biopolymers. 2004;73:16–26. doi: 10.1002/bip.10496. [DOI] [PubMed] [Google Scholar]
- 15.Chamary J.V., Hurst L.D. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- 17.Duret L. tRNA gene number and codon usage in the C.elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 18.Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.dos Reis M., Savva R., Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;34:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamary J.V., Hurst L.D. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol. Biol. Evol. 2004;21:1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- 21.Chamary J.V., Parmley J.L., Hurst L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nature Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 22.Comeron J.M. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urrutia A.O., Hurst L.D. Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics. 2001;159:1191–1199. doi: 10.1093/genetics/159.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urrutia A.O., Hurst L.D. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duret L. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 26.Kanaya S., Yamada Y., Kinouchi M., Kudo Y., Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 2001;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 27.Lavner Y., Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 28.Hartl D.L., Moriyama E.N., Sawyer S.A. Selection intensity for codon bias. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Innan H., Stephan W. Selection intensity against deleterious mutations in RNA secondary structures and rate of compensatory nucleotide substitutions. Genetics. 2001;159:389–399. doi: 10.1093/genetics/159.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konecny J., Schoniger M., Hofacker I., Weitze M.D., Hofacker G.L. Concurrent neutral evolution of mRNA secondary structures and encoded proteins. J. Mol. Evol. 2000;50:238–242. doi: 10.1007/s002399910027. [DOI] [PubMed] [Google Scholar]
- 31.Pickering B.M., Willis A.E. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Duan J., Antezana M.A. Mammalian mutation pressure, synonymous codon choice, and mRNA degradation. J. Mol. Evol. 2003;57:694–701. doi: 10.1007/s00239-003-2519-1. [DOI] [PubMed] [Google Scholar]
- 33.Duan J., Wainwright M.S., Comeron J.M., Saitou N., Sanders A.R., Gelernter J., Gejman P.V. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 34.Capon F., Allen M.H., Ameen M., Burden A.D., Tillman D., Barker J.N., Trembath R.C. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum. Mol. Genet. 2004;13:2361–2368. doi: 10.1093/hmg/ddh273. [DOI] [PubMed] [Google Scholar]
- 35.Shabalina S.A., Ogurtsov A.Y., Rogozin I.B., Koonin E.V., Lipman D.J. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 2004;32:1774–1782. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brudno M., Poliakov A., Salamov A., Cooper G.M., Sidow A., Rubin E.M., Solovyev V., Batzoglou S., Dubchak I. Automated whole-genome multiple alignment of rat, mouse, and human. Genome Res. 2004;14:685–692. doi: 10.1101/gr.2067704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogurtsov A.Y., Roytberg M.A., Shabalina S.A., Kondrashov A.S. OWEN: aligning long collinear regions of genomes. Bioinformatics. 2002;18:1703–1704. doi: 10.1093/bioinformatics/18.12.1703. [DOI] [PubMed] [Google Scholar]
- 39.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogurtsov A.Y., Shabalina S.A., Kondrashov A.S., Roytberg M.A. Analysis of internal loops within RNA secondary structure in almost quadratic time. Bioinformatics. 2006 doi: 10.1093/bioinformatics/btl083. in press. [DOI] [PubMed] [Google Scholar]
- 41.Karlin S., Mrazek J. What drives codon choices in human genes? J. Mol. Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- 42.Carlini D.B., Chen Y., Stephan W. The relationship between third-codon position nucleotide content, codon bias, mRNA secondary structure and gene expression in the drosophilid alcohol dehydrogenase genes Adh and Adhr. Genetics. 2001;159:623–633. doi: 10.1093/genetics/159.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Workman C., Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanawalt P.C. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 45.Beutler E., Gelbart T., Han J.H., Koziol J.A., Beutler B. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc. Natl Acad. Sci. USA. 1989;86:192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogurtsov A.Y., Sunyaev S., Kondrashov A.S. Indel-based evolutionary distance and mouse–human divergence. Genome Res. 2004;14:1610–1616. doi: 10.1101/gr.2450504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shabalina S.A., Ogurtsov A.Y., Kondrashov V.A., Kondrashov A.S. Selective constraint in intergenic regions of human and mouse genomes. Trends Genet. 2001;17:373–376. doi: 10.1016/s0168-9525(01)02344-7. [DOI] [PubMed] [Google Scholar]
- 48.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 49.Shabalina S.A., Ogurtsov A.Y., Lipman D.J., Kondrashov A.S. Patterns in interspecies similarity correlate with nucleotide composition in mammalian 3′UTRs. Nucleic Acids Res. 2003;31:5433–5439. doi: 10.1093/nar/gkg751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shabalina S.A., Spiridonov N.A. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akashi H. Inferring weak selection from patterns of polymorphism and divergence at ‘silent’ sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eyre-Walker A. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152:675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McVean G.A., Vieira J. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics. 2001;157:245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondrashov F.A., Ogurtsov A.Y., Kondrashov A.S. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. 2005 doi: 10.1016/j.jtbi.2005.10.020. in press. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y., Carlini D.B., Baines J.F., Parsch J., Braverman J.M., Tanda S., Stephan W. RNA secondary structure and compensatory evolution. Genes Genet. Syst. 1999;74:271–286. doi: 10.1266/ggs.74.271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.