Abstract
Besides linear RNAs, pre-mRNA splicing generates three forms of RNAs: lariat introns, Y-structure introns from trans-splicing, and circular exons through exon skipping. To study the persistence of excised introns in total cellular RNA, we used three Escherichia coli 3′ to 5′ exoribonucleases. Ribonuclease R (RNase R) thoroughly degrades the abundant linear RNAs and the Y-structure RNA, while preserving the loop portion of a lariat RNA. Ribonuclease II (RNase II) and polynucleotide phosphorylase (PNPase) also preserve the lariat loop, but are less efficient in degrading linear RNAs. RNase R digestion of the total RNA from human skeletal muscle generates an RNA pool consisting of lariat and circular RNAs. RT–PCR across the branch sites confirmed lariat RNAs and circular RNAs in the pool generated by constitutive and alternative splicing of the dystrophin pre-mRNA. Our results indicate that RNase R treatment can be used to construct an intronic cDNA library, in which majority of the intron lariats are represented. The highly specific activity of RNase R implies its ability to screen for rare intragenic trans-splicing in any target gene with a large background of cis-splicing. Further analysis of the intronic RNA pool from a specific tissue or cell will provide insights into the global profile of alternative splicing.
INTRODUCTION
Completion of the human genome sequencing project will be followed by detailed genome annotation (1–3). Most of available physical resources for cDNA clones were only partially sequenced as expressed sequence tags (ESTs). Full-length cDNAs, copied from complete mRNAs, are an important resource for identifying genes and determining their structural features, forming a basis for transcriptome analysis [reviewed in (4,5)]. In theory, excised intronic cDNA sequences generated by pre-mRNA splicing can constitute a complementary resource. As an initial approach to investigating the availability of the intronic cDNA, we examined how well the excised lariat RNAs are preserved in the total cellular RNA. We tested representative Escherichia coli exoribonucleases for their ability to remove abundant linear RNAs and to enrich lariat introns in RNA sources. A special RT–PCR protocol was subsequently used to detect and identify specific lariat introns of the target human gene transcripts.
Besides linear and lariat RNAs, pre-mRNA splicing process has the capability to produce two topologically distinct RNA species, i.e. a Y-structure branch RNA and a true circular RNA. Trans-splicing releases introns with branches, instead of lariat introns in the case of conventional cis-splicing [reviewed in (6,7)]. Two kinds of pre-mRNA trans-splicing take place naturally in eukaryotes; (i) Intergenic trans-splicing, in which a short spliced-leader (SL) sequence is transferred to pre-mRNA 3′ splice sites, and becomes a 5′- end sequence which is shared by multiple mRNAs. This SL trans-splicing takes place most commonly in trypanosomes and nematodes; (ii) Intragenic (or homotypic) trans-splicing, in which the 5′ splice site on one pre-mRNA is joined to the 3′ splice site on another pre-mRNA transcribed from the same gene. This type of trans-splicing has been reported in Drosophila and mammals [(8,9) and references therein]. Circular RNAs, without any branching of the 2′, 5′-phosphodiester bond, have been identified in several transcripts of mammalian genes (10–18). It has been suggested that circular RNA consisting of spliced exons is generated by re-splicing of excised lariat RNA in the process of exon skipping type alternative splicing (14). Removing of an intron, including the branch portion, in this process produces a complete circular RNA molecule with a 3′, 5′-phosphodiester bond. Therefore, circular RNA that consists of multiple exons could be a by-product, and thus evidence, of the exon skipping type alternative splicing.
A 3′ to 5′ exoribonuclease digests linear RNAs from their free 3′ termini, but it should also digest Y-structure branch RNA that has two 3′ termini, while the loop portions of lariat RNA and circular RNA that lack their 3′ ends must be refractory to exoribonucleases. Thus, 3′ to 5′ exoribonucleases can be used to discriminate lariat RNA and branch RNA, which are distinct intron products of cis-splicing and trans-splicing, respectively. We wish to examine which kinds of E.coli exoribonucleases can strictly distinguish these RNA structures as substrates.
Seven different 3′ to 5′ RNases have been characterized in E.coli so far; Ribonuclease II (RNase II), ribonuclease R (RNase R), polynucleotide phosphorylase (PNPase) and oligoribonuclease have been reported to function in mRNA degradation [reviewed in (19)]. The substrate specificities of these exoribonucleases are different. Oligoribonuclease is required for the complete degradation of small oligoribonucleotides (20). RNase II prefers homopolymers such as poly(A), whereas RNase R is more active on rRNAs (21). The degradation activities of PNPase and RNase II are affected by RNA secondary structures while RNase R is more effective on the structured RNA, e.g. the repetitive extragenic palindromic (REP) sequence (21,22).
In this study, we analyzed the digestion activity of three highly purified exoribonucleases, RNase R, RNase II and PNPase, with model RNA substrates prepared from in vitro splicing of human β-globin pre-mRNA, which is a well-characterized splicing substrate (23,24). We found that RNase R is an ideal ribonuclease to destroy abundant linear RNAs while the loop portions of lariat RNAs remain fully intact. Using human total RNA, we demonstrate that RNase R digestion successfully yields an RNA source that consists of lariat RNAs and circular RNAs derived from pre-mRNA splicing. An mRNA source for conventional cDNA is usually separated using oligo(dT) chromatography by the poly(A) tail on its 3′ end, whereas our method screens for lariat RNAs by the unique 2′-5′ linked loop structure. Our results indicate that this new technology can be used to construct an intronic cDNA library, which should complement the information contained in conventional (exonic) cDNA libraries generated from mature mRNAs.
MATERIALS AND METHODS
Preparation of E.coli RNases
E.coli RNase II, RNase R and PNPase genes were all cloned into the pET15b expression vector (Novagen). The enzymes were overexpressed in E.coli BL21(DE3) or Rosetta(DE3)pLysS strains (Novagen) after induction with isopropyl-β-d-thiogalactopyranoside (IPTG). The RNase R used in this work is a truncated form with deletion of the C-terminal 83 amino acids (Arg/Lys-rich region). This truncated RNase R shows no difference in activity when compared with full-length RNase R. The details of cloning, overexpression and purification of these exoribonucleases will be published elsewhere (Y. Zuo, Y. Wang and A. Malhotra, manuscript in preparation). In brief, RNase II and C-terminal truncated RNase R were chromatographically purified using the same series of four columns; Affi-gel Blue (Amersham Biosciences), Hydroxyapatite (BioRad), Mono Q (Amersham Biosciences) and Superdex S200 (Amersham Biosciences). PNPase was purified using an initial ammonium sulfate precipitation step, followed by Hydroxapatite, Mono Q and Superdex S200 column chromatography. The purified enzymes are fully active and are at least 95% pure as judged by SDS–PAGE (Coomassie blue staining).
In vitro splicing
In vitro splicing assays with HeLa cell nuclear extracts were performed as described previously (24,25). Splicing reactions with human β-globin pre-mRNA were incubated at 30°C for 1 h. The splicing products were extracted with phenol and ethanol precipitated (without tRNA as a carrier) to be used as RNA substrates of the further digestion with RNases.
RNase digestion
RNA digestion reactions with RNase II and RNase R were performed essentially as described (21,26). PNPase degradation reactions were performed in 5 mM Tris–HCl (pH 8.2), 5 mM MgCl2, 60 mM KCl and 10 mM sodium phosphate. Purified enzymes (1 µg) were added to 10 µl reaction mixtures along with splicing products that were prepared by in vitro splicing with 20 fmol of β-globin pre-mRNA. The reaction mixtures were incubated at 37°C for the indicated time. The purified enzymes (1 µg) were also used to digest 1 µg of human skeletal muscle total RNA (Clontech) in 10 µl at 37°C for 30 min followed by ethanol precipitation; the digestion was repeated three times in total. RNase-digested products were extracted with phenol/chloroform, ethanol precipitated, and analyzed by denaturing 5.5 or 9.0% PAGE or 1.0% agarose gel electrophoresis and autoradiography as described previously (24).
Debranching assays
The debranching assays were performed as described previously (27). Extracted RNAs from each RNase-digested RNA sample were treated for 30 min at 30°C with HeLa cell S100 extract under standard splicing conditions (25). The samples were analyzed using denaturing 9% PAGE and autoradiography.
Oligonucleotide-targeted RNase H digestion
To prepare branch RNA, oligonucleotide-targeted RNase H digestion was performed with the loop RNA (lariat RNA without most of the 3′ tail), which was prepared from in vitro splicing reaction with human β-globin pre-mRNA followed by RNase R digestion. The majority of RNA extracted from RNase R digestion mixture is the loop RNA. Therefore, extracted total RNA was digested by 0.16 U/µl of RNase H (Takara Mirus Bio) at 37°C for 30 min in the reaction mixture containing 40 mM HEPES–KOH (pH 7.7), 100 mM KCl, 4 mM MgCl2, 1 mM DTT, 5% glycerol and 8 µM synthetic oligonucleotide (TTGTAACCTTGA; Operon Biotechnologies). The RNase H-digested RNA was extracted with phenol/chloroform and ethanol precipitated. This RNA was used as substrate in RNase digestion reactions, carried out as described above (incubation for 15 min).
Mapping of mRNA sequence of the human dystrophin gene
To obtain the complete structure of the human dystrophin gene, its largest cDNA sequence (accession no. NM_000109) was mapped onto the human chromosome X, which spans the region of chrX: 30 897 002 to chrX: 33 117 383 (http://genome.ucsc.edu/). The exon–intron structure was defined by the alignment of mRNA sequence with the genomic sequence using the SIM4 program (28). We designed DNA primers for RT–PCR based on the obtained sequences.
Detection of target RNAs by RT–PCR
The method to detect lariat introns by RT–PCR across the branch site was reported previously (29–32). Human skeletal muscle total RNA (1 µg; Clontech) and the RNase R-digested RNA from the same source (1 µg) were used as templates for the RT–PCR. To prepare the cDNAs of dystrophin mRNAs, circular RNAs and lariat introns, the reverse transcription reactions were performed with a reverse transcriptase (SuperScript RNase H−; Invitrogen) and random hexamers in accordance with the manufacturer's instruction. The reaction mixtures (total 10 µl) containing 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM DTT, 1 mM each of dNTPs, 2.5 µM random hexamer primers, 1 U/µl Prime RNase inhibitor (Eppendorf), 10 U/µl SuperScript RNase H− Reverse Transcriptase, and total RNA (undigested) or RNase R-digest template RNAs (see above) were incubated at 30°C for 10 min, at 42°C for 120 min, at 50°C for 30 min, at 60°C for 30 min, at 99°C for 5 min, and finally kept at 4°C or lower.
These intronic cDNAs were used as templates for the first PCRs. The PCRs (in total 50 µl) containing 50 mM KCl, 10 mM Tris–HCl (pH 9.0), 0.1% Triton X-100, 2.5 mM MgCl2, 2 µM specific DNA primers (two kinds; see below), 0.05 U/µl Taq DNA polymerase (Promega), and 1 µl cDNA solutions, were incubated for 5 min at 94°C followed by multiple PCR cycles (see below); at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 30 s, with an incubation at 72°C for 10 min at the final round. These first PCR products were purified on a Sephacryl S300 column (Amersham Biosciences) and used as templates for the second nested PCR. The second PCRs were the same as the first PCRs except for using an adjacent inner set of primers. The optimal cycle numbers of the first and second PCRs were determined to reflect proportional changes in the PCR products.
The sequence of DNA primers (Operon Biotechnologies) and cycle numbers for PCRs are as follows. Detection of the dystrophin mRNA: first PCR (20 cycles) with E7-S1 primer (ACTGGAACATGCATTCAACATCGC) and E8-A1 primer (CACTTTAGGTGGCCTTGGC), second PCR (10 cycles) with E7-S2 primer (CAGGCATAGAGAAACTGCTCGATCC) and E8-A2 primer (ATTTCCACTTCCTGGATGGC). Detection of the dystrophin lariat intron 8: first PCR (25 cycles) with I8-S1 primer (ACTCTCCAACATGATATTAAGTGCC) and I8-A1 primer (TGTGCACGTAATACCTAAAAATGC), and second PCR (19 cycles) with I8-S2 primer (CATTCTGGGAGTATACCAATTTCG) and I8-A2 primer (AGAAAACATCTTGAATAGTAGCTGTCC). Detection of the dystrophin lariat intron 10: first PCR (25 cycles) with I10-S1 primer (GCCTGCTTCTGAAGAACTTGAC) and I10-A1 primer (GTTGGAATCCCAAGCACATC), second PCR (19 cycles) with I10-S2 primer (AAAGTGGTTTTGGGATTCTGC) and I10-A2 primer (GAGGAAAAAGGATGACTTGCC). Detection of the dystrophin lariat intron 24: first PCR (25 cycles) with I24-S1 primer (TTCATGCCATAATAACCATTGAAC) and I24-A1 primer (GGGAGAGGAGAGCAAAATCC), second PCR (19 cycles) with I24-S2 primer (TTTGCAAGACTGTTAGGCAGTC) and I24-A2 primer (CCAGCTGTAAAACACTGATCTAACC). Detection of the dystrophin circular RNA: first PCR (25 cycles) with E16-S1 primer (GCATGGCTGGATAACTTTGC) and E3-A1 primer (TTCGAGGAGGTCTAGGAGGC), second PCR (19 cycles) with E16-S2 primer (CGGTGTTGGGATAATTTAGTCC) and E3-A2 primer (TGAAGAGGTTCTCAATATGCTGC).
The amplified PCR products were analyzed using a 6% PAGE and visualized by ethidium bromide staining. To estimate the enrichment factor, the PCR products were quantified by densitometry (ImageQuant; Molecular Dynamics) and the concentrations of total RNA samples were measured by UV absorbance.
RESULTS
RNase R efficiently degrades linear RNA species of in vitro splicing products
As originally characterized (23), major in vitro splicing products of human β-globin mini-gene transcripts are the lariat intermediate, spliced mRNA, the first exon (exon 1), and the lariat intron, which are well separated from unspliced pre-mRNA by denaturing 5.5% PAGE (Figure 1A, lane 1). We used these RNA products to test RNA degradation activity of three E.coli 3′ to 5′ exoribonucleases, RNase R, RNase II and PNPase.
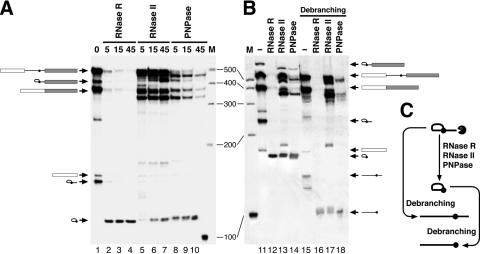
Figure 1.
(A) Digestions of the splicing products with RNase R, RNase II and PNPase. Human β-globin pre-mRNA was spliced with HeLa cell nuclear extract in vitro (lane 1) and the RNA products were further digested with each RNase for indicated incubation time (min; lanes 2–10). The RNAs were analyzed by denaturing 5.5% PAGE. The positions of splicing products and RNase-digested products are schematically indicated with their structures. DNA markers (New England Biolabs) are shown with their sizes (M). (B) Analyses of the lariat RNAs by debranching. In vitro spliced products and their RNase-digested RNA products [lanes 11–14; reactions are corresponding to lanes 1, 3, 7 and 9 in (A), respectively] were debranched with HeLa cell S100 extract (lanes 15–18). The RNAs were analyzed by denaturing 9.0% PAGE. The positions of RNA products are schematically represented with their structures. (C) Digestion pathway of the 3′ tail of lariat RNA by RNases and debranching of the products.
RNase II partially degraded a majority of the linear RNA species, which are unspliced pre-mRNA and spliced mRNA (lanes 5–7). PNPase degraded these linear RNAs more efficiently than RNase II, but some linear RNAs still remained undigested even after 45 min of incubation (lanes 8–10). In contrast, these linear RNAs are almost completely degraded by RNase R (lanes 2–4). Since substrate specificity of these RNases is different (21,22), it is difficult to compare absolute activity of these RNases. Nevertheless, RNase R appears suitable for removing abundant linear RNAs from bulk RNA populations under this condition (using same mass of the RNases).
These results are consistent with previous characterization of these exoribonucleases [reviewed in (19)]. RNase II and PNPase are sensitive to secondary structure and they pause or dissociate at stable RNA duplexes (33–35), whereas RNase R is particularly efficient at degrading RNA with extensive secondary structure (21,22). Our results thus suggest that large linear in vitro splicing products (pre-mRNA and spliced mRNA) may contain extensive secondary structure, e.g. partial duplex and hairpins.
RNase R-resistant RNA is a loop portion of lariat intron
Splicing products with a lariat structure, i.e. lariat intermediate and lariat intron, were also completely removed with RNase R. However, a novel discrete product emerged with much higher mobility (Figure 1A, lanes 2–4). This product was detected with all three RNases and is very stable even after long incubation (45 min; lanes 4, 7 and 10). Since these RNases are 3′ to 5′ exoribonucleases, we assumed that this new product is the lariat RNA lacking a 3′ tail.
To verify the new product, the same RNA samples were analyzed by denaturing 9% PAGE (Figure 1B). Electrophoresis on a higher polyacrylamide percentage gel facilitates the detection of a non-linear RNA fragment such as a lariat intron with its relatively slower mobility (23,24). As expected, we observed marked mobility shifts of the novel product as well as the lariat intron and intermediate (lanes 11–14, compare lanes 1–4).
Next we performed a debranching assay to confirm the structure and length of this novel product. HeLa cell nuclear and cytosolic S100 extracts, which contain the debranching enzyme, cleave the 2′, 5′-phosphodiester bond of lariat intron (27). Our debranching assay with S100 extract cleaved the lariat intron and generated a linearized fragment of the expected size (130 nt; Figure 1B, land 15 and Figure 1C). The novel product was also debranched and generated a linear intron fragment, which matches well with the length (94 nt) of the intron lacking the 3′ portion from the branch point (Figure 1B, lanes 16–18 and Figure 1C). Both the novel product and its linearized fragment were purified from the gel and RT–PCR was performed. The downstream 3′ portion could not be amplified and the verified sequence matched to the intron sequence (data not shown), indicating that the novel product is indeed the lariat intron without most of the 3′ tail. We found that this loop structure is highly resistant to exoribonucleases. It is remarkable that the digestion of in vitro splicing products with RNase R can isolate this particular RNA species to near homogeneity (Figure 1A, lane 4).
RNase R degrades branch RNA
A Y-structure branch intron is generated through trans-splicing, which corresponds to a lariat intron of a conventional cis-splicing [reviewed in (6,7)]. We examined whether the branch RNA is susceptible to RNase R digestion. To prepare substrate RNA, the loop portion of a lariat intron (Figure 2A, lane 1) was cleaved 8–19 nt downstream from the 5′ splice site using RNase H with a complementary oligonucleotide (Figure 2A, lane 2 and Figure 2B).
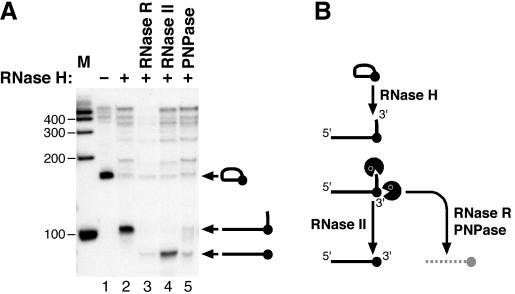
Figure 2.
(A) Digestions of the branch RNA with RNase R, RNase II and PNPase. The loop portion of a lariat intron (lane 1; corresponding to Figure 1A, lane 3) was cleaved by oligonucleotide-targeted RNase H to prepare a branch RNA (lane 2). This branch RNA was digested with each RNase for 15 min (lanes 3–5). The RNAs were analyzed by denaturing 9.0% PAGE. The positions of loop RNA, branch RNA and RNase-digested products are schematically represented by their structures. (B) Cleavage of the loop RNA by RNase H and the degradation pathways by RNases are schematically shown.
Exoribonuclease RNase II digested this branch RNA almost completely and generated a new product with faster mobility on the gel (lane 4). This RNA product was also generated, albeit much less, with the exoribonucleases RNase R and PNPase (lanes 3 and 5). Based on its estimated size, we assumed that this product is a partially degraded RNA lacking a majority of the 2′-5′ linked fragment (Figure 2B). This prediction was verified by the RT–PCR followed by sequencing. Since RNase R could digest the branch RNA most efficiently even with a short incubation (15 min), it appears to be the most suitable exoribonuclease to digest branch RNAs.
These results suggest that RNase II has lower exoribonuclease activity on undigested branched nucleotide(s), while RNase R can digest the branched nucleotides more efficiently (Figure 2B). Previously it has been demonstrated that RNase II leaves ∼10 unpaired nucleotides adjacent to a duplex structure in vitro (36). It was also shown that RNase II can generate mature 3′ termini on tRNAs in vivo (37), suggesting that it can approach as close as 4 nt from the aminoacyl stem. While RNase R can shorten RNA processively to di- and tri-nucleotides, RNase II becomes more distributive when the length of the substrate reaches ∼10 nt; this leaves an undigested core of 3–5 nt (21). It is thus possible that RNase II leaves a larger overhang when digesting one end of branch RNAs, which may further interfere with digestion from the other 3′ end (Figure 2B). Alternatively, RNase R could be digesting partially degraded branch RNA more efficiently from its 3′ ends, even when the same numbers of the 2′-5′ linked nucleotide(s) are left.
RNase R can be used to prepare an RNA source containing only lariat and circular RNAs
We have demonstrated that RNase R is capable of destroying linear and branch RNAs in vitro. Therefore in theory, extensive RNase R digestion of endogenous natural RNA can produce an RNA sample that consists of only lariat and circular RNAs. We examined this possibility by analyzing splicing products of human dystrophin pre-mRNA using the human total RNA from skeletal muscle. The human dystrophin gene is the largest known gene spanning over 3 Mb with at least 79 exons (38,39), and thus its large transcript (∼14 kb) can serve as an excellent candidate to examine validity of our method.
The general feature of a total cellular RNA is a very large amount of rRNAs. We observed typical discrete bands of 28S/18S rRNAs and small RNA species (including 5/5.8S rRNAs and tRNAs) on agarose gel, and all these rRNAs were fully degraded by RNase R (Figure 3, lanes 1 and 2).
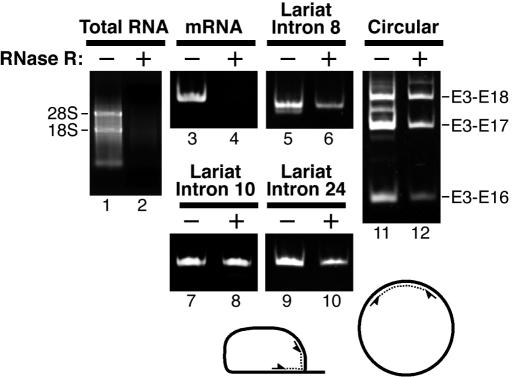
Figure 3.
RT–PCR detection of mRNA, intact lariat introns, and circular RNAs from either RNase R-digested (+) or mock-digested (−) human skeletal muscle total RNA. The RNA was directly analyzed by 1.0% agarose gel electrophoresis (lanes 1 and 2). The RNA was used for RT–PCR with primers to detect the dystrophin mRNA including exons 7 and 8 (lanes 3 and 4), three lariat introns as indicated (lanes 5–10), and the circular RNAs including multiple exons (lanes 11 and 12). The PCR products were analyzed by native 6.0% PAGE (lanes 3–12). The structures of the lariat intron and circular RNA are schematically represented and arrows indicate the positions of primers used for PCR.
Next we performed RT–PCR to detect specific mRNAs that exist in much less quantity than rRNAs. We could successfully detect human dystrophin mRNA using specific primers targeting their exon 7 and exon 8 in the human total RNA, whereas we barely detected this mRNA after RNase R digestion (lanes 3 and 4). Even after RNase R digestion, extensive amplification with more PCR cycles detected a weak signal with this primer set; however, it appears to be derived from the circular RNA including exons 7 and 8 (see below). Since the distance between the 3′ primer position in exon 8 and the 3′ terminus of the dystrophin mRNA is over 12.8 kb (38), our data suggest that RNA degradation by RNase R from the 3′ terminus is quite efficient. We conclude that RNase R has the potential to destroy a large fraction of mRNA species in a human total RNA preparation since most mRNAs are smaller than the dystrophin mRNA.
We then examined whether a specific lariat intron of dystrophin pre-mRNA exists in this RNase R-treated RNA sample. Previously it has been reported that MMLV and AMV reverse transcriptases have the ability to read through a 2′-5′ linked ribonucleotide in a template RNA (29). By taking advantage of this activity, lariat molecules of a group II intron, retrotransposon Ty1 RNA, and Drosophila pre-mRNA intron, were successfully detected in the total cellular RNA by RT–PCR across the branch site (30–32). Because of the opposing direction of the two primers, this arrangement never generates PCR products from any contaminating genomic DNA. Using this RT–PCR technique, we detected an expected lariat form of intron 8 of the dystrophin pre-mRNA in the human skeletal muscle total RNA (Figure 3, lane 5). We could detect an identical fragment from the lariat intron 8 after RNase R digestion (lane 6). Integrity of the lariat structure was also verified by different sets of primers that cover whole loop portion of the lariat (data not shown). Subcloning of the RT–PCR products followed by the sequencing revealed that the fragment contains a junction between the 5′ splice site and 24 bases upstream of the 3′ splice site. The position and surrounding sequence of the identified branch point, CUCUAUCCACUCCC (branched nucleotide is underlined), was matched to the consensus in mammalian introns (40), confirming our detection of the bona fide lariat intron 8. Using the same approach, we also detected lariat RNAs before and after RNase R digestion using primer sets targeting introns 10 and 24 of the dystrophin pre-mRNA (lanes 7–10). Quantitative analyses revealed that total RNAs and these three lariat RNAs remained ∼0.25% and ∼66.9% after RNase R treatment, respectively. We thus estimated an enrichment factor of ∼268-fold. These results indicate that the lariat introns generated by splicing of dystrophin pre-mRNA are well conserved and highly enriched in the RNase R-digested RNA source.
Lastly, we examined a rare circular RNA of dystrophin pre-mRNA using our RNA source. Since the circular RNA that contains spliced exons could be a re-spliced by-product of corresponding exon skipping, the detection of such a special RNA provides evidence for alternative splicing (14). Indeed, it has been reported that eleven (out of fourteen) kinds of multi-exon skipping in the dystrophin pre-mRNA generate circular RNAs, including the corresponding skipped exons (16). The 5′ end of the most upstream skipped exon is connected to the 3′ end of the most downstream skipped exon in the circular RNA; we thus designed the PCR primers across the connection of these exons. Using primers for exon 3 and exon 16, we detected the exon 3-16, 3-17 and 3-18 junctions, which are generated from three known circular RNAs including exons 3–16, exons 3–17 and exons 3–18 [Figure 3, lane 11; (16)]. Strikingly, all these circular RNAs from the dystrophin gene were detectable after RNase R digestion (lane 12). This result shows that the circular RNAs, even in extremely low amounts (16), are resistant to RNase R digestion, while the enormously abundant linear RNAs are almost completely digested.
DISCUSSION
It has been reported that half-lives of four mammalian nuclear introns after splicing are relatively long, ranging from 6 to 29 min, probably due to the existence of branched structures (41). This is probably the critical reason why spliced lariat introns, which must be degraded eventually in vivo, are present in total cellular RNA preparations. It is also remarkable that rare circular RNAs generated by multi-exon skipping events (14,16) are detected. Here we demonstrated that the RNA population in the RNase R-treated total RNA consists of these lariat and circular RNA molecules, preserving evidence of constitutive and alternative splicing events.
Our results suggest the potential use of RNase R-digested RNA source from human total RNA for an intronic cDNA library to study global splicing profiling. To be successful for this purpose, lariat molecules from the target genes should be represented well enough in the RNA source so that they can be detected by RT–PCR. We succeeded in detecting three introns (introns 8, 10 and 24), which were arbitrarily chosen from the dystrophin gene, in the RNase R-digested RNA prepared from human muscle total RNA. We could also detect circular RNAs including exons 3–16, 3–17 and 3–18, which were generated by multi-exon skipping from exon 2 to 17, exon 2 to 18, and exon 2 to 19, respectively (16). It has been reported that the amounts of these circular RNAs are much less than their corresponding mRNAs (16). We expect that intron lariats generated from splicing of dystrophin pre-mRNA are well represented in the RNase R-digested RNA source prepared from human muscle total RNA.
The RNase R-digested RNA source may not necessarily guarantee coverage of all the lariat introns from rare transcripts. However, this approach can be applied to the analysis of alternative splicing events using any kind of endogenous RNA from specific tissues or cell lines in which the mRNA isoforms of interest are differentially expressed. As we show in this study, such specific cellular RNAs can be treated with RNase R to remove a vast majority of linear RNAs. This enriched RNA source may facilitate high throughput cloning of the lariat structures to prepare an intronic cDNA library. Since we can directly detect excised lariat RNAs, including either introns or exons, it may be an advantage to exclusively analyze any kind of alternative splicing in which alternative initiation and termination are precluded. This technology will also be valuable for providing more branch-site mapping data that have been lacking, and are essential for developing more accurate computational gene prediction algorithms [reviewed in (42)].
Our in vitro and in vivo results demonstrate that RNase R degrades Y-structure RNA, whereas the loop portion of lariat RNAs and circular RNAs are fully resistant. This feature implies that our approach can be used to distinguish between cis-splicing and trans-splicing of a target gene. Intragenic trans-splicing of individual genes has been reported rarely [(8,9) and references therein], probably because trans-splicing would be apparent only if it would contain sequences originating from two distinguishable alleles, or intergenic, containing sequences from two different genes, e.g. a SL trans-splicing [reviewed in (6,7)]. A recent genome-wide survey suggested that the natural levels of intragenic trans-splicing are much higher than that expected previously (43). Our technique provides an effective tool to directly examine an intragenic trans-splicing event by monitoring the disappearance of the RT–PCR signal from Y-structure intron in RNase R-treated RNA source. Conversely, if the signal persists after RNase R digestion, this would indicate the generation of an intact lariat RNA and provide evidence of a conventional cis-splicing event.
This is the first report to propose the potential construction of an intronic cDNA library from an RNA source that exclusively consists of lariat and circular RNAs. We have developed a new approach that will provide complementary information for global transcriptome analysis, such as recent studies with human and mouse full-length cDNA libraries (44,45).
Acknowledgments
We are grateful to Dr M. P. Deutscher for valuable comments, encouragement and financial support. We thank Dr K. E. Rudd for critical reading of the manuscript, Dr K. Ohe for helpful suggestions, and G. Kozloski and T. Venkataraman for technical assistance. A. Mayeda was supported by a new investigator developmental grant from the Muscular Dystrophy Association, and an institutional research grant from the Sylvester Braman Family Breast Cancer Research Institute. A. Malhotra is supported by a grant from National Institute of Health (GM69972). J.W. and M.Q.Z. are supported by a grant from National Institute of Health (HG001696). Y.Z. is supported in part by a postdoctoral fellowship from the American Heart Association Florida/Puerto Rico Affiliate (0325296B). A. Mayeda and A. Malhotra are research members of the Sylvester Comprehensive Cancer Center. The Open Access publication charges for this article were waived by Oxford University Press
Conflict of interest statement. None declared.
REFERENCES
- 1.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Hayashizaki Y., Kanamori M. Dynamic transcriptome of mice. Trends Biotechnol. 2004;22:161–167. doi: 10.1016/j.tibtech.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Harbers M., Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nature Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen T.W. Trans-splicing. In: Krainer A.R., editor. Eukaryotic mRNA processing. Oxford: IRL press; 1997. pp. 310–334. [Google Scholar]
- 7.Hastings K.E. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Horiuchi T., Giniger E., Aigaki T. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes. Dev. 2003;17:2496–2501. doi: 10.1101/gad.1137303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahara T., Tasic B., Maniatis T., Akanuma H., Yanagisawa S. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol. Cell. 2005;18:245–251. doi: 10.1016/j.molcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 11.Cocquerelle C., Daubersies P., Majerus M.A., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 13.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 14.Zaphiropoulos P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl Acad. Sci. USA. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaphiropoulos P.G. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol. Cell. Biol. 1997;17:2985–2993. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surono A., Takeshima Y., Wibawa T., Ikezawa M., Nonaka I., Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum. Mol. Genet. 1999;8:493–500. doi: 10.1093/hmg/8.3.493. [DOI] [PubMed] [Google Scholar]
- 17.Li X.F., Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 1999;274:8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- 18.Gualandi F., Trabanelli C., Rimessi P., Calzolari E., Toffolatti L., Patarnello T., Kunz G., Muntoni F., Ferlini A. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J. Med. Genet. 2003;40:e100. doi: 10.1136/jmg.40.8.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deutscher M.P., Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S., Deutscher M.P. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Z.F., Deutscher M.P. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Z.F., Deutscher M.P. An important role for RNase R in mRNA decay. Mol. Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 23.Ruskin B., Krainer A.R., Maniatis T., Green M.R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 24.Mayeda A., Krainer A.R. Mammalian in vitro splicing assays. Methods Mol. Biol. 1999;118:315–321. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 25.Mayeda A., Krainer A.R. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- 26.Deutscher M.P., Marlor C.W., Zaniewski R. Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA. Proc. Natl. Acad. Sci. USA. 1984;81:4290–4293. doi: 10.1073/pnas.81.14.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruskin B., Green M.R. An RNA processing activity that debranches RNA lariats. Science. 1985;229:135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- 28.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorsch J.R., Bartel D.P., Szostak J.W. Reverse transcriptase reads through a 2′-5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 1995;23:2811–2814. doi: 10.1093/nar/23.15.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel J., Hess W.R., Borner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Z., Menees T.M. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science. 2004;303:240–243. doi: 10.1126/science.1087023. [DOI] [PubMed] [Google Scholar]
- 32.Burnette J.M., Miyamoto-Sato E., Schaub M.A., Conklin J., Lopez A.J. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarneros G., Portier C. Different specificities of ribonuclease II and polynucleotide phosphorylase in 3′ mRNA decay. Biochimie. 1990;72:771–777. doi: 10.1016/0300-9084(90)90186-k. [DOI] [PubMed] [Google Scholar]
- 34.McLaren R.S., Newbury S.F., Dance G.S., Causton H.C., Higgins C.F. mRNA degradation by processive 3′–5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J. Mol. Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- 35.Coburn G.A., Mackie G.A. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 36.Coburn G.A., Mackie G.A. Overexpression, purification, and properties of Escherichia coli ribonuclease II. J. Biol. Chem. 1996;271:1048–1053. doi: 10.1074/jbc.271.2.1048. [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Deutscher M.P. Maturation pathways for E.coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 38.Roberts R.G., Coffey A.J., Bobrow M., Bentley D.R. Exon structure of the human dystrophin gene. Genomics. 1993;16:536–538. doi: 10.1006/geno.1993.1225. [DOI] [PubMed] [Google Scholar]
- 39.Nishio H., Takeshima Y., Narita N., Yanagawa H., Suzuki Y., Ishikawa Y., Minami R., Nakamura H., Matsuo M. Identification of a novel first exon in the human dystrophin gene and of a new promoter located more than 500 kb upstream of the nearest known promoter. J. Clin. Invest. 1994;94:1037–1042. doi: 10.1172/JCI117417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burge C.B., Tuschl T., Sharp P.A. Splicing of precusors to mRNAs by the spliceosomes. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA world, 2nd edition. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 41.Clement J.Q., Maiti S., Wilkinson M.F. Localization and stability of introns spliced from the Pem homeobox gene. J. Biol. Chem. 2001;276:16919–16930. doi: 10.1074/jbc.M005104200. [DOI] [PubMed] [Google Scholar]
- 42.Zhang M.Q. Computational prediction of eukaryotic protein-coding genes. Nature Rev. Genet. 2002;3:698–709. doi: 10.1038/nrg890. [DOI] [PubMed] [Google Scholar]
- 43.Dixon R.J., Eperon I.C., Hall L., Samani N.J. A genome-wide survey demonstrates widespread non-linear mRNA in expressed sequences from multiple species. Nucleic Acids Res. 2005;33:5904–5913. doi: 10.1093/nar/gki893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I & II Team. Analysis of the mouse transcriptome based on functional annotation of 60 770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 45.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., et al. Complete sequencing and characterization of 21 243 full-length human cDNAs. Nature Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]