Abstract
Heat shock protein (Hsp)90 is emerging as an important therapeutic target for the treatment of cancer. Two analogues of the Hsp90 inhibitor geldanamycin are currently in clinical trials. Geldanamycin (GA) and its analogues have been reported to bind purified Hsp90 with low micromolar potency, in stark contrast to their low nanomolar antiproliferative activity in cell culture and their potent antitumor activity in animal models. Several models have been proposed to account for the ≈100-fold-greater potency in cell culture, including that GA analogues bind with greater affinity to a five-protein Hsp90 complex than to Hsp90 alone. We have determined that GA and the fluorescent analogue BODIPY-GA (BDGA) both demonstrate slow, tight binding to purified Hsp90. BDGA, used to characterize the kinetics of ligand–Hsp90 interactions, was found to bind Hsp90α with koff = 2.5 × 10−3 min−1, t1/2 = 4.6 h, and Ki* = 10 nM. It was found that BDGA binds to a functional multiprotein Hsp90 complex with kinetics and affinity identical to that of Hsp90 alone. Also, BDGA binds to Hsp90 from multiple cell lysates in a time-dependent manner with similar kinetics. Therefore, our results indicate that the high potency of GA in cell culture and in vivo can be accounted for by its time-dependent, tight binding to Hsp90 alone. In the broader context, these studies highlight the essentiality of detailed biochemical characterization of drug–target interactions for the effective translation of in vitro pharmacology to cellular and in vivo efficacy.
Keywords: benzoquinone ansamycin, time-dependent inhibition, BODIPY-geldananmycin
Heat shock protein (Hsp)90 is a ubiquitous, highly expressed molecular chaperone protein capable of sensing cellular stress (1). In cells, Hsp90 functions as a multiprotein chaperone complex, with cochaperones that include Hsp70, Hsp40, and Hop (2). Upon ATP binding and hydrolysis, this intermediate complex forms a mature chaperone complex containing p23, which catalyzes the conformational maturation of “client protein” substrates (3, 4). Multiple client proteins of the Hsp90 chaperone complex are involved in signal-transduction pathways, cell-cycle regulation, and apoptosis pathways commonly deregulated in cancer (5, 6). Although essential for cellular viability, the pharmacological inhibition of this chaperone has emerged as an attractive approach for inhibition of tumorigenesis (7–10).
The natural product geldanamycin (GA), a benzoquinone ansamycin, binds to Hsp90, inhibits its ATPase activity (11), and decreases cellular levels of client proteins involved in cancer cell survival, such as mutated p53, mutated B-Raf, Akt, Bcr-Abl, and ErbB2 (10). GA has potent antiproliferative activity in many cell lines in culture (12) and inhibits tumor growth in mouse xenograft models (10). Two GA analogues, 17-allylamino,17-demethoxygeldanamycin (17-AAG) and 17-dimethylaminoethylamino,17-demethoxygeldanamycin (17-DMAG), are currently in multiple clinical trials for the treatment of cancer (7, 13, 14).
GA binds to the N-terminal ATP-binding domain of Hsp90 and inhibits ATP binding and ATP-dependent chaperone activities (15). The inhibitory potency and affinity of benzoquinone ansamycins for the isolated Hsp90 protein have been determined by several methods and shown to be in the low micromolar range (8, 11, 16). However, these compounds exhibit low nanomolar cellular antiproliferative activity (11, 12, 17, 18). Several explanations have been described for the markedly increased potency in cells. Chiosis et al. (17) have proposed that the physicochemical properties of the ansamycins results in its intracellular accumulation from cell culture media, leading to highly potent antiproliferative activity (11, 12, 17, 18). Kamal et al. (19) have provided biochemical and cellular evidence that the ansamycins bind to and inhibit an Hsp90 multiprotein complex with much higher affinity than to Hsp90 alone (17, 19).
The purpose of this study is to characterize further the binding and inhibitory activity of GA with purified Hsp90 protein and the Hsp90 chaperone complex to understand better the cellular and in vivo activity of this class of compounds.
Results
Recombinant full-length Hsp90, Hsp70, Hsp40, Hop, and p23 were expressed in Escherichia coli and purified to homogeneity (Fig. 1). Protein identity was confirmed by N-terminal sequencing and molecular mass confirmed by liquid chromatography MS. The in vitro chaperone activity of these proteins was characterized by following the method of Walerych et al. (20), in which the luciferase is heat-denatured in the presence of Hsp90 and allowed to refold, with addition of Hsp70, Hsp40, and ATP at ambient temperature, followed by quantitation of refolding as a measurement of luciferase activity. By using the purified proteins, the time-dependent refolding of active luciferase was found to be greatly diminished by removing one of the proteins (Hsp70 or Hsp40) or the substrate ATP (Fig. 2). This strict requirement for the presence of all three proteins of the chaperone complex and the substrate indicate that the three chaperone proteins form a functional complex in vitro. This result is consistent with the findings reported by Walerych et al. (20).
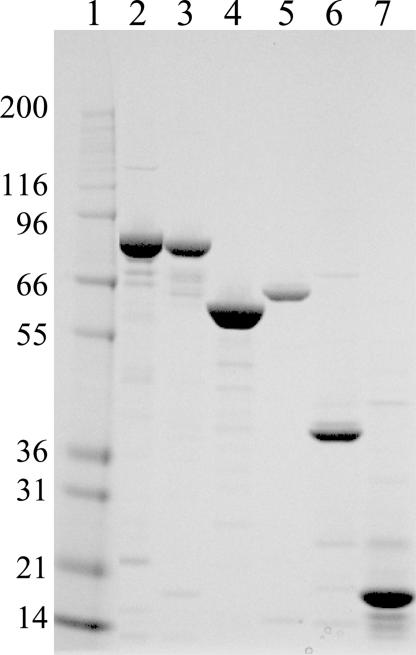
Fig. 1.
SDS/PAGE of the purified proteins Hsp90, Hsp70, Hsp40, Hop, and p23. Resolution of purified protein by 4–12% Bis-Tris SDS/PAGE in Mops buffer run at 200 V for 50 min. Protein loading: 2 μg per lane. Lane 1, molecular mass markers; lane 2, Hsp90α; lane 3, Hsp90β; lane 4, Hop; lane 5, Hsp70; lane 6, Hsp40; lane7, p23.
Fig. 2.
Time course of luciferase refolding in the presence of Hsp90 complex chaperone proteins. Hsp90α (2.5 μM) was incubated with 0.25 μM luciferase at 50°C for 8 min before diluting 6-fold into renaturation buffer containing 0.5 mM ATP, 2 μM Hsp70, and 1 μM Hsp40 (●). Reactions run in the absence of ATP (○), Hsp70 (□), or Hsp40 (▵) exhibit no substantial refolding activity.
To assess the inhibitory activity of GA against Hsp90 in the absence and presence of cochaperones, multiple assay formats were established to detect Hsp90 ATPase activity. In all of these formats, very low ATPase-specific activity was measured; in all cases, kcat < 1 h−1 (data not shown). This result was observed for Hsp90 alone and in multiple combinations with cochaperones (Hsp70, Hsp40, Hop, and p23), by following protocols of Pratt and Toft (21). The lack of robust ATPase activity, consistent with reports for the human isozymes (19, 22), makes establishment of a multiple-turnover assay for detailed characterization of inhibitors impractical. Additionally, analysis of very low catalytic activity could easily be confounded by ATPase activity from protein contaminants in the Hsp90 preparation. For these reasons, we established assays to characterize GA binding to Hsp90.
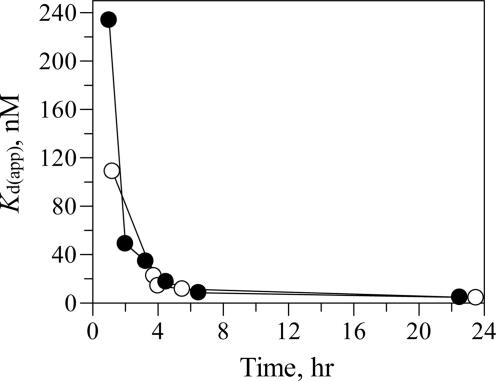
The binding affinity of a fluorescently labeled analogue of GA, 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-geldanamycin (BODIPY-geldanamycin or BDGA), to Hsp90α was determined by measuring the changes in the fluorescence anisotropy of the ligand in the presence of varying concentrations of Hsp90, as described in ref. 23. The Kd(app) values determined for Hsp90–BDGA binding were found to depend on the protein–ligand incubation time, such that the Kd(app) > 1,000 nM, with a very short incubation time (6 min) and a Kd(app) = 4.6 nM after 24 h incubation (Table 1). The Kd(app) value determined after a short protein–ligand incubation time is not representative of the true equilibrium dissociation constant because of slow binding that approaches equilibrium over several hours. To determine whether the observed time-dependent binding is due to the fluorescent group attached to GA, titrating concentrations of unlabeled GA were incubated with Hsp90 for varying times, followed by addition of BDGA for 30 min and BDGA fluorescence anisotropy measured. Although BDGA binding to Hsp90 is under preequilibrium conditions after 30 min, its fluorescence anisotropy is used as a probe to quantify the concentration of free Hsp90, from which GA-Hsp90 Kd(app) values are calculated, as described in detail in Materials and Methods. GA was found to have the same potency and time-dependent binding to Hsp90 as its fluorescent analogue, indicating that the BODIPY fluorescent group does not affect Hsp90–GA-binding interactions (Table 1). Because the fluorescent BDGA and unlabeled GA were found to possess the same apparent binding affinity and time-dependence of binding, the fluorescent ligand was used for detailed characterization of the time-dependence of binding to Hsp90 as a means of characterizing the Hsp90–GA interactions.
Table 1.
Kd(app) values for ligand binding to Hsp90α alone vs. protein–ligand incubation time
| Time, hr | Kd(app), nM* | ||
|---|---|---|---|
| BDGA | GA | Compound 1 | |
| 0.1 | >1,000 | ND | ND |
| 1 | 45 | 80 | 104 |
| 2 | 21 | 58 | 100 |
| 24 | 4.6 | 9.0 | 170 |
*Kd(app) values are an average, n = 2. Compound 1 is 4-butyl-6-[4-(2-methyl-1,3-thiazol-4-yl)-5-isoxazolyl]-1,3-benzenediol. ND, not determined.
BDGA binding to Hsp90α was then characterized by monitoring fluorescence anisotropy as a function of incubation time at different concentrations of Hsp90. The fluorescence anisotropy of BDGA increases from r0 = 0.04 to rb = 0.17 vs. time, reflecting a decrease in the rotational diffusion of BDGA in solution upon binding to Hsp90. These data are fit to a pseudo-first-order rate equation, Eq. 8 (Fig. 3A and B). The kobs values obtained from the time course fit to the data are replotted vs. the concentration of Hsp90 ([Hsp90]) (Fig. 3C). The curvature of the plotted points approaching a maximal value is indicative of a two-step binding model, where
 |
Fig. 3.
Kinetic analysis of the time-dependence of BDGA binding to Hsp90α. BDGA binding to Hsp90α was measured by monitoring the fluorescence anisotropy of BDGA (10 nM) as a function of incubation time in the presence of varying concentrations of the protein. BDGA unbound in solution and bound to Hsp90α has approximate anisotropy values of 0.04 and 0.17, respectively. Data were fit to a pseudo-first-order rate equation, Eq. 8, to determine kobs values. (A) [Hsp90]: 1.5 μM (▾), 1.25 μM (▿), 0.90 μM (▴), 0.70 μM (▵), 0.50 μM (■), 0.30 μM (□), 0.10 μM (●), 0 μM (○). (B) [Hsp90]: 0.10 μM (▴), 0.075 μM (▵), 0.050 μM (■), 0.025 μM (□), 0.0125 μM (●), 0 μM (○). (C) Replot of kobs vs. [Hsp90]. Data fit to the equation for a two-step protein–ligand-binding model, Eq. 1. Kinetic constants derived from the data are summarized in Table 2.
In this model, Hsp90 and BDGA rapidly form an encounter complex with association and dissociation rate constants k3 and k4, respectively. The binding of BDGA to Hsp90 induces a time-dependent conformational change in the enzyme that results in a much-higher-affinity Hsp90–BDGA* complex, with forward and reverse rate constants of k5 and k6. The equilibrium dissociation constant for the encounter complex is Ki = k4/k3 and for the Hsp90-BDGA* complex is Ki* = Ki k6/(k5 + k6) (24). This model is simplified by assuming that k6 is much slower than k4, and, therefore, the overall dissociation rate of Hsp90–BDGA* to Hsp90 + BDGA (also referred to as koff) is equivalent to k6. The data in the plot of kobs vs. [Hsp90] is fit to the equation
and the average values of these constants are shown in Table 2. BDGA forms an encounter complex with the enzyme with a Ki = 450 nM, which equilibrates to an Hsp90–BDGA* complex with Ki* = 10 nM. The dissociation rate of the tightly bound Hsp90–BDGA* to Hsp90 + BDGA is very slow; koff = k6 = 2.5 × 10−3 min−1 with a t1/2 = 4.6 h.
Table 2.
Kinetic constants for BDGA binding to Hsp90 (α and β) alone and in the presence of cochaperone proteins
| Kinetic parameter | Enzyme | |||
|---|---|---|---|---|
| Hsp90α alone | Hsp90α with cochaperones | Hsp90β alone | Hsp90β with cochaperones | |
| Ki, nM | 450 ± 70 | 860 ± 290 | 510 ± 150 | 1100 ± 290 |
| k5, min−1 | 0.10 ± 0.0061 | 0.14 ± 0.021 | 0.10 ± 0.0084 | 0.14 ± 0.021 |
| k6, min−1 × 103 | 2.5 ± 0.20 | 2.7 ± 0.26 | 5.7 ± 0.48 | 6.0 ± 0.60 |
| t1/2, h | 4.6 | 4.2 | 2.0 | 1.9 |
| Ki*, nM | 10 ± 1.8 | 15 ± 5.7 | 28 ± 8.3 | 45 ± 14 |
All values are an average, n = 2.
Time-dependence of BDGA-binding experiments were also performed with Hsp90β in an identical manner, in the absence and presence of 1.0 μM cochaperones Hsp70, Hsp40, HOP, and p23 and the results summarized in Table 2. The isoform-specific differences observed are small (within 3-fold) for the kinetic constants of BDGA binding to Hsp90α and -β. More importantly, the kinetics constants for BDGA binding to Hsp90 are unaffected by the presence of the cochaperones Hsp70, Hsp40, Hop, and p23. This result is observed for both the Hsp90α and -β isoforms. Also, the kinetics of BDGA binding to Hsp90 in the presence of a single cochaperone (Hsp70, Hsp40, Hop, or p23) and in the presence of the two cochaperone proteins Hsp70 and Hsp40 has also been characterized. Under all of these conditions, the kinetics and affinity of BDGA binding to Hsp90 were found to be identical. Because it was shown that Hsp90, Hsp70, and Hsp40 form a functional chaperone complex, it can be concluded that the kinetics of BDGA binding to Hsp90 as a single protein is the same as binding of the ligand to Hsp90 in the context of the full chaperone complex.
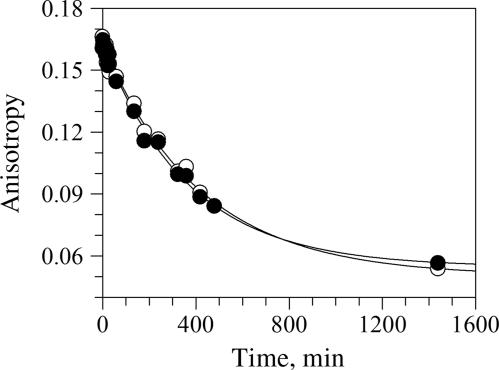
The determination of the kinetic constant k6 for BDGA–Hsp90 binding is based on the y-intercept of the plot of kobs vs. [Hsp90] (Fig. 3C). Because determination of the k6 value could vary substantially because of small data variations, it was important to determine k6 (equivalent to koff) by an independent method. For this determination, BDGA was incubated with Hsp90α for 24 h under conditions for which essentially all BDGA is bound to the protein. The sample was then diluted 100-fold into a solution with excess GA, and the BDGA–Hsp90α rate of dissociation was measured by the time-dependent change in anisotropy under conditions where there is no appreciable BDGA–Hsp90α reassociation (Fig. 4). The data fit well to a monoexponential decay, with average koff = 2.5 × 10−3 min−1, average t1/2 = 4.5 h (n = 2). The values from this dilution experiment are in close agreement with those determined from progress-curve analysis.
Fig. 4.
Determination of the BDGA–Hsp90α dissociation rate (koff). BDGA was incubated with Hsp90α under conditions so that essentially all BDGA is bound to the protein. The sample is then diluted 100-fold into a solution with excess GA, and the BDGA–Hsp90α rate of dissociation measured by the time-dependent change in anisotropy under conditions where there is no appreciable BDGA–Hsp90α reassociation. ○ and ● are replicates of the same experiment. The data are fit to a monoexponential decay; average koff = 2.5 × 10−3 min−1, average t1/2 = 4.5 h (n = 2).
The time-dependent BDGA–Hsp90 binding was then examined by measuring BDGA binding to Hsp90 in lysates from cancer cells (SKOV-3) and normal proliferating human umbilical vein endothelial cells (HUV-EC) from culture. Kd(app) values were determined for BDGA–Hsp90 binding as a function of incubation time. It was found that the BDGA–Hsp90 Kd(app) values decrease ≈40-fold over a 24-h incubation time (Fig. 5), with kinetics consistent with those observed by using purified Hsp90 (Table 1), indicative of time-dependent binding. Also, the BDGA Kd(app) values determined at 24 h with HUV-EC lysate (4.3 ± 0.6 nM) and with SKOV-3 lysate (4.7 ± 1.0 nM) are consistent with those determined by using purified Hsp90 (4.6 ± 1.5 nM). Thus, the observation of time-dependent binding of BDGA to purified Hsp90 is consistent with the observed time-dependent binding of the ligand to Hsp90 in cell lysates. Additionally, the affinity and time-dependence of binding were found to be similar between the lysates from cancer and from normal proliferating cells.
Fig. 5.
Measurement of BDGA binding to Hsp90 from cell lysates. BDGA was incubated in the presence of varying amounts of cell lysate for 1–24 h at room temperature, followed by measurement of fluorescence anisotropy. Kd(app) values were determined for BDGA–Hsp90 binding from the titration data of fluorescence anisotropy vs. [cell lysate] by using the integrated rate equation, Eq. 2. (○) Lysate from proliferating human umbilical vein endothelial cells (HUV-EC); (●) lysate from SKOV-3 cells.
Discussion
The benzoquinone ansamycins are an important class of Hsp90 inhibitors that possess potent antitumor activity in preclinical models and may emerge as efficacious therapeutic agents for the treatment of cancer (5). More generally, Hsp90 is an attractive therapeutic antitumor target because inhibition of its chaperone activity results in lower cellular levels of multiple client proteins that are critical for cancer-cell survival (10). Yet, our biochemical understanding of the inhibitory activity of the benzoquinone ansamycins, exemplified by GA, is inconsistent with its cellular antiproliferative and in vivo antitumor activity.
Geldanamycin, and its analogues 17-AAG and 17-DMAG, have been reported by many groups to have inhibitory activity and binding affinity in the range of 0.3–10 μM (8, 11, 16). This moderate potency is in contrast to the low nanomolar antiproliferative activity of the compounds in multiple cell lines in culture that are due to Hsp90 inhibition (11, 12, 17, 18). To effectively develop additional Hsp90-directed compounds as antitumor agents, it is important to understand better how the observed moderate biochemical potency of the current compounds translates into very high cellular potency.
Two important models have been proposed to explain this discrepancy. Kamal et al. (19) have provided biochemical and cellular evidence that the ansamycins bind to and inhibit an Hsp90 multiprotein complex consisting of Hsp90, Hsp70, Hsp40, Hop, and p23 with much higher affinity than to Hsp90 alone. Those authors also found that Hsp90 in tumor cells is present in a multiprotein complex in a much higher fraction than it is in normal cells. Chiosis and colleagues (11) have proposed that the physicochemical properties of the ansamycins results in its intracellular accumulation from cell culture media, leading to highly potent antiproliferative activity. This group has observed that nanomolar concentrations of the inhibitor added to cell media result in micromolar intracellular concentrations. An aspect that is common to both these models is that the benzoquinone ansamycins possess only micromolar affinity to Hsp90 as a single protein.
The studies reported here demonstrate that BDGA (a fluorescent analogue of GA amenable to kinetic binding studies) and GA both possess micromolar affinity to Hsp90 if the protein–ligand incubation time is short (<0.5 h), consistent with previous reports. These Kd values, however, are inaccurate because they are determined under preequilibrium conditions. BDGA binding to purified Hsp90α approaches equilibrium at incubation times >10 h, with a Ki* = 10 nM, a potency more consistent with the cellular antiproliferative activity of this class of compounds. Time-dependent binding is not a characteristic common to all Hsp90 inhibitors, because we found that a recently reported compound (Compound 1: 4-butyl-6-[4-(2-methyl-1,3-thiazol-4-yl)-5-isoxazolyl]-1,3-benzenediol) (25) possesses submicromolar affinity but is not slow-binding (Table 1).
The BDGA–Hsp90 time course with recombinant Hsp90 is consistent with the time-dependence of BDGA binding to Hsp90 in cell lysates. The endogenous Hsp90 in cell lysates may be associated with multiple endogenous cochaperone proteins, but BDGA binds to the endogenous Hsp90 with low nanomolar affinity similar to that observed with purified protein. Thus, the time-dependent binding observed with purified Hsp90 is consistent with that observed when using endogenous Hsp90 from cell lysate.
In contrast to the report of Kamal et al. (19), we find that BDGA binds to Hsp90 alone and Hsp90 in a chaperone complex with the same affinity and time-dependence of binding. Hsp90 was incubated with various combinations of the cochaperones Hsp70, Hsp40, Hop, and p23, and it was found that, under all conditions, BDGA bound with similar characteristics. Specifically, the binding of BDGA to Hsp90 in the presence of Hsp70 and Hsp40 was found to possess the same characteristics as Hsp90 alone (data not shown). The mixture of these three purified proteins was shown to possess ATP-dependent chaperone activity that also depended on the presence of each of the proteins. Hence, these purified proteins can form a functional chaperone complex that binds BDGA with characteristics similar to Hsp90 alone.
Interestingly, it was found that BDGA binds to Hsp90 in lysates from cancer cells with similar affinity to Hsp90 in lysates from normal proliferating cells. This is also in contrast to the findings of Kamal et al. (19), who reported that 17-AAG has much greater affinity for Hsp90 from tumor cells than from normal cells. Two differences exist between the Kamal studies and those presented here. The Kamal group reported IC50 values for 17-AAG, whereas this study characterize affinity of GA and BDGA. All three of these benzoquinone ansamycins are similar in structure and may be expected to have similar binding characteristics, consistent with their similar cellular potencies. Subtle structural differences between these ligands may, however, account for some of the observed differences. Also, the Kamal group measured 17-AAG affinity by its ability to compete with the binding of biotin–GA to Hsp90 in various forms. A complication in interpreting differences in 17-AAG affinity by this method is that a change in affinity of the biotin–GA ligand for Hsp90 would affect measured 17-AAG affinity. It is unclear from the Kamal et al. studies whether the observed affinity differences are due to changes in 17-AAG affinity, biotin–GA affinity, or both.
The results reported here indicate that BDGA and GA are slow, tight-binding inhibitors of the Hsp90 protein and do not possess different affinity for an Hsp90 chaperone complex. As has been observed for many pharmacological agents, the affinity or inhibitory activity of the agent against a single protein target is sufficient to account for cellular on-target activities. Although detailed biochemical studies have demonstrated that the chaperone activity of Hsp90 requires cochaperone proteins (1), the current studies provide evidence that identification of Hsp90 inhibitors as pharmacological agents may be pursued by using Hsp90 as a single protein.
The tight-binding and slow-dissociation characteristics of GA–Hsp90 binding may account for an important pharmacological characteristic of the benzoquinone ansamycins. It has been reported that these agents accumulate intracellularly in cell culture and are retained longer in xenograft tumors than in normal murine tissues upon i.v. administration (26). Unlike most pharmacological targets, Hsp90 is present intracellularly at very high concentrations (≈1–4% of total cellular protein). Inhibitors of this target that are tight binding with a slow dissociation rate, such as the benzoquinone ansamycins, would be expected to accumulate at the intracellular site of the target simply because of the law of mass action. If these binding characteristics of the ligand are responsible for its accumulation intracellularly and in tumors, then Hsp90 inhibitors with different structures and the same binding characteristics should also accumulate. It will be important to characterize the time-dependence of binding of Hsp90 inhibitors of other structural classes, because this biochemical attribute may have significant impact on the pharmacological profile of these compounds in patients.
Materials and Methods
BDGA was synthesized as reported in ref. 27. Source 15Q, Sephadex G-25, Superdex 200, and Superdex 75 were purchased from Amersham Pharmacia Bioscience. Ceramic hydroxyapatite type I was purchased from Bio-Rad. The Smt3 hydrolase (Ulp1) and tobacco etch virus (TEV) proteases were prepared in-house. Bovine gamma globulin (BGG), Steady-Glo Luciferase Assay System, and luciferase were purchased from Panvera (Madison, WI). Ni-NTA agarose, DNA purification, and agarose gel band extraction kits were from Qiagen (Valencia, CA). All other laboratory chemicals and materials were of standard laboratory grade.
Cloning, Expression, and Protein Purification.
The full-length cDNA of human Hsp90 complex subunits (Hsp90β, Hsp70, Hsp40, Hop, and p23) were cloned into the Gateway entry vector pENTR-SD-TOPO. The subunit genes were subcloned into the Gateway bacterial expression vector pDEST14 with an N-terminal His6-Smt3 tag gene (protein sequence cleavable by Ulp1 protease). All subunits were expressed in the E. coli strain BL21(DE3), containing the vector pRR692. The Hsp90β construct was grown to log phase at 30°C and induced with 1 mM isopropyl β-d-thiogalactoside (IPTG) at 18°C and harvested 21 h after induction. Hsp70, Hsp40, Hop, and p23 His6-Smt3 fusions were grown to log phase, induced at 29°C, and harvested 5 h after induction. Hsp90α was expressed as an N-terminal maltose-binding protein (MBP) tag fusion with a tag-subunit TEV cleavage site by using the Gateway entry vector pENTR-D-TOPO. Expression experiments were carried out within the BL21(DE3) strain with pRR692 vector. Cells were grown to log phase at 30°C, induced at 18°C, and harvested after 18 h.
Lysates from E. coli cells expressing His6-Smt3-Hsp90β, His6-Smt3-Hop, His6-Smt3-Hsp70, His6-Smt3-Hsp40, His6-Smt3-P23, and His6-MBP-TEV-Hsp90α were centrifuged and the overexpressed recombinant proteins captured on Ni-NTA agarose and then cleaved on-column with either Ulp1 protease or TEV protease. Recombinant proteins were recovered in the flow-through fractions, whereas N-terminal tags His6-Smt3 or His6-MBP-Tev remained on the column. Proteins in flow-through fractions were purified by Source15Q chromatography, followed by Superdex 200 or Superdex 75 size-exclusion chromatography. The purity of all of the isolated proteins, estimated by visual inspection of the Coomassie-stained gels, was 80–95% (Fig. 1). Mass determination by liquid chromatography MS and N-terminal sequencing of all proteins matched predicted molecular mass and sequence from DNA sequence. As expected, the expression of N-terminal-tagged protein, followed by cleavage with either Ulp1 protease or TEV protease, resulted in isolated protein with no additional amino acids on the native sequence. The one exception was Hsp90α, which contains an additional Gly on the N terminus. The final yield for all proteins was ≈0.5–1.5 mg/g of E. coli cell paste. Additional detail regarding cloning, expression, and protein purification is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Chaperone Assay.
The chaperone assay was performed by following the protocol of Walerych et al. (20), with modifications described below. Hsp90α, 2.5 μM and luciferase, 0.25 μM in denaturation buffer (25 mM Tris, pH 7.5, 8 mM MgSO4, 0.01% BGG, and 10% glycerol) were incubated at 50°C for 8 min. It was determined that this incubation time was sufficient to fully inactivate luciferase. Five-microliter samples of the denatured mix was cooled for 3 min at room temperature and then diluted into 25 μl of renaturation buffer (25 mM Tris, pH 7.5, 8 mM MgSO4, 0.01% BGG, 10% glycerol, 0.5 mM ATP, 2 mM DTT, 5 mM KCl, 2 μM Hsp70, and 1 μM Hsp40). The renaturation reaction was incubated at room temperature for 150 min, followed by dilution of 5 μl of the renatured sample into 45 μl of luciferin reagent (from Steady-Glo Luciferase Assay System), and mixed on a shaker in the dark for 4 min before reading the luminescence signal on an Envision plate reader (PerkinElmer Life Sciences). Controls were performed in an identical manner in the absence of ATP, Hsp70, or Hsp40 in the renaturation step.
Binding of BDGA to Purified Hsp90 and Hsp90 Complex Measured by Fluorescence Anisotropy.
Hsp90 was titrated into 10 nM BDGA in assay buffer [20 mM Hepes, pH 7.5, 0.1 mg/ml BGG, 2 mM DTT, 50 mM KCl, 5 mM MgCl2, 20 mM Na2MoO4, and 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)], and fluorescence anisotropy (r) was measured at varying BDGA–Hsp90 incubation times (up to 48 h) at ambient temperature. Fluorescence was measured in an Envision plate reader (PerkinElmer Life Sciences), excitation = 485 nm, emission = 535 nm. The apparent dissociation constant for BDGA, Kd(app), was determined by fitting the anisotropy data to the integrated rate equation, (see Eq. 2), that assumes a single Hsp90-binding site for BDGA, as described in ref. 28.
 |
 |
 |
In Eq. 2, r is the anisotropy at a given concentration of BDGA and Hsp90, rb is the anisotropy of the bound BDGA, and ro is the anisotropy of the unbound BDGA. These calculated dissociation constants, and those for competing ligands described below, are apparent dissociation constants (Kd(app)), because they are not necessarily determined under protein–ligand equilibrium conditions.
For fluorescence anisotropy competition studies, the competing ligand (2–5,000 nM) was incubated with 50 nM Hsp90 in assay buffer for varying incubation times (0.1–24 h). BDGA (10 nM) was then added (total volume of 40 μl), the sample was incubated for 30 min, and fluorescence anisotropy was measured. A control was used for each assay, consisting of BDGA in the absence of Hsp90. The apparent dissociation constant for the competing ligand (CL), Kd(app)CL, was obtained by fitting the data to the following model and using the following equations:
From the law of mass action and the binding isotherm, Eq. 5 was employed with terms defined in Eqs. 6 and 7 (28).
Kd(app)CL is obtained from the fitting with [BDGA]t = 10 nM, [Hsp90]t = 25 nM dimeric protein, and Kd(app) = 162 nM for Hsp90α after 30 min (determined in independent experiments).
Determination of Kinetic Constants for Time-Dependent Binding.
The change in fluorescence anisotropy of 10 nM BDGA was measured in the presence of Hsp90 (12.5–900 nM) over 24 h. The kobs was calculated by fitting the progress curves to a pseudo-first-order rate equation:
The kobs value from each progress curve was then graphed against [Hsp90] and fit to Eq. 1. The kinetic constants k5 and k6 are defined according to a two-step BDGA–Hsp90-binding model, described in Results. The values calculated for k6 were confirmed by a dilution experiment in which 600 nM Hsp90 was preincubated with 500 nM BDGA for 24 h, followed by diluting 100-fold in assay buffer containing 500 nM GA. The curves were then fit to a pseudo-first-order rate equation, Eq. 8, where kobs = koff.
Binding of BDGA to Hsp90 in Cell Extracts Measured by Fluorescence Anisotropy.
SKOV-3 and human umbilical vein endothelial cells (HUV-EC) were grown to 70% confluence (proliferating conditions). Cells were trypsinized, washed, and resuspended in lysis buffer [20 mM Hepes, pH 7.4, 50 mM KCl, 5 mM MgCl2, 0.01% Nonidet P-40, 2 mM DTT, 0.1 mg/ml BSA, protease inhibitors (EDTA free), and phosphatase inhibitors (cocktails I and II from Calbiochem)]. Pelleted cells were frozen in lysis buffer at −80°C overnight. The pellets in lysis buffer were thawed, centrifuged for 15 min at 14,000 × g and 4°C, and the supernatant was collected.
The supernatant was added to assay buffer (20 mM Hepes, pH 7.4, 7.0 nM BDGA, 0.1 mg/ml BSA, 50 mM KCl, 5 mM MgCl2, 0.01% Nonidet P-40, and 2 mM DTT). Fluorescence anisotropy was measured on an Analyst plate reader (Molecular Devices), exc. = 485 nm, em. = 535 nm. The concentration of Hsp90 in cell lysate was determined by incubating BDGA in titrating concentrations of lysate for 24 h under conditions where [BDGA] > Kd(app) for Hsp90–BDGA binding. Under these conditions, the EC50 for the titration is equivalent (within 2-fold) to [Hsp90]/2. It was subsequently confirmed that the [BDGA] used in this determination was greater than the Kd(app) after 24-h incubation. Kd(app) values for BDGA binding to Hsp90 in lysates was determined from the fluorescence anisotropy data as a function of [Hsp90] and fit to the integrated rate equation, Eq. 2.
Supplementary Material
Acknowledgments
We thank Kevin Duffy and colleagues for synthesis, isolation, and confirmation of structure of the compounds 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-geldanamycin (BDGA), and 4-butyl-6-[4-(2-methyl-1,3-thiazol-4-yl)-5-isoxazolyl]-1,3-benzenediol.
Abbreviations
- BDGA
4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-geldanamycin
- CL
competing ligand
- GA
geldanamycin
- Hsp
heat shock protein
- TEV
tobacco etch virus.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Pratt W. B. Proc. Soc. Exp. Biol. Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 2.Pearl L. H., Prodromou C. Curr. Opin. Struct. Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 3.Young J. C., Hartl F. U. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosano H., Stensgard B., Charlesworth M. C., McMahon N., Toft D. J. Biol. Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs J. S. Expert. Opin. Investig. Drugs. 2005;14:569–589. doi: 10.1517/13543784.14.6.569. [DOI] [PubMed] [Google Scholar]
- 6.Kamal A., Boehm M. F., Burrows F. J. Trends Mol. Med. 2004;10:283–290. doi: 10.1016/j.molmed.2004.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goetz M. P., Toft D., Reid J., Ames M., Stensgard B., Safgren S., Adjei A. A., Sloan J., Atherton P., Vasile V., et al. J. Clin. Oncol. 2005;23:1078–1087. doi: 10.1200/JCO.2005.09.119. [DOI] [PubMed] [Google Scholar]
- 8.Roe S. M., Prodromou C., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. J. Med. Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 9.Workman P. Mol. Cancer Ther. 2003;2:131–138. [PubMed] [Google Scholar]
- 10.Neckers L. Trends Mol. Med. 2002;8:S55–S61. doi: 10.1016/s1471-4914(02)02316-x. [DOI] [PubMed] [Google Scholar]
- 11.Panaretou B., Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte T. W., Neckers L. M. Cancer Chemother. Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 13.Grem J. L., Morrison G., Guo X. D., Agnew E., Takimoto C. H., Thomas R., Szabo E., Grochow L., Grollman F., Hamilton J. M., et al. J. Clin. Oncol. 2005;23:1885–1893. doi: 10.1200/JCO.2005.12.085. [DOI] [PubMed] [Google Scholar]
- 14.Thomas X., Campos L., Le Q. H., Guyotat D. Hematology. 2005;10:225–235. doi: 10.1080/10245330500093120. [DOI] [PubMed] [Google Scholar]
- 15.Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 16.Chiosis G., Rosen N., Sepp-Lorenzino L. Bioorg. Med. Chem. Lett. 2001;11:909–913. doi: 10.1016/s0960-894x(01)00099-3. [DOI] [PubMed] [Google Scholar]
- 17.Chiosis G., Huezo H., Rosen N., Mimnaugh E., Whitesell L., Neckers L. Mol. Cancer Ther. 2003;2:123–129. [PubMed] [Google Scholar]
- 18.Neckers L. Clin. Cancer Res. 2002;8:962–966. [PubMed] [Google Scholar]
- 19.Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M. F., Fritz L. C., Burrows F. J. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 20.Walerych D., Kudla G., Gutkowska M., Wawrzynow B., Muller L., King F. W., Helwak A., Boros J., Zylicz A., Zylicz M. J. Biol. Chem. 2004;279:48836–48845. doi: 10.1074/jbc.M407601200. [DOI] [PubMed] [Google Scholar]
- 21.Pratt W. B., Toft D. O. Exp. Biol. Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 22.McLaughlin S. H., Smith H. W., Jackson S. E. J. Mol. Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 23.Kim J., Felts S., Llauger L., He H., Huezo H., Rosen N., Chiosis G. J. Biomol. Screen. 2004;9:375–381. doi: 10.1177/1087057104265995. [DOI] [PubMed] [Google Scholar]
- 24.Copeland R. A. Evaluation of Enzyme Inhibitors in Drug Discovery. A Guide for Medicinal Chemists and Pharmacologists. New York: Wiley; 2005. pp. 141–149. [PubMed] [Google Scholar]
- 25.Drysdale M. J., Mock B. W., Finch H., Webb P., McDonald E., James K. E., Cheung K. M., Matthews & Lloyd T. P. Isoxazole Compounds as Inhibitors of Heat Shock Proteins. 2004:1–180. (Eur. Pat. Off. No. WO 2004/072051 A1)
- 26.Eiseman J. L., Lan J., Lagattuta T. F., Hamburger D. R., Joseph E., Covey J. M., Egorin M. J. Cancer Chemother. Pharmacol. 2005;55:21–32. doi: 10.1007/s00280-004-0865-3. [DOI] [PubMed] [Google Scholar]
- 27.Llauger-Bufi L., Felts S. J., Huezo H., Rosen N., Chiosis G. Bioorg. Med. Chem. Lett. 2003;13:3975–3978. doi: 10.1016/j.bmcl.2003.08.065. [DOI] [PubMed] [Google Scholar]
- 28.Lai Z., Auger K. R., Manubay C. M., Copeland R. A. Arch. Biochem. Biophys. 2000;381:278–284. doi: 10.1006/abbi.2000.1998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.