Abstract
Mice engineered to lack GM2/GD2 synthase (GalNAc-T), with resultant deficit of GM2, GD2, and all gangliotetraose gangliosides, were originally described as showing a relatively normal phenotype with only a slight reduction in nerve conduction. However, a subsequent study showed that similar animals suffer axonal degeneration, myelination defects, and impaired motor coordination. We have examined the behavior of cerebellar granule neurons from these neonatal knockouts in culture and have found evidence of impaired capacity for Ca2+ regulation. These cells showed relatively normal behavior when grown in the presence of physiological or moderately elevated K+ but gradually degenerated in the presence of high K+. This degeneration in depolarizing medium was accompanied by progressive elevation of intracellular calcium and onset of apoptosis, phenomena not observed with normal cells. No differences were detected in cells from normal vs. heterozygous mice. These findings suggest that neurons from GalNAc-T knockout mice are lacking a calcium regulatory mechanism that is modulated by one or more of the deleted gangliosides, and they support the hypothesis that maintenance of calcium homeostasis is one function of complex gangliosides during, and perhaps subsequent to, neuronal development.
Keywords: GM1 ganglioside, ganglioside-deficient neurons, cerebellar granule neurons, neuritogenesis, intracellular calcium
Gangliosides are the major sialoglycoconjugates of the vertebrate central nervous system (1, 2) and have been proposed as important determinants in neuronal differentiation (3–5). Early support for this idea came from study of ganglioside storage disease, in which pyramidal neurons of the cerebral cortex were shown to sprout ectopic dendrites from the “meganeurite” ganglioside storage area with formation of aberrant synapses (6). The gangliotetraose family, consisting of GM1 and its oligosialo derivatives (GD1a, GD1b, GT1b, GQ1b, etc.), are the predominant forms in the neuron and are biosynthesized by a series of Golgi-localized enzymes that add sugars sequentially to the membrane-anchoring ceramide moiety (7, 8). One approach to the study of their mechanistic roles has been development of genetically altered mice or cell lines that overexpress or lack one or more specific ganglioside. Neuro-2a cells overexpressing GD3 and complex gangliosides of the b-series underwent spontaneous neuritogenesis and cholinergic differentiation (9), whereas blockade of GM3 synthase with antisense vector reduced neuritogenesis in cerebellar granule neurons (CGN).† On the other hand, embryonic stem cells deficient in GD3 synthase could be induced to express a complex neurite network, suggesting that b-series gangliosides are not essential for neuronal differentiation of uncommitted precursor cells (10). NG-CR72 cells, mutants of the NG108–15 line deficient in GM1 synthase, were found to respond positively to dendritogenic agents with prolific neurite outgrowth but negatively to axonogenic stimuli with apoptosis (11). The latter effect was attributed to loss of Ca2+ homeostasis because of ganglioside deficiency with resulting vulnerability to the Ca2+-elevating effect of axonogenic agents.
At the in vivo level, mice lacking complex gangliosides because of disruption of the GM2/GD2 synthase (UDP-N-acetyl-d-galactosamine:GM3/GD3 N-acetyl-d-galactosaminyltransferase = GalNAc-T; EC 2.4.1.92) gene were reported to show slight reduction in neural conduction velocity but no major histological or behavioral abnormalities (12). However, subsequent studies of similar mice revealed decreased myelination plus axonal degeneration (13) and impaired motor coordination (14). In view of our finding of altered Ca2+ regulation in NG-CR72 cells (11), it seemed appropriate to investigate that property in neurons from mutant mice of the above type. We find that CGN from such mice gradually degenerate when subjected to elevated K+, under conditions that cause no damage and are in fact beneficial to similar cells from normal mice. These depolarizing conditions induce progressively higher Ca2+ levels in the mutant cells after a number of days in culture, indicating loss of some aspects of Ca2+ regulation that we hypothesize is because of ganglioside deficit.
Materials and Methods
Mice engineered with a disrupted gene for GM2/GD2 synthase, on C57BL/6 background, were kindly supplied by Ronald Schnaar (Departments of Pharmacology and Neuroscience, The Johns Hopkins University School of Medicine, Baltimore, MD). Heterozygotes were mated, and cerebellar granule neurons from 6- to 7-day-old pups were prepared in culture as previously described for rat CGN (15), with some modifications. Each cerebellum was dissociated and cultured separately, for later correlation with genetic status. After removal of meninges, each cerebellum was finely minced with a razor and digested with 0.25% trypsin for 30 min at 36°C. To this mixture was added an equal volume of CGN culture medium consisting of DMEM supplemented with 10% heat-inactivated FBS, 50 μg/ml gentamycin, 50 units/ml penicillin, and 50 μg/ml streptomycin (tissue culture items purchased from GIBCO). The resulting tissue was triturated, overlaid on 4% BSA in DMEM, and centrifuged at 100 × g for 5 min. The resulting pellet was suspended in the above culture medium and the cells seeded at a density of 2 × 105/cm2 onto 96-well plastic culture plates or 25-mm glass coverslips placed in 6-well plates (for Ca2+ measurement and apoptosis determination); both plastic and glass were precoated with poly-l-lysine (1 mg/ml overnight at room temperature). The cells were cultured overnight at 37°C in an incubator with 5% CO2/95% humidified air, and the next day the media were replaced with fresh medium containing 10% FBS, N2 supplements (16), 50 ng/ml insulin, 80 μM 5-fluoro-2′-deoxyuridine in DMEM, the above additives, and the desired amount of KCl. Small volumes (approximately one-tenth of starting volume) of additional medium were added every 2 days.
After 5 days in vitro (DIV), cells were photographed with a Nikon Diaphot microscope at ×400 magnification to assess degeneration. Cell death was characterized with terminal deoxynucleotidyltransferase-mediated UTP end labeling assay using Apoptag Plus Fluorescein in Situ, Apoptosis Detection Kit, according to the manufacturer's instructions (Intergen, Purchase, NY). Quantification of cell loss was achieved with 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay after 7 DIV as described (17, 18). For determination of equilibrium levels of intracellular Ca2+ ([Ca2+]i), cells were first grown on the glass coverslips in 25 mM KCl with above medium, and each day, beginning on 3 DIV, fura-2 (Molecular Probes) dissolved in DMSO was added to give a concentration of 5 μM; 250 μM sulfinpyrazone was also present. The cells were incubated at 37°C for 60 min and then washed with buffered solution containing 5 mM KCl, 140 mM NaCl, 5 mM Ca2+, 20 mM 3-[N-morpholino]propanesulfonic acid], 1 mM MgSO4, 10 mM glucose, and 250 μM sulfinpyrazone. They were stabilized in the same solution for 5 min before Ca2+ measurements, which were begun by placing region of interest markers on 20 cells in one field. Nine frames of each field were recorded over a period of 20 seconds and averaged to rule out random instrument fluctuations. Fluorescence was measured at 510 nm after dual excitation at 350 and 380 nm; the procedure was repeated in 2 more fields (n = 3). The cells were then sequentially transferred to similar media containing 35 mM and 55 mM KCl, stabilized for 5 min, and similar measurements made for each in turn. [Ca2+]i was calculated from the equation [Ca2+]i = Kd[(R − Rmin)/(Rmax − R)](F0/FS) (19) where r = F350/F380, F0/FS = 5.52, Rmin = 0.35, Rmax = 2.68, and Kd = 224 nM. Differences were analyzed statistically by the unpaired Student's t test. A MiraCal Interline Digital Ratio Imaging System, with Olympix cooled charge-coupled device interline camera and xenon light source, was used.
Animal genotype was identified by PCR analysis of DNA isolated from brain tissue. The 5′ and 3′ primers were 5′-TAC CAG GCC AAC ACA GCA-3′ and 5′-CAG GTC CAG GGG CGT CTT-3′, respectively. The resultant PCR products were applied to a 0.7% agarose gel that was developed electrophoretically in TAE buffer (0.8 mM Tris-acetate/40 μM Na2-EDTA, pH 8.5) for 2.5 h. Bands were revealed and photographed under UV light.
Ganglioside deficiency was determined by analyzing the ganglioside pattern in the cerebral hemispheres of each brain. Total lipids were extracted and partitioned according to Folch et al. (20), and gangliosides were isolated from the aqueous-methanol upper phase by reverse phase chromatography on Sep-Pak cartridges (Waters–Millipore) (21). After evaporation to dryness, the residues were dissolved in small volumes of chloroform/methanol (1:1) and portions equivalent to 0.8 mg protein applied to a 20 × 20-cm high-performance thin-layer chromatography plate coated with silica gel 60 (EM Science). Separation was effected with the solvent system chloroform-methanol-0.25% aqueous KCl (5:4:1) and ganglioside bands revealed with resorcinol spray (21). Total levels of ganglioside sialic acid were determined by resorcinol assay (22).
Results and Discussion
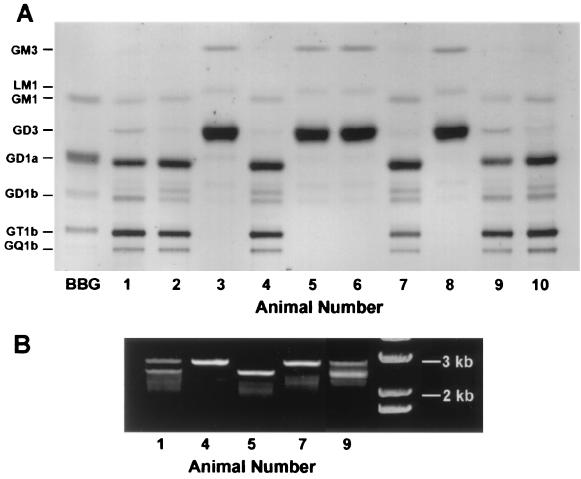
In a typical experiment, 10 mouse pups were separately processed for culturing of CGN, identification of the GalNAc-T gene with PCR analysis, and ganglioside pattern determination in brain tissue. The latter revealed four pups (nos. 3, 5, 6, and 8) as knockouts with inactivated GalNAc-T gene (2.5 kb) that resulted in the deficiency of GM2, GD2, and all gangliotetraose gangliosides (Fig. 1). This deficiency was accompanied by marked increase of GD3 and GM3, as previously reported for these mutant mice (12, 13). We also noted increase of a ganglioside running just ahead of GM1, possibly corresponding to LM1 (IV3NeuAc-nLcOse4Cer), a member of the neolactotetraose family (23, 24). Among the nonknockouts, there were four normals (nos. 2, 4, 7, and 10) showing a single PCR band (2.9 kb), and two heterozygotes (nos. 1 and 9) showing double PCR bands at 2.5 and 2.9 kb (although this litter departed from the expected Mendelian distribution, overall we obtained twice as many heterozygotes as knockout or wild type). The heterozygotes possessed virtually all of the gangliotetraose gangliosides present in wild type and in addition showed a modest increase in GD3 along with reduction of a ganglioside running just ahead of GD1b. It was of some interest that total cerebral ganglioside content of the knockouts was 4.84 ± 1.02 μg sialic acid/mg protein that was only marginally less than the value for heterozygotes (6.17 ± 0.40) but significantly less than normals (6.91 ± 1.00, P < 0.05).
Figure 1.
Ganglioside pattern and genotype analysis of one representative litter. (A) Thin-layer chromatogram of gangliosides purified from the cerebra of 10 littermate pups. Isolated material corresponding to 0.8 mg protein was applied at the origin, and the plate developed in chloroform-methanol-0.25% aqueous KCl (5:4:1); ganglioside bands were revealed with resorcinol spray. Pups nos. 3, 5, 6, and 8 were deficient in complex gangliosides and showed an excess of GM3 and LM1 together with a very large accumulation of GD3. Pups nos. 1 and 9 were heterozygotes (see below) and contained complex gangliosides similar to wild type (nos. 2, 4, 7, and 10) with modest elevation of GD3 accompanied by reduction of a ganglioside running just ahead of GD1b. BBG, bovine brain gangliosides (standards). (B) Genotype identification. DNA isolated from the above brain tissues was subjected to PCR and the resulting products developed in a 0.7% agarose gel. The 2.9- and 2.5-kb bands represent wild-type and mutant GalNAc-T alleles, respectively. Pups nos. 4 and 7 were identified as wild type, nos. 1 and 9 as heterozygotes, and no. 5 as knockout. The far right column is DNA ladder standards (GIBCO).
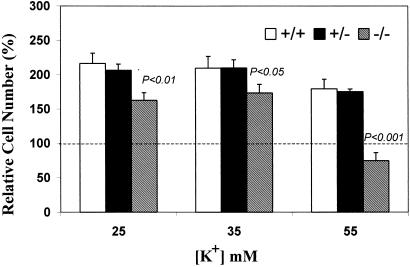
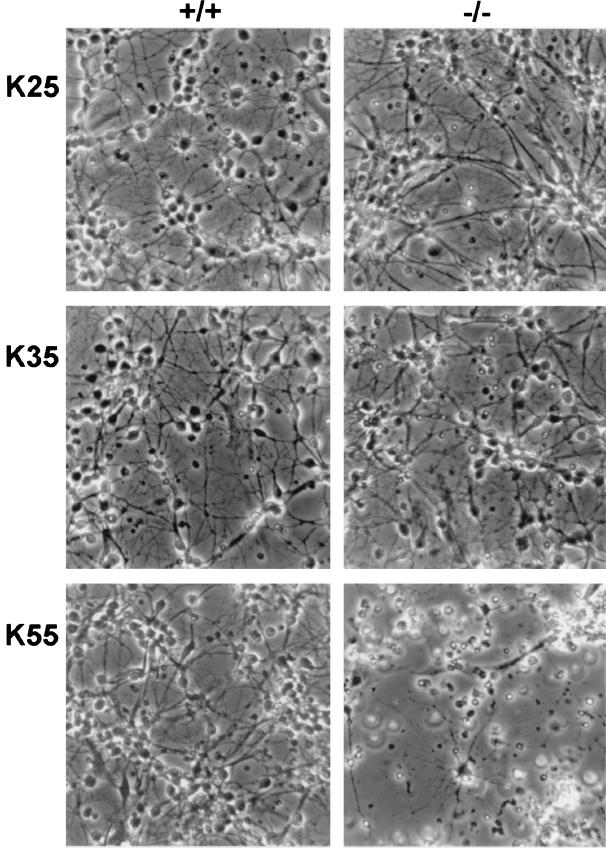
Quantification of cell survival, carried out after 7 DIV with MTT assay on cells in 96-well plates, revealed that CGN from genetically normal mice and heterozygotes survived well in depolarizing levels of KCl—significantly better, in fact, than cells grown in 5 mM KCl (Fig. 2). This was consistent with earlier reports demonstrating enhanced survival of rat CGN in the presence of elevated K+ (18, 25). On the other hand, similar treatment of CGN from the knockout mice showed somewhat attenuated survival improvement at KCl concentrations of 25 and 35 mM and striking loss of cells in both relative and absolute terms at 55 mM KCl. Evidence of deteriorating cell morphology was evident from phase contrast microscopy (Fig. 3), even at 25 and 35 mM KCl, despite the relatively modest MTT results at those concentrations. Microscopic evidence of cell destruction was most evident at 55 mM KCl for the mutated CGN. Application of the terminal deoxynucleotidyltransferase-mediated UTP end labeling assay revealed apoptosis as a cause of cell death (Fig. 4), analogous to that previously seen with NG-CR72 cells, a mutant line lacking GM1-synthase (11). The above study was repeated with two additional litters with similar results.
Figure 2.
Cell quantification after exposure to elevated K+. Cerebellar granule neurons from mutant (−/−), heterozygote (+/−), and normal (+/+) mice were cultured in 96-well plastic plates for 7 days in varying concentrations of KCl, as described. Results are expressed relative to MTT values obtained with cells from the same animal grown in 5 mM KCl. There was no significant difference of absolute MTT values between the 3 genotypes at 5 mM KCl (not shown). Normal and heterozygote cell survival increased at all concentrations of KCl, whereas mutant cell survival decreased markedly at 55 mM KCl. P values compare CGN from knockout mice with those from wild type. (Bars = SD; n = 3.)
Figure 3.
Morphological comparison of CGN exposed to elevated K+. Cells were grown 5 DIV, as described, in media containing variable amounts of KCl. Representative fields were photographed at ×400.
Figure 4.
Calcium-induced apoptosis of cerebellar granule neurons from knockout mice, grown in high KCl. Cells from normal (+/+) and mutant (−/−) mice were cultured on coverslips as described in 55 mM KCl for 5 days, then fixed and stained with ApopTag kit. Two fields for each treatment are shown. High KCl induced apoptosis in knockout but not normal neurons.
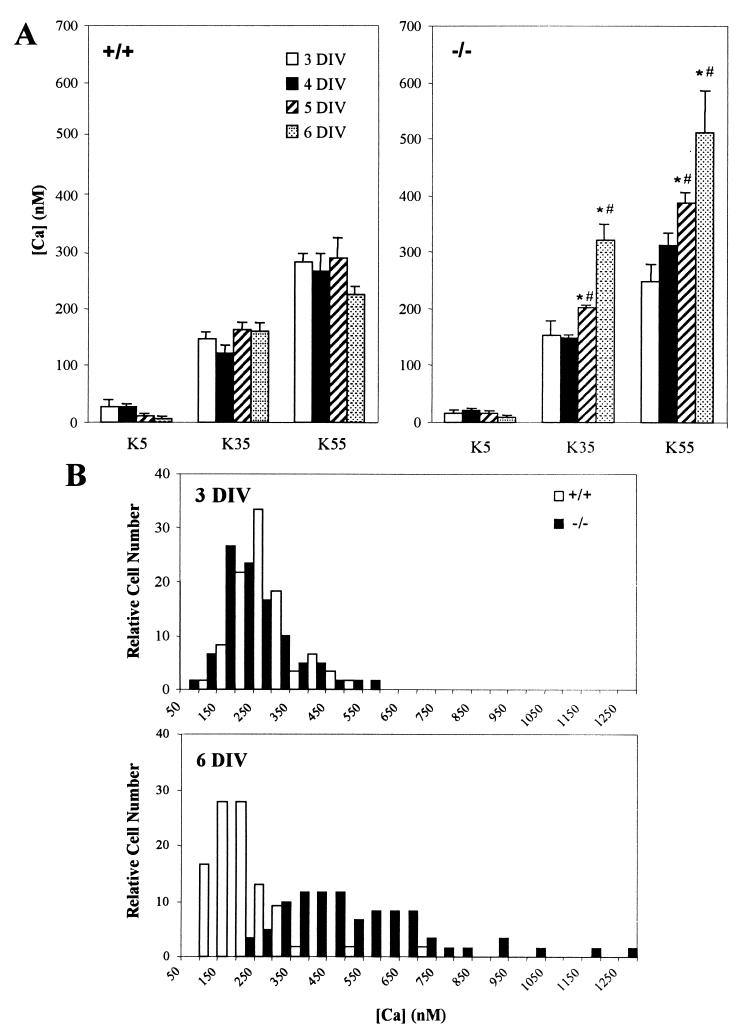
Calcium measurements revealed significant differences between CGN from wild-type and knockout mice (Fig. 5A). Cells were grown in medium containing 25 mM KCl for the periods shown, then placed sequentially in buffered KCl of different concentrations for [Ca2+]i measurement. Normal CGN showed no increase of [Ca2+]i at any time point resulting from sequential testing with the 3 KCl concentrations, whereas mutant cells revealed significant [Ca2+]i increases in such testing at 5 and 6 DIV. [Ca2+]i was significantly higher in mutant than in normal cells at 6 DIV with elevated KCl, whereas no difference was seen at any time with physiological K+ (5 mM). Fig. 5B shows the distribution of cells with respect to [Ca2+]i when the cells were placed in 55 mM KCl; at 6 DIV, virtually all mutant cells had higher [Ca2+]i than normal cells, whereas such difference was not observed after 3 DIV. These findings may relate to the report that voltage-dependent Ca2+ channels appear in cultured CGN after 3 DIV (26). The pronounced reduction in survival of mutant cells at 55 mM KCl (Fig. 2) thus correlates with elevated levels of [Ca2+]i.
Figure 5.
(A) Calcium measurements in mutant and normal CGN subjected to different K+ concentrations. After 3–6 DIV, [Ca2+]i was measured ratiometrically with fura-2 fluorescence after placing cells in different concentrations of KCl. Whereas the normal (+/+) cells (Left) showed no increase at any DIV with rising K+ concentrations, mutant (−/−) cells (Right) showed progressive increase of [Ca2+]i with development at 35 and 55 mM KCl (*, P < 0.05 comparing [Ca2+]i at 5 and 6 DIV with that at 3 DIV for the same KCl). In addition, [Ca2+]i levels were higher in the mutant than the normal cells at 35 and 55 mM KCl after 5 and 6 DIV (#, P < 0.05) comparing [Ca2+]i in mutant cells with those in normal cells at same DIV and KCl). (Bars = SD; n = 3.) Note that the low levels of [Ca2+]i at 5 mM KCl are in general agreement with those reported for similar cultures of rat CGN (50). (B) Frequency distribution of [Ca2+]i in normal and knockout CGN at 55 mM KCl after 3 and 6 DIV. Whereas little difference was seen at 3 DIV, virtually all of the mutant cells possessed higher [Ca2+]i than normal cells at 6 DIV. Data are presented as percentages in each group of ∼60 cells measured.
Both the beneficial and detrimental effects of depolarizing concentrations of K+ are likely attributable to elevation of [Ca2+]i, shown to occur with rat CGN grown in elevated K+ (18, 25). Although other studies reported that mouse CGN survive equally well at normal and elevated K+ concentrations (26, 27), we have found that under currently used conditions, normal mouse CGN also showed enhanced survival over time in the presence of higher K+ (Fig. 2). Our results suggest there may be a critical transition between approximately 320–390 nM [Ca2+]i when the beneficial effects of Ca2+ elevation give way to apoptosis inception. In comparing the timing of the differentiation and innervation of postmitotic CGN in vivo with the development of K+ dependence in vitro, it was suggested that depolarization of CGN in culture mimics the influence of physiological stimulation in vivo through excitatory amino acid receptors (25). The deleterious effects of 55 mM K+ to CGN from ganglioside-deficient (knockout) mice suggest the mutant cells have lost a Ca2+ regulatory mechanism that is modulated by one or more of the deleted gangliosides. Possible existence of such mechanism(s) was previously suggested in the demonstration that GM1 and other gangliotetraose gangliosides protected CGN in culture against glutamate toxicity (28) and Neuro-2a cells subjected to Ca2+ ionophore toxicity (17). Calcium modulatory effects of gangliosides have been observed in several other studies, involving both exogenous (29–36) and endogenous (18, 37–44) gangliosides. GM1 has been the predominant endogenous ganglioside to be studied from this standpoint, mainly in the context of its plasma membrane locus.
GM1 has also been shown to occur in the nuclear envelope of CGN (15, 45), where it is elevated during axon outgrowth and was proposed to function as modulator of nuclear Ca2+ after onset of axonogenesis (5, 46, 47). Recent study of NG-CR72 cells, a mutant of the NG108–15 line deficient in GM1 synthase, revealed striking vulnerability to Ca2+-elevating axonogenic agents but not to dendritogenic stimuli that exerted no observable effect on [Ca2+]i (11). That these cells could be rescued from apoptosis by a membrane-permeant form of GM1, but not by GM1 itself, suggested the protective effect in that system operated at the nuclear membrane. Those GM1-deficient neuroblastoma cells bear some resemblance to the mutant CGN of this study in that both appear relatively normal until challenged with Ca2+-elevating agents. That such challenge may have physiological relevance is suggested by the recent demonstration of spontaneous and frequent Ca2+ sparks in primary neuronal cultures as indication of a general phenomenon of intraneuronal calcium “noise” (48). This phenomenon suggests that aberrant Ca2+ regulation resulting from ganglioside deficiency may contribute to the axonal degeneration observed in the knockout mice and the significant deficits in their motor behavior at maturity (13). Studies are currently in progress to differentiate the Ca2+-modulating effects of gangliosides in the nuclear and plasma membranes during development.
Acknowledgments
The assistance of Dr. Ronald Schnaar in establishing the breeding colony of knockout mice and of Dr. Richard Howells in PCR analysis is gratefully acknowledged. This work was supported by National Institutes of Health Grant NS33912.
Abbreviations
- [Ca2+]i
intracellular Ca2+
- CGN
cerebellar granule neurons
- DIV
days in vitro
- MTT
3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide. Ganglioside nomenclature is that of Svennerholm (49)
Footnotes
Zeng, G., Gao, L., Li, D. D., Tokuda, A. & Yu, R. K. (1998) Ann. NY Acad. Sci. 845, 431 (abstr.).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011523698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011523698
References
- 1.Ledeen R W. In: Biosynthesis, Metabolism, and Biological Effects of Gangliosides. Margolis R U, Margolis R K, editors. New York: Plenum; 1989. pp. 43–83. [Google Scholar]
- 2.Yu R K, Saito M. In: Structure and Location of Gangliosides. Margolis R U, Margolis R K, editors. New York: Plenum; 1989. pp. 1–42. [Google Scholar]
- 3.Yates A J. Neurochem Pathol. 1986;5:309–329. doi: 10.1007/BF02842941. [DOI] [PubMed] [Google Scholar]
- 4.Schengrund C-L. Brain Res Bull. 1990;24:131–141. doi: 10.1016/0361-9230(90)90297-d. [DOI] [PubMed] [Google Scholar]
- 5.Ledeen R W, Wu G, Lu Z-H, Kozireski-Chubak D F, Fang Y. Ann NY Acad Sci. 1998;845:161–175. doi: 10.1111/j.1749-6632.1998.tb09669.x. [DOI] [PubMed] [Google Scholar]
- 6.Purpura D P, Suzuki K. Brain Res. 1976;116:1–21. doi: 10.1016/0006-8993(76)90245-6. [DOI] [PubMed] [Google Scholar]
- 7.van Echten G, Sandhoff K. J Biol Chem. 1993;268:5341–5344. [PubMed] [Google Scholar]
- 8.Maccioni H J, Daniotti J L, Martina J A. Biochim Biophys Acta Lipids Lipid Metab. 1999;1437:101–118. doi: 10.1016/s1388-1981(99)00002-5. [DOI] [PubMed] [Google Scholar]
- 9.Kojima N, Kurosawa N, Nishi T, Hanai N, Tsuji S. J Biol Chem. 1994;269:30451–30456. [PubMed] [Google Scholar]
- 10.Kawai H K, Sango K, Mullin K A, Proia R L. J Biol Chem. 1998;273:19634–19638. doi: 10.1074/jbc.273.31.19634. [DOI] [PubMed] [Google Scholar]
- 11.Wu, G., Lu, Z.-H., Xie, X., Li, L. & Ledeen, R. W. (2000) J. Neurochem., in press. [DOI] [PubMed]
- 12.Takamiya K, Yamamoto A, Furukawa K, Yamashiro S, Shin M, Okada M, Fukumoto S, Haraguchi M, Takeda N, Fujimura K, S, et al. Proc Natl Acad Sci USA. 1996;93:10662–10667. doi: 10.1073/pnas.93.20.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheikh K A, Sun J, Liu Y, Kawai H, Crawford T O, Proia R L, Griffin J W, Schnaar R L. Proc Natl Acad Sci USA. 1999;96:7532–7537. doi: 10.1073/pnas.96.13.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald M P, Gutshall H, Kawai H, Proia R L, Crawley J N. Soc Neurosci Abstr. 1999;25:1117. [Google Scholar]
- 15.Kozireski-Chubak D F, Wu G, Ledeen R W. Dev Brain Res. 1999;115:201–208. doi: 10.1016/s0165-3806(99)00062-0. [DOI] [PubMed] [Google Scholar]
- 16.Bottenstein J E, Sato G H. Proc Natl Acad Sci USA. 1979;760:514–517. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura K, Wu G, Ledeen R W. J Neurosci Res. 1992;31:245–253. doi: 10.1002/jnr.490310205. [DOI] [PubMed] [Google Scholar]
- 18.Wu G, Lu Z-H, Nakamura K, Spray D C, Ledeen R W. J Neurosci Res. 1996;44:243–254. doi: 10.1002/(SICI)1097-4547(19960501)44:3<243::AID-JNR5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 19.Grynkiewicz G, Poenie M, Tsien R Y. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 20.Folch J, Lees M, Sloan Stanley G H. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 21.Ledeen R W, Yu R K. Methods Enzymol. 1982;83:139–191. doi: 10.1016/0076-6879(82)83012-7. [DOI] [PubMed] [Google Scholar]
- 22.Svennerholm L. Biochim Biophys Acta. 1957;24:604–611. doi: 10.1016/0006-3002(57)90254-8. [DOI] [PubMed] [Google Scholar]
- 23.Ando S, Kon K, Isobe M, Yamakawa T. J Biochem (Tokyo) 1973;73:893–895. doi: 10.1093/oxfordjournals.jbchem.a130152. [DOI] [PubMed] [Google Scholar]
- 24.Li Y T, Mansson J E, Vanier M T, Svennerholm L. J Biol Chem. 1973;248:2634–2636. [PubMed] [Google Scholar]
- 25.Gallo V, Kingsbury A, Balàzs R, Jorgensen O S. J Neurosci. 1987;7:2203–2213. doi: 10.1523/JNEUROSCI.07-07-02203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng L, Juurlink B H J, Hertz L. Dev Brain Res. 1991;63:1–12. doi: 10.1016/0165-3806(91)90061-m. [DOI] [PubMed] [Google Scholar]
- 27.Mogensen H S, Jorgensen O S. Int J Dev Neurosci. 2000;18:61–68. doi: 10.1016/s0736-5748(99)00105-7. [DOI] [PubMed] [Google Scholar]
- 28.De Erausquin G A, Manev H, Guidotti A, Costa E, Brooker G. Proc Natl Acad Sci USA. 1990;87:8017–8021. doi: 10.1073/pnas.87.20.8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G, Vaswani K K, Lu Z-H, Ledeen R W. J Neurochem. 1990;55:484–491. doi: 10.1111/j.1471-4159.1990.tb04161.x. [DOI] [PubMed] [Google Scholar]
- 30.Spoerri P E, Dozier A K, Roisen F J. Dev Brain Res. 1990;56:177–188. doi: 10.1016/0165-3806(90)90080-i. [DOI] [PubMed] [Google Scholar]
- 31.Guerold B, Massarelli R, Forster V, Freysz L, Dreyfus H. J Neurosci Res. 1992;32:110–115. doi: 10.1002/jnr.490320113. [DOI] [PubMed] [Google Scholar]
- 32.Doherty P, Ashton S V, Skaper S D, Leon A, Walsh F S. J Cell Biol. 1992;117:1093–1099. doi: 10.1083/jcb.117.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan Z, Stokes B T, Brocklyn J R V, Yates A J. Biochim Biophys Acta. 1992;1136:315–318. doi: 10.1016/0167-4889(92)90123-s. [DOI] [PubMed] [Google Scholar]
- 34.Hilbush B S, Levine J M. J Biol Chem. 1992;267:24789–24795. [PubMed] [Google Scholar]
- 35.Isasi S C, Bianco I D, Fidelio G D. Life Sci. 1995;57:449–456. doi: 10.1016/0024-3205(95)00278-e. [DOI] [PubMed] [Google Scholar]
- 36.Müthing J, Maurer U, Weber-Schürholz S. Carbohydr Res. 1998;307:147–157. doi: 10.1016/s0008-6215(98)00028-7. [DOI] [PubMed] [Google Scholar]
- 37.Dixon S J, Stewart D, Grinstein S, Spiegel S. J Cell Biol. 1987;105:1153–1161. doi: 10.1083/jcb.105.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu G, Ledeen R W. J Neurochem. 1991;56:95–104. doi: 10.1111/j.1471-4159.1991.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 39.Wu G, Ledeen R W. Prog Brain Res. 1994;101:101–112. doi: 10.1016/s0079-6123(08)61942-1. [DOI] [PubMed] [Google Scholar]
- 40.Milani D, Minozzi M-C, Petrelli L, Guidolin D, Skaper S D, Spoerri P E. J Neurosci Res. 1992;33:446–475. doi: 10.1002/jnr.490330313. [DOI] [PubMed] [Google Scholar]
- 41.Carlson R O, Masco D, Brooker G, Spiegel S. J Neurosci. 1994;14:2272–2281. doi: 10.1523/JNEUROSCI.14-04-02272.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouy H, Detterre P, Debré P, Bismuth G. J Immunol. 1994;152:3271–3281. [PubMed] [Google Scholar]
- 43.Wang Y, Tsui Z, Yang F. FEBS Lett. 1999;457:144–148. doi: 10.1016/s0014-5793(99)01024-8. [DOI] [PubMed] [Google Scholar]
- 44.Fang Y, Wu G, Xie X, Lu Z-H, Ledeen R W. Neurochem Res. 2000;25:931–940. doi: 10.1023/a:1007596223484. [DOI] [PubMed] [Google Scholar]
- 45.Wu G, Lu Z-H, Ledeen R W. J Neurosci. 1995;15:3739–3746. doi: 10.1523/JNEUROSCI.15-05-03739.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu G, Lu Z-H, Ledeen R W. J Neurochem. 1995;64:1419–1422. doi: 10.1046/j.1471-4159.1995.65031419.x. [DOI] [PubMed] [Google Scholar]
- 47.Kozireski-Chuback D, Wu G, Ledeen R W. J Neurosci Res. 1999;55:107–118. doi: 10.1002/(SICI)1097-4547(19990101)55:1<107::AID-JNR12>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 48.Melamed-Book N, Kachalsky S G, Kaiserman I, Rahamimoff R. Proc Natl Acad Sci USA. 1999;96:15217–15221. doi: 10.1073/pnas.96.26.15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svennerholm L. J Neurochem. 1963;10:613–623. doi: 10.1111/j.1471-4159.1963.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 50.Ciardo A, Meldolesi J. J Neurochem. 1991;56:184–191. doi: 10.1111/j.1471-4159.1991.tb02579.x. [DOI] [PubMed] [Google Scholar]