Abstract
Purpose
To overcome radiation resistance in esophageal adenocarcinoma by tumor-specific apoptotic gene targeting using tumor necrosis factor-related apoptosis-inducing ligand (TRAIL).
Methods and Materials
Adenoviral vector Ad/TRAIL-F/RGD with a tumor specific human telomerase reverse transcription promoter was used to transfer TRAIL gene to human esophageal adenocarcinoma and normal human lung fibroblastic cells (NHLF). Activation of apoptosis was analyzed using western blot, FACS, and TUNEL assay. A human esophageal adenocarcinoma mice model was treated with intratumoral injections of Ad/TRAIL-F/RGD plus local radiotherapy.
Results
The combination of Ad/TRAIL-F/RGD and radiotherapy increased cell-killing effect in all esophageal adenocarcinoma cell lines but not in NHLF cells. This combination also significantly reduced clonogenic formation (p < 0.05) and increased sub-G1 DNA accumulation in cancer cells (p < 0.05). Activation of apoptosis by Ad/TRAIL-F/RGD plus radiotherapy was demonstrated by activation of caspase-9, -8, -3 and cleaved PARP in vitro and TUNEL assay in vivo. Combined Ad/TRAIL-F/RGD and radiotherapy dramatically inhibited tumor growth and prolonged mean survival in esophageal adenocarcinoma model to 31.6 days from 16.7 days for radiotherapy alone and 21.5 days for Ad/TRAIL-F/RGD alone (p< 0.05).
Conclusions
The combination of tumor-specific TRAIL-gene targeting and radiotherapy enhances the effect of suppressing esophageal adenocarcinoma growth and prolonging survival.
Keywords: Tumor-specific targeting, TRAIL gene therapy, Radiation resistance, Esophageal adenocarcinoma, Apoptosis
INTRODUCTION
Esophageal cancer is one of the least studied yet deadliest cancers worldwide (1). Recently, the incidence of esophageal cancer in the United States increased dramatically, particularly adenocarcinomas in the lower esophagus (2). The American Cancer Society estimates that in the year 2005, there will be 14,520 new cases of this cancer and 13,570 attributable deaths (3). Most patients present with locally advanced disease, and local regional failure is one of the main causes of poor quality of life and an increased death rate. The current standard treatment for locally advanced esophageal cancer is chemotherapy concurrent with radiotherapy. Radiotherapy is crucial for tumor local control, however, the local regional control rate is less than 50%, and the 5-year overall survival rate is only 26% (4). In a dose-escalation study in esophageal cancer comparing 50.4 Gy and 64.8 Gy radiation, both given concurrent with chemotherapy, the higher-dose did not increase the 2-year survival rate or local regional control rate (5). These findings indicate that esophageal cancer is resistant to radiation and radiation resistance is a major reason for treatment failure in this patient population. The study also showed that dose escalation was associated with higher normal tissue toxicity and a higher mortality rate (5).
Apoptosis is an important pathway of cell death following radiotherapy, chemotherapy, and apoptotic gene therapy in cancers, including esophageal cancer (6–8). A major mode of resistance to antitumor therapy, including radiotherapy and chemotherapy, is insensitivity to apoptosis induction (9). Apoptosis is controlled by a variety of genes, some with proapoptotic functions and others with antiapoptotic function (6). The balance between these competing activities determines cell fate (10). Radiation and proapoptotic genes trigger apoptosis at different levels and in different pathways (6, 11), which suggests that the two therapies should be synergistic or additive. In addition, the toxicity profiles associated with radiotherapy and gene therapy are different. Therefore, the combination of standard radiotherapy plus apoptotic gene therapy may enhance apoptosis induction, therefore overcoming tumor resistance to radiation and improving the therapeutic ratio.
The tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), a proapoptotic gene, might be very useful in gene therapy for radiation-resistant esophageal cancer. It specifically kills malignant cells but spares the normal tissues by binding with death receptors in cancer cell membrane and triggering apoptosis through a mitochondria-independent signal transduction pathway (12–14). This pathway is different from the one involved in radiation-induced apoptosis (15–18). To develop an efficient system for delivering the TRAIL gene with minimal effect on normal cells, we constructed the adenoviral vector Ad/gTRAIL, which expresses the green fluorescent protein (GFP)/TRAIL fusion protein under the control of the human telomerase reverse transcript (hTERT) promoter (19). GFP functions as a marker for the successful delivery of the TRAIL gene, and hTERT ensures the cancer-specific expression of TRAIL (most cancer cells overexpress hTERT, whereas normal cells express a minimal level of hTERT). Our preliminary data showed that Ad/gTRAIL elicited a high level of gene expression and apoptosis in tumor cells but not in normal cells, including normal human hepatocytes, the most common normal cells affected by TRAIL (19). In a mouse breast cancer study, we showed that intratumoral delivery of Ad/gTRAIL induced apoptosis in the tumor cells, inhibited tumor growth, and prolonged animal survival (20). In addition, Ad/gTRAIL had a bystander effect (i.e., it killed cancer cells close to the cells transduced by TRAIL) and sensitized cancer cells to chemotherapy in chemotherapy-resistant human breast cancer (21–22). Preliminary data obtained in a breast cancer system also showed that TRAIL protein plus radiotherapy had a synergistic effect on an established breast cancer xenograft (23).
However, we found the heterogeneity of human cancer cells to be a problem, because some cancer cells, particularly primary cancer cells, may not have an adenoviral receptor in their membranes, thus hindering adenoviral vector delivery. To create a more efficient gene delivery system for clinical use in esophageal cancer using local intratumoral injections, we inserted the gene sequence of an adenovirus fiber (F/RGD) into the construct to produce a new adenoviral vector system, Ad/TRAIL-F/RGD. The new adenoviral vector can deliver the therapeutic gene into cancer cells via 1) the adenoviral receptor or 2) attachment of the modified adenovirus fiber (F/RGD) to cell membrane. GFP was removed because a gene marker was no longer needed. The hTERT promoter targets cancer cells, however, the promoter may not be strong enough to express the large amount of TRIAL required to kill cancer cells in patients with esophageal cancer. To increase promoter strength, we coexpressed GAL4VP16 fusion protein and a very strong transcription factor for the GT (GAL4 DNA-binding site plus TATA box) promoter. GT is a very strong promoter that can be activated only by GAL4VP16. Therefore, hTERT controls the specific expression of GAL4VP16, which in turn activates the GT promoter, which initiates TRIAL expression in tumors. Preliminary data have shown that Ad/TRAIL-F/RGD can deliver the therapeutic gene more efficiently than Ad/gTRAIL (24).
We hypothesized that the combination of Ad/TRAIL-F/RGD plus radiotherapy would enhance cell killing of esophageal adenocarcinoma cells and prolong survival in mice bearing human esophageal adenocarcinoma. In this study, we evaluated the efficacy of Ad/TRAIL-F/RGD combined with radiotherapy in vitro in three esophageal adenocarcinoma cell lines and in normal human lung fibroblast cells (NHLF), and in vivo in an esophageal adenocarcinoma model. Our results showed that the combination of Ad/TRAIL-F/RGD and radiotherapy significantly inhibited the growth of human esophageal adenocarcinoma cells in vitro and in vivo. This finding provides the experimental evidence for the potential clinical use of this combination.
METHODS AND MATERIALS
Cell lines and cell culture
Three human esophageal adenocarcinoma cancer cell lines were used. Seg-1 cells were obtained from David Beer (University of Michigan, Ann Arbor, MI). SKGT-4 and SKGT-5 cells were obtained from David Schrump (Surgery Branch, National Cancer Institute, Bethesda, MD). Three cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, 1% glutamine, and 1% antibiotics and cultured at 37°C in a humidified incubator containing 5% CO2. Normal human lung fibroblast (NHLF) cells purchased from Clonetics (San Diego, CA) were cultured in medium recommended by the manufacturer.
Adenovectors
Both Ad/CMV-GFP, an adenoviral vector with a GFP under the control of the CMV promoter, and Ad/gTRAIL, an adenoviral vector with a GFP marker and TRAIL gene under the control of the hTERT promoter, have been described previously (19, 21). We recently modified the Ad/gTRAIL vector and constructed a new adenoviral vector system by removing the GFP marker gene and adding the integrin-binding modify RGD sequence in the H1 loop of adenoviral fiber (see introduction for details), namely Ad/TRAIL-F/RGD (24). The expansion, purification, titration, and quality analyses of all of these vectors were performed at the vector core facility at The University of Texas M. D. Anderson Cancer Center as described previously (19, 21, 24). The titer used in this study was determined by the absorbency of the dissociated virus at A260 nm [1 A260 nm unit = 1012 viral particles (VP)/ml], and the titer used to determine additive information was determined with a plaque assay. Particle infection unit ratios were usually between 30:1 and 100:1. Thus, the multiplicity of infection (MOI) of 1,000 VP was equivalent to an MOI of 10 to 30 infectious units. Unless otherwise specified, Ad/CMV-GFP was used as a vector control, and phosphate buffered saline (PBS) was used as a mock control.
Growth inhibition and XTT assay
The inhibition of tumor-cell growth caused by Ad/TRAIL-F/RGD alone, Ad/gTRAIL alone, radiotherapy alone, or radiotherapy in combination with Ad/TRAIL-F/RGD, or Ad/CMV-GFP was analyzed by quantitatively determining cell viability using an improved XTT assay (Cell Proliferation Kit II; Roche Molecular Biochemicals, Indianapolis, IN). Briefly, cells were plated in 96-well microtiter plates at 1 × 103 cells/well in 100 μl of medium. One day late, the medium was removed from each well and replaced with a 100-μl aliquot of medium containing Ad/TRAIL-F/RGD, Ad/gTRAIL, or Ad/CMV-GFP vector at various MOIs. After a 24-h incubation with the adenoviral vectors, cells in the 96-well plates were irradiated with various doses of γ-radiation in a 137Cs unit (model E-0103; U. S. Nuclear Corp., Burbank, CA) at room temperature. Cells were then incubated at 37°C in a humidified atmosphere containing were quantified by XTT 5% CO2. Three days after incubation, cell growth and viability assay. Briefly, the culture medium was removed, and 50 μl of an XTT reaction mixture was added to each well with fresh medium to a final concentration of 0.3 mg/ml per well. Cells were then incubated for 2 h at 37°C. The absorbance was measured at a wavelength of 450 nm against a reference wavelength of 630 nm in a microplate reader (Model MRX; Dynatech Laboratories, Chantilly, VA). The percentage of viable cells was calculated in terms of the absorbency of treated cells compared with the absorbency of untreated control cells. Each experiment was performed in quadruplicate and repeated at least twice.
Cell clonogenic survival
Cells were cultured in 10-cm dishes in normal culture medium overnight and were then treated with Ad/TRAIL-F/RGD or Ad/CMV-GFP at various MOIs (500 VP for Seg-1 cells and 5000 VP for SKGT-5 cells). Cells treated with PBS alone were used as the control. After 24-h exposure to Ad/TRAIL-F/RGD or Ad/CMV-GFP, cells were irradiated in 10-cm dishes with various doses of radiation in a 137Cs unit at room temperature. The irradiated cells were then seeded in triplicate onto 10-cm dishes at a density of 1,000 cells per dish to yield 50 to 200 colonies per dish. The cells were then cultured in an incubator containing 5% CO2 at 37°C for 14 days. Individual colonies (> 50 cells/colony) were fixed and stained with a solution containing 0.25% crystal violet and 10% ethanol for 10 min. The colonies were counted with an imaging system (FluorChem 8800 imaging system; Alpha Innotech, San Leandro, CA) using a visible light source. Each experiment was performed in triplicate and repeated at least twice.
Flow cytometry assay
Cells (2 × 105) were plated into 6-well plates 1 day before treatment. These cells were then treated with Ad/TRAIL-F/RGD, Ad/gTRAIL, or Ad/CMV-GFP at various MOIs. Cells treated with PBS alone were used as the control. After 24 h of exposure to Ad/TRAIL-F/RGD or Ad/CMV-GFP, the cells were irradiated with 2 to 5 Gy of radiation in a 137Cs unit at room temperature. Two or 3 days after treatment, floating and attached cells were harvested and washed with PBS. One part was used for the analysis of GFP expression, which involved determining the percentage of GFP-positive cells using a fluorescent activated cell sorting (FACS) system (Becton-Dickinson, Mansfield, MA). The second part of the sample, which was fixed by 70% ethanol overnight and stained with propidium iodide before analysis, was used to quantify the apoptotic cells. This was done using flow cytometry, which measured the sub-G0/G1 cellular DNA content using Cell Quest software (Becton-Dickinson, San Jose, CA).
Western blot analysis
Cells were cultured overnight in 10-cm dishes in normal culture medium and then treated with Ad/TRAIL-F/RGD, Ad/gTRAIL, or Ad/CMV-GFP at an MOI of 500 VP. After 24-h exposure to Ad/TRAIL-F/RGD or Ad/CMV-GFP, the cells were irradiated with 5 Gy of radiation in a 137Cs unit at room temperature. Two or 3 days after the treatment, cells were washed with cold PBS and lysed in Laemmli’s lysis buffer. Equal amounts of protein and lysate were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to enhanced chemiluminescence (ECL) membranes (Hybond; Amersham Corp., Arlington Heights, IL). These membranes were then blocked with a buffer containing 5% fat-free milk and PBS with 0.05% Tween 20 for 1 h or overnight at 4°C, washed three times with PBS with 0.05% Tween 20, and incubated with primary antibodies for at least 1 h at room temperature. Rabbit anti-human TRAIL, caspase-3 and caspase-9 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit antihuman caspase-8, mouse antihuman poly (ADP-ribose) polymerase (PARP), and mouse antihuman TRAIL were obtained from BD Pharmingen (San Diego, CA). After a second washing with PBS and 0.05% Tween 20, membranes were incubated with peroxidase-conjugated secondary antibodies and developed with an ECL kit (Amersham Bioscience, Buckinghamshire, United Kingdom). ß-actin was used as a loading control.
Animal study
All of the animals were maintained, and animal experiments were performed under National Institutes of Health and institutional guidelines established for the Animal Core Facility at M. D. Anderson Cancer Center. The animals used in this study were female Nu/nu mice (4–6 weeks old) purchased from Charles River Laboratories (Wilmington, MA).
Prior to tumor cell inoculation, the mice were subjected to 3.5 Gy of total body irradiation from a 137Cs radiation source (model E-0103; U. S. Nuclear Corp.). Seg-1 cells were used to establish subcutaneous tumors in mice. Briefly, 3 x 106 cells were injected into the right hind legs of each mouse. When the average size of tumors reached 5 to 7 mm in diameter, the mice were randomly divided into 6 groups of 10 mice each. Mice were then treated according to the following schedule: on day 1, each mouse was intratumorally injected with Ad/TRAIL-F/RGD, Ad/CMV-GFP, or PBS at a dose of 5 x 1010 VP/tumor in a volume of 0.1 ml; on day 3, tumors were locally irradiated with a vertical 60Co- γ radiation beam from a Theratron 780C Cobalt unit (Theratronics International, Ltd., Kanata, ON, Canada) at a dose of 5 Gy/tumor. Each mouse was restrained in a custom-designed jig with anesthesia and positioned in the radiation field such that only the tumor xenograft implanted on the hind legs was exposed to the radiation beam, and the rest of the body was shielded by a lead block. On day 5, each mouse was intratumorally injected with adenoviral vector same as day 1. Mice were ear-tagged so that data obtained from individual animals could be traced. Tumor dimensions were measured 3 times per week using a digital caliper. Tumor volume was calculated using the equation V (mm3) = a x b2/2, where a is the largest diameter and b is the smallest diameter. The mice were killed at the animal core facility in accordance with our institutional policy when the tumor size (>1000 mm3 or 10% of body weight) suggested that the mice would die within 1 week. The survival times of mouse were estimated on the basis of the date of killing.
Immunohistochemistry
Tumor sections fixed in 4 μm paraformaldehyde were rehydrated and then incubated with a 1:200 of rabbit polyclonal antibody to TRAIL (Santa Cruz Biotechnology) for 60 min. The slides were stained with DakoCytomation Envision + System-HRP (DAB) using Dakoautostainer (DakoCytomation Co., Carpinteria, CA) to detect the expression of TRAIL. Counterstaining was performed with haematoxylin, and the slides were covered with Kaiser’s glycerin-gelatin. A brown color indicated TRAIL expression. The number of TRAIL positive cells was counted under a light microscope (×400 magnification) in randomly chosen fields and calculated as a percentage of at least 1,000 scored cells.
TUNEL assay
To detect apoptotic cells in tumor, we used in situ cell death detection kit, POD (Roche Applied Science, Indianapolis, IN). The staining was performed according to the manufacturer’s instructions, counterstained with haematoxylin, and viewed under a light microscope. A brown color indicated apoptotic nuclei as visualized using DAB substrate. Apoptotic cells were counted under a light microscope (×400 magnification) in randomly chosen fields, and the apoptosis index was calculated as a percentage of at least 1,000 scored cells.
Statistical analysis
Differences among the treatment groups were analyzed by analysis of variance (AOV) using statistical software (StatSoft, Tulsa, OK). A difference was considered statistically significant when the P value was 0.05 or less. Differences in tumor growth in vivo among the treatment groups were assessed by AOV with a repeated measurement module. AOV was performed to determine statistical significance between each treatment group by using the SAS procedure with the SAS version 6.12 software. Survival was assessed by using the Kaplan-Meier method.
RESULTS
Ad/TRAIL-F/RGD exhibited a strong cell-killing effect of in human esophageal adenocarcinoma cell lines
The functionality of Ad/TRAIL-F/RGD was first evaluated in the human esophageal adenocarcinoma cell line Seg-1. Treatment of Seg-1 cells with Ad/TRAIL-F/RGD at an MOI of 1,000 VP after 2 to 5 days resulted in massive cell death as shown by cell density and morphology (Fig. 1A). In contrast, infecting the Seg-1 cells with Ad/CMV-GFP (marker gene only) at the same dose and time had no noticeable effect on cell density, and Ad/gTRAIL (vector with TRAIL but without RGD) killed cancer cells only after 5 days and to a lesser degree. Cells treated with PBS were used as the mock control.
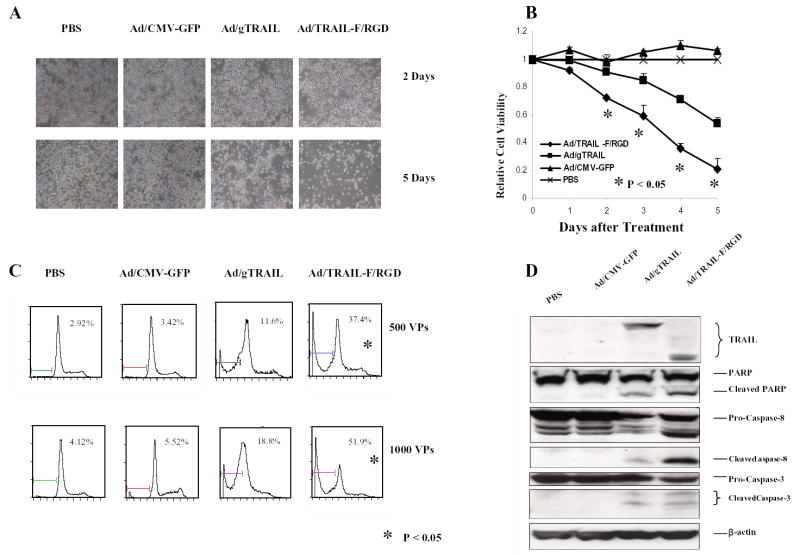
Fig. 1.
Ad/TRAIL-F/RGD is more effective than Ad/gTRAIL in human esophageal adenocarcinoma cell lines. A, Seg-1 cells treated with Ad/TRAIL-F/RGD, Ad/gTRAIL, and Ad/CMV-GFP at an MOI of 1,000 VP for 5 days. Cells treated with PBS were used as a control. There was a strong apoptotic effect of Ad/TRAIL-F/RGD compared with Ad/gTRAIL or Ad/CMV-GFP by cell density and morphology. B, Cell viability determined by the XTT assay in Seg-1 cells 5 days after cells were treated with various vectors at MOI of 1,000 VP. Cells treated with PBS were used as the control, with their viability set at 1. Each experiment was performed in quadruplicate and repeated at least twice. Values are means of quadruplicate assay results. Bars, standard deviation. C, Flow cytometric analysis of apoptosis (sub-G1) in Seg-1 cells at third day after various vector treatment at the indicated MOIs. D, TRAIL expression and activation of the apoptotic signal transduction pathway in Seg-1 3 days after various vector treatments at 500 MOIs using western blot analysis. P<0.05 in Figure 1B and 1C as comparison between Ad/TRAIL-F/RGD and PBS, Ad/gTRAIL, Ad/CMV-GFP respectively and combined.
The cell-killing effect of Ad/TRAIL-F/RGD was analyzed by measuring cell viability via XTT assay (Fig. 1B) and by quantifying apoptotic cells via flow cytometry (Fig. 1C). After 2 to 5 days, treatment of Seg-1 cells with Ad/TRAIL-F/RGD dramatically reduced cell viability compared with Ad/gTRAIL or Ad/CMV-GFP at the same MOIs (P < 0.05) (Fig. 1B). The result was supported by a flow cytometry study showing treatment of Seg-1 cells with Ad/TRAIL-F/RGD at MOIs of 500 VP and 1,000 VP significantly increased the number of apoptotic cells compared with Ad/gTRAIL or Ad/CMV-GFP (P <0.05) (Fig. 1C). The induction of apoptosis by Ad/TRAIL-F/RGD was confirmed by western blot analysis. (Fig.1D). Caspase-8, Caspase-3, and PARP cleavage were observed in cells treated with Ad/gTRAIL or Ad/TRAIL-F/RGD. However, in comparison with Ad/gTRAIL, treatment with Ad/TRAIL-F/RGD notably increased the cleavage of Caspase-8 , Caspase-3, and PARP. Taken together, our data demonstrated that Ad/TRAIL-F/RGD is more effective than Ad/gTRAIL in cell killing and apoptosis induction in human esophageal adenocarcinoma cells.
Dose-dependent TRAIL gene expression specifically suppresses human esophageal adenocarcinoma cells
To assess transgene expression of TRAIL in various cells, we infected esophageal adenocarcinoma cell Seg-1, SKGT-4, and SKGT-5, and NHLF cells with Ad/CMV-GFP or Ad/gTRAIL (contains both TRAIL gene and GFP under control of hTERT) at MOIs of 0 VP to 1,000 VP. Treatment with Ad/CMV-GFP and Ad/gTRAIL resulted in near similar levels of GPF-positive cells in all esophageal adenocarcinoma cell lines but not in NHLF (Fig. 2A, 2B). There was a significant difference in the number of GFP-positive cells in NHLF cells with Ad/gTRAIL versus Ad/CMV-GFP (P <0.05). More than 80% of NHLF cells treated with Ad/CMV-GFP at 1,000 VP were positive for GFP, whereas only 5% of NHLF cells treated with Ad/gTRAIL were positive for GFP (Fig. 2B), indicating tumor-specific transgene expression under control of the hTERT promotor in Ad/gTRAIL.
Fig. 2.
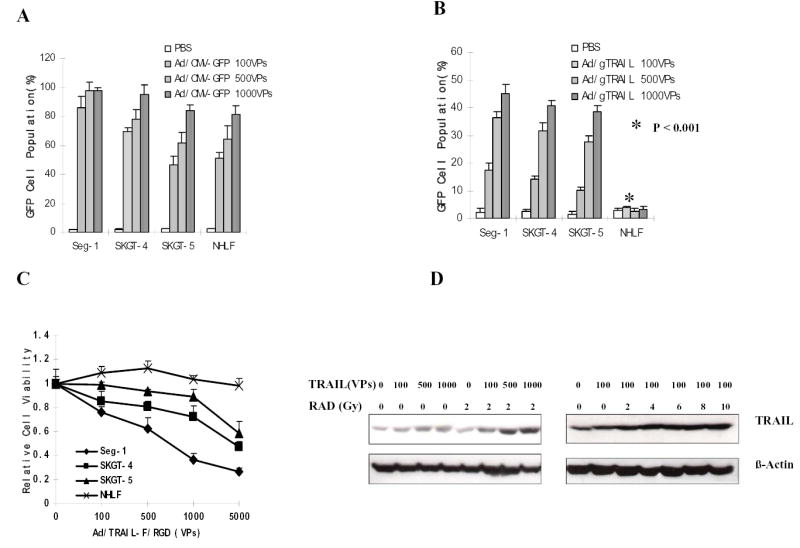
Dose-dependent TRAIL gene expression specifically suppresses human esophageal adenocarcinoma cells but not in NHLF. A or B, Results of flow cytometry analysis of transgene expression in GFP cell population. Four cell lines were treated with Ad/CMV-GFP or Ad/gTRAIL at MOIs ranging from 0 to 1,000 VP and harvested 2 days after treatment. Levels of GFP in the cells were determined by using FACS. C, Cell viability determined by the XTT assay in different cell lines at the fourth day after treatment with Ad/TRAIL-F/RGD at MOIs ranging from 0 to 5,000 VP. D, Level of TRAIL expression in Seg-1 cells after treatment with different doses of Ad/TRAIL-F/RGD plus/minus radiotherapy, as determined by western blots analysis. β-actin was used as a loading control.
To obtain further evidence that the hTERT promotor can drive specific expression of the TRAIL gene in esophageal adenocarcinoma cells but not in normal cells, we used the XTT assay to compare cell viability after treatment with Ad/TRAIL-F/RGD in all esophageal adenocarcinoma cells and in normal cells. Treatment with Ad/TRAIL-F/RGD resulted in a dose-dependent cell-killing effect at MOIs of 0 VP to 5,000 VP in all cancer cell lines tested but not in NHLF cells (Fig. 2C). Seg-1 cells were the most sensitive followed by SKGT-4 cells, and SKGT-5 cells were the least sensitive. NHLF cells were not sensitive. The level of transgene TRAIL expression in esophageal adenocarcinoma cells was also found to have a dose-dependent relationship with Ad/TRAIL-F/RGD by western blot analysis (Fig. 2D), and radiation appeared to increase TRAIL expression.
The cell-killing effect of the combination of Ad/TRAIL-F/RGD and radiotherapy is enhanced in esophageal adenocarcinoma cells but not in NHLF cells
Treatment with ionizing radiation (0–10 Gy) alone led to dose-dependent inhibition of cell growth in the 3 esophageal adenocarcinoma cell lines and NHLF cells, according to results of the XTT assay (Fig. 3). We then studied whether the combination of ionizing radiation and Ad/TRAIL-F/RGD could profoundly inhibit the growth of these cells. Cells treated with PBS and Ad/CMV-GFP were used as mock and vector controls, respectively. An improved inhibitory effect on cell growth was observed in the 3 esophageal adenocarcinoma cell lines, which were treated with various doses of Ad/TRAIL-F/RGD and ionizing radiation (Fig. 3A, 3C,and 3D). In contrast, in NHLF cells, no difference were observed among Ad/TRAIL-F/RGD plus ionizing radiation, Ad/CMV-GFP, or PBS, indicating that Ad/TRAIL-F/RGD did not sensitize the NHLF cells to radiation (Fig. 3B).
Fig. 3.
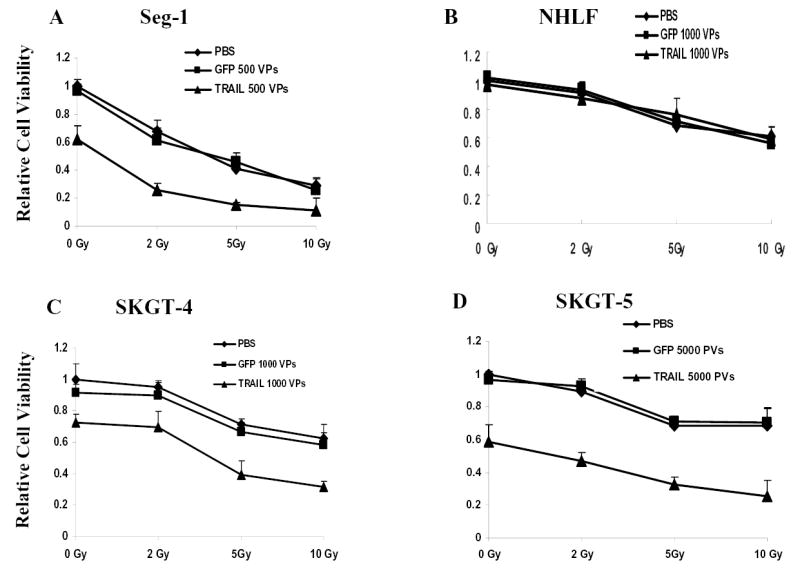
Enhanced cell-killing effects of Ad/TRAIL-F/RGD plus radiotherapy in esophageal adenocarcinoma cells but not in NHLF by XTT assay. Four cell lines were treated with Ad/TRAIL-F/RGD at the indicated MOIs and radiation at the indicated doses. Cell viability was determined by the XTT assay 4 days after gene therapy and 3 day after radiotherapy. Cells treated with PBS were used as the control, with their viability set at 1. Each experiment was performed in quadruplicate and repeated at least twice. The values shown are the means of quadruplicate assay results. Bars, standard deviation. TRAIL: Ad/TRAIL-F/RGD, GFP: Ad/CMV-GFP (vector control).
To confirm the effect of TRAIL gene therapy combined with radiotherapy, we conducted a clonogenic formation assay in two esophageal adenocarcinoma cell lines after treatment with radiotherapy, Ad/TRAIL-F/RGD, or both. PBS and Ad/CMV-GFP were again used as mock and vector controls, respectively. Compared with the control group or the groups receiving single-agent treatment, the group receiving the combination of Ad/TRAIL-F/RGD plus radiotherapy had significantly reduced clonogenic formation (Fig. 4, P < 0.05). In Seg-1 cells, radiation alone at 2 Gy inhibited 55.4% of colony formation and Ad/TRAIL-F/RGD alone at an MOI of 500 VP suppressed 32.3% of colony formation. However, the combination of radiation and Ad/TRAIL-F/RGD treatment inhibited 90.5% of colony formation . A similar inhibition of colony formation was observed in the SKGT-5 cells. The different percentages of inhibition shown by the XTT and clonogenic assays were caused by the different cell inhibition mechanisms by these two assays.
Fig. 4.
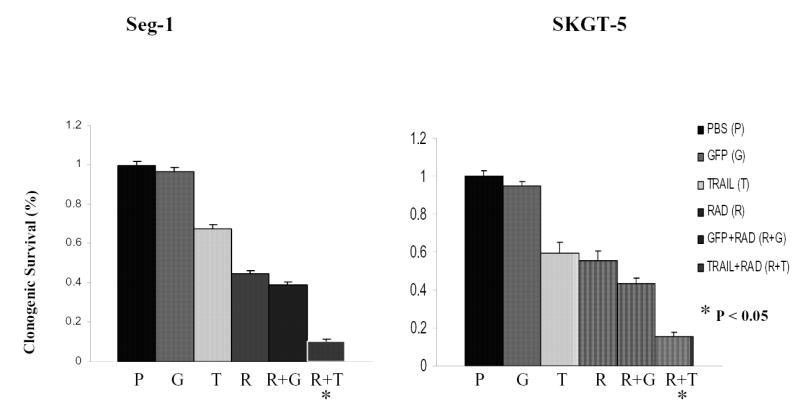
The influence of Ad/TRAIL-F/RGD on radiosensitivity was examined on the basis of the clonogenic survival of cell lines after exposure to PBS, Ad/CMV-GFP (GFP), Ad/TRAIL-F/RGD (TRAIL), radiation (RAD), Ad/CMV-GFP plus radiotherapy (GFP+RAD), Ad/TRAIL-F/RGD and radiotherapy (TRAIL+RAD). The MOIs for the cells were 500 VP for Seg-1 cells and 5,000 VP for SKGT-5 cells. Each experiment was performed in triplicate and repeated at least twice. Values are means of the results of triplicate assays. Bars, standard deviation. P<0.05 in Figure 5 as comparison between Ad/TRAIL-F/RGD plus radiation and PBS, Ad/CMV-GFP, Ad/TRAIL-F/RGD, radiation, Ad/CMV-GFP plus radiation respectively and combined.
The combination of TRAIL gene therapy and radiotherapy improved apoptosis induction and activation of the apoptotic signal transduction pathway in esophageal adenocarcinoma cells but not in NHLF cells
To confirm that Ad/TRAIL-F/RDG had no toxic effect in NHLF cells, we used a FACS assay to evaluate apoptosis induction of Ad/TRAIL-F/RDG cells by quantifying the sub-G1 population in NHLF after treatment with Ad/TRAIL-F/RDG alone, Ad/CMV-GFP alone, radiotherapy alone and the combination of each of these with radiotherapy. As shown in Fig. 5A, only background levels of apoptosis were observed in NHLF cells after treatment with either of these six treatments.
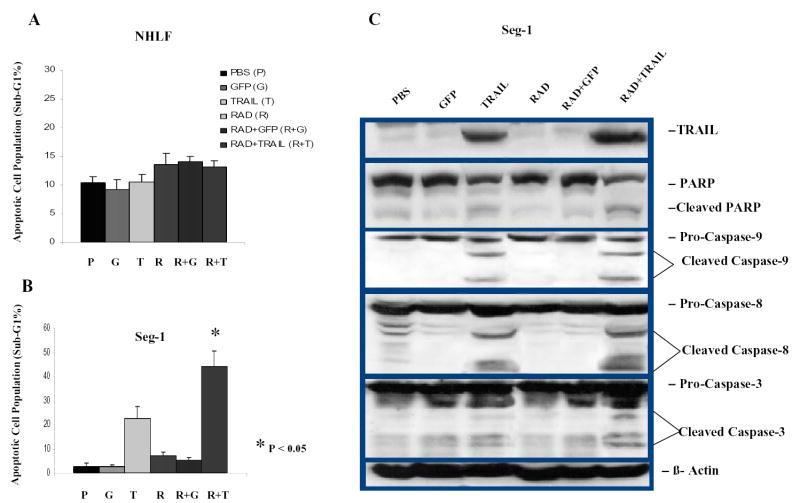
Fig. 5.
Improved apoptosis induction and activation of the apoptotic signal transduction pathway by the combination of TRAIL gene therapy and radiotherapy but not in NHLF in vitro. A, FACS analysis was used to quantify apoptotic cells 4 days after 1 of 6 treatments in NHLF. B, FACS was used to quantify apoptotic cells 4 days after 1 of 6 treatments in Seg-1 cells. C, Western blot analysis of TRAIL expression and cleavage of PARP, caspase-8, caspase-9, and caspase-3 in Seg-1 cells 4 days after Ad/TRAIL-F/RGD (TRAIL) treatment combined with radiotherapy. Cells were treated with PBS (lane 1), Ad/CMV-GFP (GFP; MOI of 500 VP) (lane 2), Ad/TRAIL-F/RGD (MOI 500 VP) (lane 3), radiotherapy (2 Gy) (lane 4), Ad/CMV-GFP (MOI 500 VP) plus radiotherapy (2 Gy) (lane 5), and Ad/TRAIL-F/RGD (MOI of 500 VP) plus radiotherapy (2 Gy) (lane 6). ß-Actin was used as a loading control. P<0.05 in Figure 5B as comparison between Ad/TRAIL-F/RGD plus radiation and PBS, Ad/CMV-GFP, Ad/TRAIL-F/RGD, radiation, Ad/CMV-GFP plus radiation respectively and combined.
To test whether the enhanced cell killing produced by the combination treatment is caused by apoptosis, we quantified the sub-G1 population of Seg-1 after treatment with TRAIL gene therapy plus radiotherapy by FACS. As shown in Fig. 5B, Ad/TRAIL-F/RGD alone induced 22.8% sub-G1 accumulation and radiotherapy alone (2 Gy) induced less than 7.0%. However, the combination of these treatments resulted in substantially increased sub-G1 accumulation (44.0%, P <0.05). These data suggest that apoptosis is significantly enhanced by the combination of TRAIL gene therapy and radiotherapy.
To further elucidate the apoptotic signal transduction pathway used by TRAIL gene therapy plus radiotherapy, we performed western blot analysis to evaluate the activation of caspase-8, caspase-3, and caspase-9 and the cleavage of PARP after treatment with PBS, Ad/CMV-GFP alone, Ad/TRAIL-F/RDG alone, radiotherapy alone, radiotherapy plus Ad/CMV-GFP, and radiotherapy plus Ad/TRAIL-F/RDG. Seg-1 cells were harvested 4 days after treatment with Ad/TRAIL-F/RGD or Ad/CMV-GFP and 3 days after radiotherapy, and the activation of caspase-8, caspase-3, and caspase-9 and the cleavage of PARP were analyzed. Treatment with Ad/TRAIL-F/RGD alone resulted in the cleavage of caspase-8, caspase-3, caspase-9, and PARP, as expected (Fig. 5C). Ad/TRAIL-F/RGD plus radiotherapy resulted in cleavage of caspase-8, caspase-3, caspase-9, and PARP that was slightly increased over that produced by treatment with Ad/TRAIL-F/RGD alone (Fig. 5C). This result suggests that apoptosis is involved in cell death produced by the combination of TRAIL gene therapy and radiotherapy. Taken together, these data suggest that the combination treatment improves apoptosis in esophageal adenocarcinoma cells but not in NHLF cells.
The combination of Ad/TRAIL-F/RGD and radiotherapy enhances apoptosis induction in vivo
We assessed TRAIL protein expression by immunohistochemical analysis of tumor specimens from the 6 treatment groups. We observed strong expression of TRAIL protein in tumor tissues obtained from animals that were treated with Ad/TRAIL-F/RGD plus radiotherapy or with Ad/TRAIL-F/RGD alone (Fig. 6A, B). We did not observe TRAIL expression in tumors that were treated with PBS, radiotherapy alone, Ad/CMV-GFP alone, or Ad/CMV-GFP plus radiotherapy. The percentage of TRAIL positive cells in tumor tissues treated with Ad/TRAIL-F/RGD plus radiotherapy was obviously higher (30.2%) than that of cells in tumor tissues treated with Ad/TRAIL-F/RGD alone (21.7%, P =0.00238). This result is also consistent with our results in vitro (Fig. 2D), in which radiotherapy increased TRAIL gene expression. To assess apoptosis induction in vivo, we performed terminal deoxynucleotidyl transferase-mediated dUTP labeling (TUNEL) staining on tumor sections 3 days after radiation in all 6 treatment groups (Fig. 6A). Brown color indicates apoptotic nuclei as visualized using the DAB substrate. Apoptotic cells were counted under a light microscope in randomly chosen fields, and the apoptosis index was calculated as a percentage of at least 1,000 scored cells. As shown in Fig. 6C, the combination of Ad/TRAIL-F/RGD and radiotherapy resulted in a significantly higher apoptotic index (27.4%) compared with Ad/TRAIL-F/RGD alone (18.1%), radiation alone (10.4%), and radiotherapy plus Ad/CMV-GFP (11.6%) after subtracting the background level of 2.9% (P =0.00117).
Fig. 6.
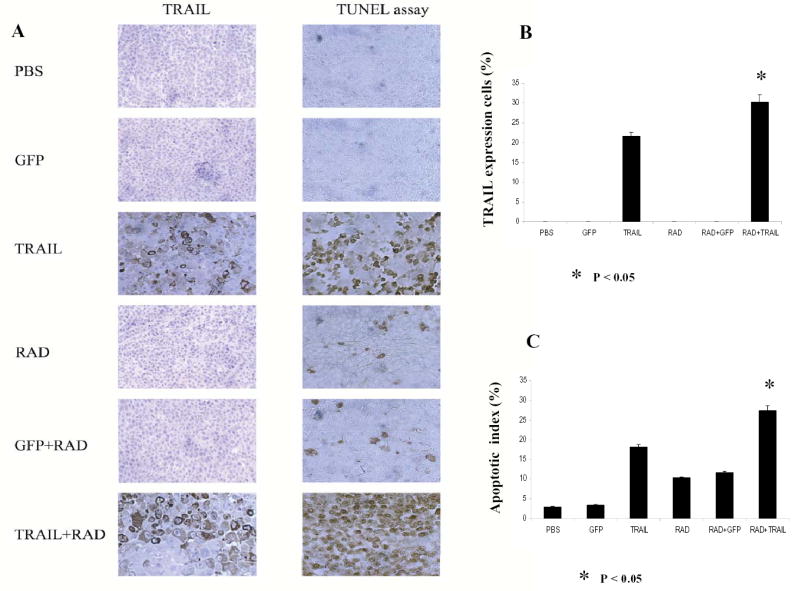
Ad/TRAIL-F/RGD (TRAIL) combined with radiotherapy enhanced apoptosis, inhibited tumor growth in vivo and prolonged mice survival. Seg-1 cells were grown as xenograft tumors in nude mice, and mice bearing tumors of 80 to 100 mm3 were treated with PBS, Ad/CMV-GFP alone (GFP), TRAIL alone, radiotherapy (RAD) alone, GFP +RAD, or TRAIL+RAD. Tumor volumes were then measured. A, Representative fields of TRAIL expression by immunohistochemistry (left panel) and tumor cell apoptosis by TUNEL assay (right panel) in Seg-1 tumors 3 days after receiving various treatments. B, The percentage of TRAIL-positive tumor cells as counted under a light microscope. C, Apoptotic index in the various treatment groups as determined by counting at least 1,000 cells per sample under a light microscope. The apoptosis index was calculated as a percentage of total number of cells scored. Bars, SE. D, Tumor growth delay curves. Data are presented as the mean ± standard error. ANOVA was performed using the SAS procedure and SAS software (version 6.12) to determine the statistical significance of the differences on tumor growth between each treatment group. P<0.05 in Figure 6B,C, D as comparison between Ad/TRAIL-F/RGD plus radiation and PBS, Ad/CMV-GFP, Ad/TRAIL-F/RGD, radiation, Ad/CMV-GFP plus radiation respectively and combined. E, Animals alive as a function of time after treatment. We used the Kaplan-Meier method for our survival analysis.
Treatment with Ad/TRAIL-F/RGD plus radiotherapy inhibits tumor growth in vivo and prolongs mouse survival
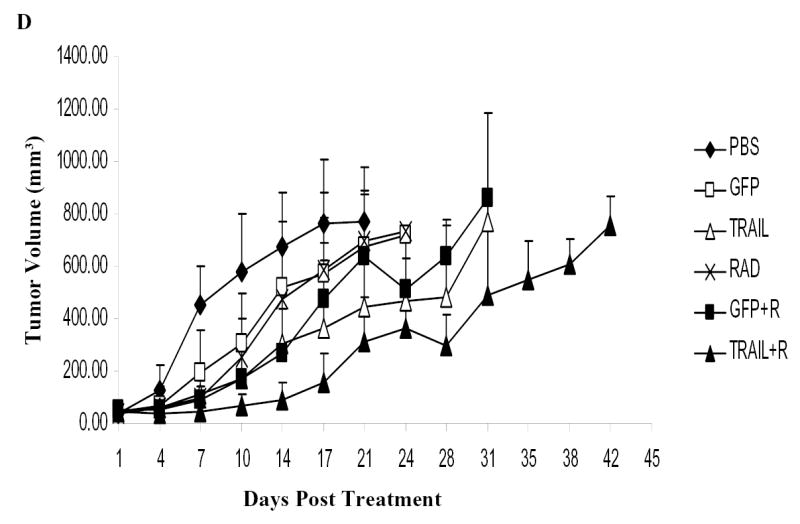
We tested the therapeutic efficacy of the combination of Ad/TRAIL-F/RDG plus radiotherapy in vivo in a human esophageal adenocarcinoma xenograft mouse model. As shown in Fig. 6D, radiotherapy alone had a minimal inhibitory effect and Ad/TRAIL-F/RDG alone had a moderate inhibitory effect on tumor growth at the tested doses; however, the combination of Ad/TRAIL-F/RGD and radiotherapy significantly inhibited tumor growth compared with PBS alone, radiotherapy alone, Ad/TRAIL-F/RDG alone, Ad/CMV-GFP alone, and Ad/CMV-GFP plus radiotherapy (day 14 and day 17, P <0.05).
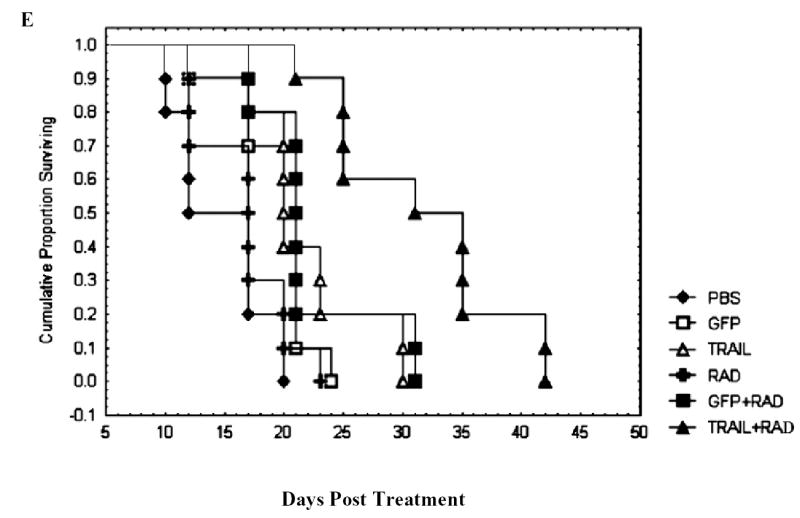
The mouse survival data also showed a significant prolongation of survival associated with the combination of Ad/TRAIL-F/RDG and radiotherapy, with a mean survival duration of 31.6 days compared with 21.5 days for Ad/TRAIL-F/RGD alone, 22.2 days for radiotherapy plus Ad/CMV-GFP, 16.7 days for radiotherapy alone, 19.6 days for Ad/CMV-GFP alone, and 14.7 days for PBS alone (P =0.00001) (Fig. 6E). Treatment with Ad/TRAIL-F/RDG alone significantly lengthened survival time compared with treatment with PBS alone, Ad/CMV-GFP alone, and radiotherapy alone (P =0.00497). Furthermore, the combination of Ad/TRAIL-F/RGD and radiotherapy prolonged survival duration significantly compared with Ad/TRAIL-F/RGD alone (P =0.00261) and compared with radiotherapy plus Ad/CMV-GFP (P=0.00359). We observed no marked treatment-related toxicities affecting the weight of mice or their general behavior.
DISCUSSION
Radiation resistance causes treatment failure in locally advanced esophageal cancer. Tumor control does not increase with radiation dose escalation, although toxicities are increased. We explored the possibility of radiation sensitization by apoptotic gene targeting and overcoming radiation resistance in esophageal adenocarcinoma by using intratumoral delivery of tumor-specific apoptotic gene targeting. Such treatment is possible in esophageal cancer patients because intratumoral injections can be accomplished in the clinical setting by endoscopy. Reversal of radiation resistance may significantly improve local control of esophageal cancer, which will translate into better quality of life and improved survival for patients.
In this study, tumor-specific TRAIL gene targeting achieved by using Ad/TRAIL-F/RDG was found to sensitize esophageal adenocarcinoma cells to radiation, with significantly inhibited tumor growth and prolonged survival duration of mice bearing a human esophageal adenocarcinoma xenograft. When radiotherapy was used alone (5 Gy), the tumor growth inhibition was minimal, and there was no survival benefit; the mean survival duration was 16.7 days in mice receiving radiotherapy only, compared with 14.7 days for those treated with PBS. These data suggest that the esophageal adenocarcinoma cell line Seg-1 is resistant to this dose of radiation. Ad/TRAIL-F/RDG alone moderately improved the tumor inhibition and prolonged the survival duration in mice. However, the combination of Ad/TRAIL-F/RDG and radiotherapy dramatically prolonged mean survival duration (31.6 days). The combination of Ad/TRAIL-F/RDG and radiotherapy may serve as a novel approach to increase the therapeutic ratio of radiotherapy in esophageal adenocarcinoma, which are radiation resistant. These data confirmed our hypothesis that Ad/TRAIL-F/RDG can overcome radiation resistance in esophageal adenocarcinoma. The preliminary data from our lung cancer model supports this conclusion (25).
Moreover, no observable toxicities were noted in the combination treatment group compared with the control group with respect to weight and general behavior. In terms of toxicity, radiotherapy commonly causes acute adverse effects, including esophagitis, pneumonitis, and chronic conditions such as esophageal fibrosis, esophageal stricture, and fistula; these complications are the major dose-limiting toxicities of radiotherapy. Gene therapy, in contrast, causes different toxicity profiles, with common systemic reactions such as low-grade fever (26), which do not overlap with radiotherapy-induced adverse effects. To minimize potential toxicities, we developed a tumor-specific adenoviral vector–mediated TRAIL gene delivery system by expressing the TRAIL gene from the hTERT promoter via GAL4 gene regulatory components that can augment transgene expression from the tumor-specific promoter without losing target specificity (19, 27, 28). As our data shown, TRAIL under control of tumor-specific hTERT induced apoptosis, inhibited tumor cell growth, sensitized esophageal cancer cells to radiotherapy, but spared normal cells such as NHLF cells (Fig. 1–3, and Fig. 5). Similar spared effect has also been shown in normal human bronchial epithelial cells using Ad/TRAIL-F/RDG (25) Therefore, combined TRAIL gene therapy and radiotherapy may improve the therapeutic efficacy of esophageal cancer with tolerable toxicities.
At M. D. Anderson, we participated in a phase II clinical study to explore the possibility of improving the therapeutic ratio via radiation sensitization using intratumoral delivery of tumor necrosis factor alpha (TNFα) gene therapy through endoscopic injections in patients with esophageal cancer. These patients are treated with concurrent chemotherapy and 50.4 Gy radiotherapy followed by surgical resection (29). The primary concerns about TNFα gene therapy are the systemic toxicity, therapeutic efficacy, and lack of tumor specificity (30). Therefore, a new approach to sensitize esophageal cancer to radiation is needed. In another study, 20% to 30% of patients who underwent indication chemoradiotherapy achieved pathologic complete response (pCR) at the time of surgery (i.e., all the tumor cells had been eliminated by induction chemotherapy and radiotherapy). The clinical outcome for patients who achieved pCR was significantly better than that of patients who did not achieve pCR (31). However, the remaining 70% to 80% of patients had residual disease after induction chemotherapy plus radiotherapy and were at high risk for recurrence and distant metastasis. Surgical esophagectomy is recommended for this group of patients. However, the surgical mortality rate associated with esophagectomy is 5%–10% (up to 25% in some institutions), and quality of life is compromised for patients who survive a long time after the surgery. Improving induction treatment using novel approach such TRAIL gene targeting to achieve pCR will be crucial for improving outcome.
It has been reported that incorporation of the integrin-binding motif Arg-Gly-Asp (RGD)-containing peptide in the HI loop of the adenoviral vector fiber protein provided the ability of the adenovirus to utilize an alternative receptor during cell entry, that results in efficient transduction in cells resistant to conventional adenoviral vectors, particularly primary tumor cells (32, 33). In the current study, we inserted a gene sequence containing the integrin-binding RGD motif sequence into the HI loop of the adenoviral fiber. Compared with Ad/gTRAIL without RGD, Ad/TRAIL-F/RDG significantly improved cancer inhibition effect and apoptosis induction without losing its tumor specificity (Fig. 1).
Although TRAIL-mediated gene therapy has shown promise, certain cancer cells, especially those that make up highly malignant tumors, are TRAIL resistant. The mechanisms of resistance to TRAIL-induced apoptosis involve dysfunctions of DR4 and DR5, which are essential for TRAIL binding to cells; a defect of either the adaptor protein Fas-associated death domain or caspase 8, which are essential for the TRAIL-induced apoptotic signal transduction pathway; overexpression of Bcl-2; or a lack of Bax or Bak (34). As shown by the results of our clonogenic assay, SKGT-5 cells are resistant to Ad/TRAIL-F/RDG therapy. When given at an MOI of 5,000 VP, Ad/TRAIL-F/RDG inhibited only 40% of colony formation. In contrast, 35% inhibition of colony formation in Seg-1 cells can be achieved with Ad/TRAIL-F/RDG at an MOI of only 500 VP. However, the combination of Ad/TRAIL-F/RDG and radiotherapy (5 Gy) inhibited 86% of colony formation in SKGT-5 cells (Fig. 4). These data suggest that radiotherapy is useful in overcoming TRAIL resistance in gene therapy.
Several mechanisms explain the observed enhanced therapeutic effect of the TRAIL gene plus radiotherapy. First, ionizing radiation may improve the transfection/transduction efficiency and transgene integration of gene therapy, as shown in our study and as shown by other (35–37). Fig. 2D showed that the level of transgene TRAIL expression in esophageal cancer cells is associated in a dose-dependent manner with Ad/TRAIL-F/RGD, as illustrated by western blot analysis and radiotherapy appeared to increase TRAIL expression. Results of our immunohistochemical analysis in vivo also showed that radiation increased TRAIL expression in tumor cells (Fig.6A, 6B). Next, gene therapy and radiotherapy target different phases of the cell cycle, with the S phase of the cell cycle being most responsive to gene therapy and the M and G2 phases being more radiosensitive (38). In addition, it has been reported that radiation and chemotherapy can cause up regulation of the expression of the DR4 or DR5 proteins or of both (23, 39, 40). By expressing more of the TRAIL receptor, cells may become more sensitive to TRAIL therapy. Further experimentation is needed to confirm this mechanism in radiotherapy.
The most important source of the improved antitumor effect of gene therapy plus radiotherapy may be that TRAIL gene therapy and radiotherapy activate distinct apoptotic pathways, which, when jointly triggered, result in an amplified response (6,11). Radiation damages DNA and triggers p53-mediated transcriptional activation of Bax, Bak, Noxa, and Puma, resulting in the mitochondrial damage by breakdown of the mitochondrial membrane potential and the release of cytochrome c (15–17). Cytoplasmic cytochrome c forms a complex with Apaf-1 and dATP, leading to activation of caspase-9 (18). Activated caspase-9 subsequently activates caspase-3 and then again activates the cascade of caspases. In contrary to radiation-induced apoptosis, TRAIL binds with its cell surface receptors and then mediates the recruitment of the FADD adapter molecule to the receptor complex. This complex directly activates caspase 8 (12) and subsequently activates caspase 3, resulting in activation of the cascade of caspases (13). In addition, caspase 8 can also activate Bid, which triggers cytochrome c release, with subsequent activation of caspase-9 and -3, thereby strongly amplifying the initial apoptotic signal (14). Triggering both pathways simultaneously explains the effective therapeutic response seen in vitro and in vivo, as our data showed. These pathways could be both p53 dependent and p53 independent.
In summary, our data showed that tumor-specific TRAIL gene targeting can overcome radiation resistance in esophageal adenocarcinoma cells and thus improve survival while sparing normal cells. Ad/TRAIL-F/RGD may improve the therapeutic ratio of radiotherapy, which is crucial for radiation-resistant esophageal cancer. In addition, radiotherapy may also overcome TRAIL resistance in highly malignant cancer cells.
Acknowledgments
The authors thank Dr. Raymond E. Meyn for his advice and review of the manuscript. The authors are grateful to Dr. James D. Cox and Dr. Jack A. Roth for their support of this project. We thank the Department of Scientific Publications and Barbara E. Lewis for their assistance in the preparation of this manuscript.
Footnotes
Grant support: This research was supported by a Radiology Society of North America (RSNA) Research Scholar Award (to J.Y.C.), a Career Developmental Award of The University of Texas Lung Cancer SPORE (to J.Y.C.), National Cancer Institute grants RO1 CA 092487-01A1 and RO1 CA 098582-01A1 (to B. F.), and a grant from the institutional research startup fund (to R. M. C).
The paper was presented in part at the 47th Annual Meeting of the American Society of Therapeutic Radiology and Oncology, October 16-20, 2005, Denver, Colorado.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2251. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Willett CG. Radiation dose escalation in combined-modality therapy for esophageal cancer. J Clin Oncol. 2002;20:1151–1153. doi: 10.1200/JCO.2002.20.5.1151. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 4.Cooper JS, Guo MD, Herskovic A, et al. Chemoradiotherapy of locally advanced esophageal cancer: Long-term follow-up of a prospective randomized trial (RTOG 85-01) JAMA. 1999;281:1623–1631. doi: 10.1001/jama.281.17.1623. [DOI] [PubMed] [Google Scholar]
- 5.Minsky BD, Pajak TF, Ginsberg RJ, et al. INT 0123 (Radiation Therapy Oncology Group 94-05) phase III trial of combined-modality therapy for esophageal cancer: High-dose versus standard-dose radiation therapy. J Clin Oncol. 2002;20:1167–1174. doi: 10.1200/JCO.2002.20.5.1167. [DOI] [PubMed] [Google Scholar]
- 6.Belka C, Jendrosek V, Pruschy M, et al. Apoptosis-modulating agents in combination with radiotherapy–current status and outlook. Int J Radiat Oncol BiolPhys. 2004;58:542–554. doi: 10.1016/j.ijrobp.2003.09.067. [DOI] [PubMed] [Google Scholar]
- 7.Moreira LF, Naomoto Y, Hmada M, et al. Assessment of apoptosis in oesophageal carcinoma preoperatively treated by chemotherapy and radiotherapy. Anticancer Res. 1995;15(2):639–644. [PubMed] [Google Scholar]
- 8.Raouf A, Evoy D, Carton E, et al. Spontaneous and inducible apoptosis in oesophageal adenocarcinoma. Br J Cancer. 2001;85(11):1781–1786. doi: 10.1054/bjoc.2001.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher DE. Apoptosis in cancer therapy: crossing the threshold. Cell. 1994;78:539–542. doi: 10.1016/0092-8674(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 10.Hetts SW. To die or not to die: an overview of apoptosis and its role in disease [Review] JAMA. 1998;279:300–307. doi: 10.1001/jama.279.4.300. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira CG, Epping M, Kruyt FA, et al. Apoptosis: target of cancer therapy. Clin Cancer Res. 2002;8:2024–2034. [PubMed] [Google Scholar]
- 12.Muzio M, Chinnaiyan AM, Kischkel FC, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 13.Stennicke HR, Jurgensmeier JM, Shin H, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 14.Luo X, Budihardjo I, Zou H, et al. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 15.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 16.Oda E, Ohki R, Murasawa H, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 17.Wei MC, Zong WX, Cheng EH, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Nijhawan D, Budihardjo I, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 19.Lin T, Gu J, Zhang L, et al. Targeted expression of green fluorescent protein/tumor necrosis factor-related apoptosis-inducing ligand fusion protein from human telomerase reverse transcriptase promoter elicits antitumor activity without toxic effect on primary human hepatocytes. Cancer Res. 2002;62:3620–3625. [PubMed] [Google Scholar]
- 20.Lin T, Huang X, Gu J, et al. Long-term tumor-free survival from treatment with the GFP-TRAIL fusion gene expressed from the hTERT promoter in breast cancer cells. Oncogene. 2002;21:8020–8028. doi: 10.1038/sj.onc.1205926. [DOI] [PubMed] [Google Scholar]
- 21.Kagawa S, He C, Gu J, et al. Antitumor activity and bystander effects of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) gene. Cancer Res. 2001;61:3330–3338. [PubMed] [Google Scholar]
- 22.Lin T, Zhang L, Davis J, et al. Combination of TRAIL gene therapy and chemotherapy enhances anticancer and antimetastasis effects in chemosensitive and chemoresistant breast cancers. Mol Ther. 2003;8:441–448. doi: 10.1016/s1525-0016(03)00203-x. [DOI] [PubMed] [Google Scholar]
- 23.Chinnaiyan AM, Prassad U, Shankar S, et al. Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci USA. 2000;97:1754–1759. doi: 10.1073/pnas.030545097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob D, Davis J, Zhu H, et al. Suppressing orthotopic pancreatic tumor growth with a fiber-modified adenovector expressing the TRAIL gene from the human telomerase reverse transcriptase promoter. Clin Cancer Res. 2004;10:3535–3541. doi: 10.1158/1078-0432.CCR-03-0512. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Cheung RM, Komaki R, Fang B, Chang JY. Radiotherapy sensitization by tumor-specific TRAIL gene targeting improves survival of mice bearing human non-small cell lung cancer. Clin Cancer Res. 2005;11(18):6657–6668. doi: 10.1158/1078-0432.CCR-04-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swisher SG, Roth JA, Komaki R, et al. Induction of p53-regulated genes and tumor regression in esophageal cancer patients after intratumoral delivery of adenoviral p53 (INGN 201) and radiation therapy. Clin Cancer Res. 2003;9:93–101. [PubMed] [Google Scholar]
- 27.Koch PE, Guo ZS, Kagawa S, et al. Augmenting transgene expression from carcinoembryonic antigen (CEA) promoter via a GAL4 gene regulatory system. Mol Ther. 2001;3:278–283. doi: 10.1006/mthe.2001.0273. [DOI] [PubMed] [Google Scholar]
- 28.Fang B, Ji L, Bouvet M, et al. Evaluation of GAL4/TATA in vivo. Induction of transgene expression by adenovirally mediated gene codelivery. J Biol Chem. 1998;273:4972–4975. doi: 10.1074/jbc.273.9.4972. [DOI] [PubMed] [Google Scholar]
- 29.Senzer N, Mani S, Rosemurgy A, et al. TNFerade biologic, an adenovector with a radiation-inducible promoter, carrying the human tumor necrosis factor alpha gene: a phase I study in patients with solid tumors. J Clin Oncol. 2004;22:592–601. doi: 10.1200/JCO.2004.01.227. [DOI] [PubMed] [Google Scholar]
- 30.Debs RJ, Fuchs HJ, Philip R, et al. Immunomodulatory and toxic effects of free and liposome-encapsulated tumor necrosis factor alpha in rats. Cancer Res. 1990;50:375–380. [PubMed] [Google Scholar]
- 31.Ajani JA, Walsh G, Komaki R, et al. Preoperative induction of CPT-11 and cisplatin chemotherapy followed by chemoradiotherapy in patients with locoregional carcinoma of the esophagus or gastroesophageal junction. Cancer. 2004;100:2347–2354. doi: 10.1002/cncr.20284. [DOI] [PubMed] [Google Scholar]
- 32.Dmitriev I, Krasnykh V, Miller CR, et al. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham TJ, Tzeng E, Shears LL, et al. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther 2004 Nov 19; [Epub ahead of print]. [DOI] [PubMed]
- 35.Jain PT, Gewirtz DA. Sustained enhancement of liposome-mediated gene delivery and gene expression in human breast tumour cells by ionizing radiation. Int J Radiat Biol. 1999;75:217–223. doi: 10.1080/095530099140672. [DOI] [PubMed] [Google Scholar]
- 36.Stevens CW, Zeng M, Cerniglia GJ. Ionizing radiation greatly improves gene transfer efficiency in mammalian cells. Hum Gene Ther. 1996;7:1727–1734. doi: 10.1089/hum.1996.7.14-1727. [DOI] [PubMed] [Google Scholar]
- 37.Zeng M, Cerniglia GJ, Eck SL, et al. High-efficiency stable gene transfer of adenovirus into mammalian cells using ionizing radiation. Hum Gene Ther. 1997;8:1025–1032. doi: 10.1089/hum.1997.8.9-1025. [DOI] [PubMed] [Google Scholar]
- 38.Simons JW, Marshall FF. The future of gene therapy in the treatment of urologic malignancies. Urol Clin North Am. 1998;25:23–38. doi: 10.1016/s0094-0143(05)70430-4. [DOI] [PubMed] [Google Scholar]
- 39.Gibson SB, Oyer R, Spalding AC, et al. Increased expression of death receptors 4 and 5 synergizes the apoptosis response to combined treatment with etoposide and TRAIL. Mol Cell Biol. 2000;20:205–212. doi: 10.1128/mcb.20.1.205-212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagane M, Pan G, Weddle JJ, et al. Increased death receptor 5 expression by chemotherapeutic agents in human gliomas causes synergistic cytotoxicity with tumor necrosis factor-related apoptosis-inducing ligand in vitro and in vivo. Cancer Res. 2000;60:847–853. [PubMed] [Google Scholar]