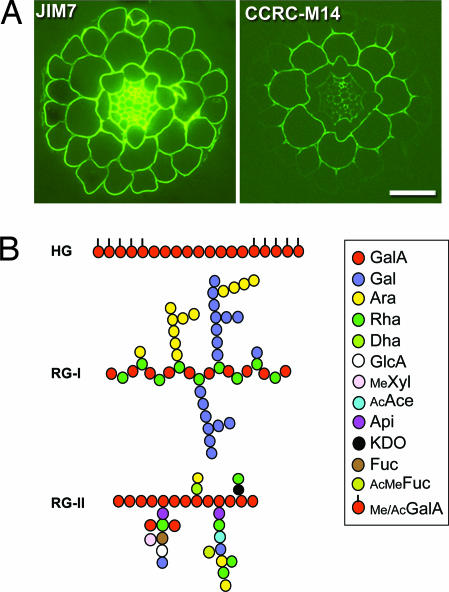
The polysaccharide-rich plant cell wall surrounding the protoplast is a complex, diverse, and dynamic entity of fundamental importance in plant growth and development (1). Walls are also of immense economic value, contributing to many agro-industrial processes, and are the major renewable energy resource (biomass) on our planet. The description of the biosynthetic processes involved in the assembly of the noncellulosic polysaccharides of the wall has, until the 21st century, been slow to unfold. This is especially true of the triad of pectic polysaccharides, which are widely distributed in primary walls (Fig. 1A; ≈10–35%) throughout the plant kingdom. The most abundant is the pectic homogalacturonan (HG), ≈70% of pectin, a homopolymer of (1–4)-α-d-galacturonic acid (GalA) residues that may be methylesterified and acetylated (2). Rhamnogalacturonan I (RG-I), ≈35% of pectin, belongs to a family of polysaccharides with a repeating backbone of (1–2)-α-l-Rha-(1–4)-α-d-GalA, with Rha residues further substituted with other oligo/polysaccharides. RG-II, ≈10% of pectin, has an HG backbone substituted with several structurally different oligosaccharide side chains (2) (Fig. 1B). This structural complexity imparts diverse physical and biochemical properties on pectins that are associated with important biological and industrial functions (2, 3). Thus, there are major efforts devoted to manipulating the quality and quantity of pectin and other wall polysaccharides through genetic manipulation and conventional breeding. This process would be greatly accelerated if we understood the mechanisms and control of the biosynthetic steps during their assembly and deposition into the wall, processes whose elucidation has been hampered by difficulties in identifying the biosynthetic genes and by the pleiotropic effects of many wall mutants. More than 50 glycosyltransferases (GTs) are predicted to be required for pectin synthesis (2), but until now, genes for only two putative pectin biosynthetic GTs, QUA1 (4) and NpGUT1 (5), have been identified; however, the enzymatic function of their encoded proteins has yet to be established. Thus, the work of Sterling et al. (6) in a recent issue of PNAS represents a significant advance, because it is the first functional identification of an Arabidopsis pectin homogalacturonan galacturonosyltransferase (GAUT1) using biochemical and functional genomic approaches.
Fig. 1.
Distribution and structure of pectin in plants. (A) Immunofluorescence micrographs showing the distribution of pectin epitopes in roots of Arabidopsis using monoclonal antibodies. Transverse sections of 5-day-old seedlings were taken ≈8 mm from the root apex. JIM7 and CCRC-M14 bind to HG and to undefined RG I epitopes, respectively (refs. 2–3, and A. Swennes and M. G. Hahn, personal communication). (Scale bar, 25 μm.) Micrographs courtesy of Glenn Freshour and Michael Hahn (University of Georgia, Athens). (B) Schematic representation of the three pectic polysaccharides (HG, RG-I, and RG-II). (Modified with permission from ref. 3.)
Because purification of GalAT to homogeneity proved elusive, Sterling et al. (6) took advantage of the rich genomic and bioinformatic resources available in Arabidopsis to identify two GT-like proteins present in partially purified fractions enriched for HG:GalAT activity. Both GTs encoded predicted Golgi-located type II membrane proteins. Expression of truncated constructs of these genes in human embryonic kidney 293 cells identified GAUT1, which was able to transfer GalA from UDP-GalA onto HG acceptors. Furthermore, anti-GAUT1 antibodies immunoprecipitated HG:GalAT activity from a partially purified protein fraction of Arabidopsis. GAUT1 belongs to a family of 25 genes in Arabidopsis that consist of 15 GAUT and 10 GAUT-like (GATL) genes (6, 7) clustered in CAZy GT8 family (http://afmb.cnrs-mrs.fr/CAZY/index.html) with homologues in other dicots, grasses (rice), and the moss Physcomitrella. Not only is this discovery important for understanding pectin biosynthesis, but it will also require us to reevaluate the types of mechanisms by which other noncellulosic polysaccharides are assembled in higher plants.
The key enzymes in wall biogenesis are the polysaccharide/glycan synthases and GTs (both classified within the GT class of carbohydrate modifying enzymes; http://afmb.cnrs-mrs.fr/CAZY/index.html) that catalyze formation of the bonds between adjacent monosaccharides from activated nucleotide-monosaccharide donors. The Arabidopsis genome contains 414 different GT genes representing 34 of the 60 GT families (http://afmb.cnrs-mrs.fr/CAZY/index.html). Many glycan synthases are type III integral membrane proteins with multiple transmembrane spanning domains, whereas the GTs are type II membrane proteins with a single transmembrane domain. The simple (unbranched) polysaccharides cellulose and callose are synthesized by glycan synthases at the plasma membrane (8). These enzymes repetitively transfer the same type of monosaccharide residue to the nonreducing end of a growing glycan chain. In contrast, complex (branched) noncellulosic polysaccharides are synthesized within the endoplasmic reticulum/Golgi complex (9, 10), using glycan synthases to form the backbone and then a number of GTs to form the side chains by each transferring single monosaccharide residues, or so it was thought. To date, all biosynthetic enzymes for the structural plant polysaccharide backbones have been type III integral membrane proteins belonging to either the CAZy GT2 family [e.g., cellulose (CESAs) (8), heteromannans (CSLAs) (11), and (1,3,1,4)-β-d-glucans (12) (and also includes fungal chitin, bacterial curdlan, and mammalian and bacterial hyaluronans)] or the GT48 family [e.g., callose (GSLs) (9)], whereas the type II GTs have been restricted to creating the branches of the matrix polysaccharides (9, 10). Now, with the identification by Sterling et al. (6) of GAUT1, the first demonstration in plants that a type II GT can elaborate a structural polysaccharide backbone, a new paradigm is provided. However, whether single GAUT proteins are capable of repetitive action to form a backbone chain or whether some form of transferase complex is required remains unresolved. Two lines of evidence might suggest the involvement of an enzyme complex. (i) Apart from GAUT1, the only other predicted GT identified by Sterling et al. (6) in the partially purified Arabidopsis GalAT active fraction was GAUT7, another member of the GAUT1-related gene family. Perhaps GAUT 7 is part of a pectin biosynthetic complex? (ii) In vitro studies have established that GalAT transfers GalA from UDP-GalA onto endogenous acceptors to produce products in the mass range of 100 to ≥500 kDa (13, 14). These detergent-solubilized GalAT fractions preferentially transfer to the nonreducing end of small exogenous (degree of polymerization > 9) oligogalacturonide (OGAs) acceptors (15–18). High-molecular-weight pectin is less favored as an acceptor, suggesting a requirement for additional factors, protein complex integrity, and/or membrane organization for sustained synthesis. It is well established that GT complexes are involved in other biosynthetic systems, e.g., for coordinated synthesis of the hypermannosylated N-linked polysaccharides on yeast glycoproteins (19) and for the assembly of the repeating disaccharide units of glycosaminoglycans (GAGs; refs. 20 and 21). Similarly, the correct assembly of cellulose-synthesizing rosette complexes at the plasma membrane requires groups of three coordinately expressed CESA proteins and probably other non-CESA proteins for cellulose microfibril synthesis (8, 22, 23).
Many other questions regarding pectin biosynthesis remain. For example, what are the roles of the other members of the GAUT1-related gene family? Significantly, two genes previously identified as putative pectin or wall biosynthetic genes, QUA1 (6) and PARVUS/GLZ1 (7, 24, 25), are also members of the GAUT1-related gene family, i.e., GAUT8 and GATL1, respectively. QUA1 has been proposed to be either a GalAT or an XylT, but biochemical proof of QUA1 activity has been elusive (25). The identification of GAUT1 as an HG:GalAT (6) adds weight to the possibility that QUA1/GAUT8 is a GalAT, although confirmation awaits verification of enzymatic activity. The identification of GATL1/PARVUS/GLZ1 in the GAUT1-related gene family is also interesting because some of the parvus/glz1 mutant phenotypes (dwarfism) are consistent with a defect in pectin synthesis. Because GalA levels in the parvus/glz1 mutant have not been reported, the role, if any, of GATL1/PARVUS/GLZ1 in pectin synthesis remains to be established. Questions remain with regard to the assembly of the various pectin backbones with at least four GalATs proposed to be required (2): one for HG, a second for RG-I, and at least two for RG-II side chains. However, it is possible that the RGI backbone, the only plant polysaccharide with a repeating disaccharide backbone (Fig. 1B), is assembled by a different mechanism. In other organisms, both polysaccharide synthases (for hyaluronans; ref. 26) and GTs (for GAGs; refs. 20 and 21) are able to elaborate such backbones either by a single “dual-action” enzyme (synthase) or by a pair of “single-action” enzymes (transferases).
The identification of GAUT1 and the GAUT1-related gene family provides the molecular tools to begin to unravel pectin synthesis. Their role in the biosynthesis of pectin backbones can now be tested, and the coordinate synthesis of the branches by GTs from other CAZy families can be explored. Furthermore, phenotypic analyses of mutants of the GAUT1-related gene family should also help define the roles of these proteins in pectin synthesis and the role of pectin in the plant. Then, of course, there are the nonglycosyl substituents (methylesters and acetyl groups): are they added postpolymerization or during backbone synthesis? Again, we can be informed by the processes of sulfation of GAG backbones in mammals and bacteria (27). What controls chain initiation and elongation? And so it goes on.
Acknowledgments
I thank Prof. B. A. Stone (LaTrobe University, Melbourne, Australia) and Dr. M. A. Doblin (University of Melbourne, Melbourne, Australia) for providing valuable input.
Conflict of interest statement: No conflicts declared.
See companion article on page 5236 in issue 13 of volume 103.
References
- 1.Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., et al. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 2.Ridley B. L., O’Neill M. A., Mohnen D. Phytochemistry. 2001;57:929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 3.Willats W. G. T., McCartney L., Mackie W., Knox J. P. Plant Mol. Biol. 2001;47:9–27. [PubMed] [Google Scholar]
- 4.Bouton S., Leboeuf E., Mouille G., Leydecker M. T., Talbotec J., Granier F., Lahaye M., Höfte H., Truong H.-N. Plant Cell. 2002;14:2577–2590. doi: 10.1105/tpc.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwai H., Masaoka N., Ishii T., Satoh S. Proc. Natl. Acad. Sci. USA. 2002;99:16319–16324. doi: 10.1073/pnas.252530499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sterling J. D., Atmodjo M. A., Inwood S. E., Kumar Kolli V. S., Quigley H. F., Hahn M. G., Mohnen D. Proc. Natl. Acad. Sci. USA. 2006;103:5236–5241. doi: 10.1073/pnas.0600120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lao N. T., Long D., Kiang S., Coupland G., Shoue D. A., Carpita N. C., Kavanagh T. A. Plant Mol. Biol. 2003;53:687–701. doi: 10.1023/B:PLAN.0000019074.60542.6c. [DOI] [PubMed] [Google Scholar]
- 8.Doblin M., Kurek I., Jacob-Wilk D., Delmer D. Plant Cell Physiol. 2002;43:1407–1420. doi: 10.1093/pcp/pcf164. [DOI] [PubMed] [Google Scholar]
- 9.Doblin M., Vergara C., Read S., Newbigin E., Bacic A. In: The Plant Cell Wall. Rose J., editor. Oxford: Blackwell; 2003. pp. 183–222. [Google Scholar]
- 10.Keegstra K., Raikhel N. Curr. Opin. Plant Biol. 2001;4:219–224. doi: 10.1016/s1369-5266(00)00164-3. [DOI] [PubMed] [Google Scholar]
- 11.Dhugga K., Barreiro R., Whitten B., Stecca K., Hazebroek J., Randhawa G., Dolan M., Kinney A., Tomes D., Nichols S., Anderson P. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- 12.Burton R. A., Wilson S. M., Hrmova M., Harvey A. J., Shirley N. J., Medhurst A., Stone B. A., Newbigin E. J., Bacic A., Fincher G. B. Science. 2006 doi: 10.1126/science.1122975. in press. [DOI] [PubMed] [Google Scholar]
- 13.Sterling J., Quigley H. F., Orellana A., Mohnen D. Plant Physiol. 2001;127:360–371. doi: 10.1104/pp.127.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doong R. L., Liljebjelke K., Fralish G., Kumar A., Mohnen D. Plant Physiol. 1995;109:141–152. doi: 10.1104/pp.109.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doong R. L., Mohnen D. Plant J. 1998;13:363–374. [Google Scholar]
- 16.Akita K., Ishimizu T., Tsukamoto T., Ando T., Hase S. Plant Physiol. 2002;130:374–379. doi: 10.1104/pp.005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii T. Plant Cell Physiol. 2002;43:1386–1389. doi: 10.1093/pcp/pcf150. [DOI] [PubMed] [Google Scholar]
- 18.Scheller H. V., Doong R. L., Ridley B. L., Mohnen D. Planta. 1999;207:512–517. [Google Scholar]
- 19.Lussier M., Sdicu A.-M., Bussey H. Biochim. Biophys. Acta. 1999;1426:323–334. doi: 10.1016/s0304-4165(98)00133-0. [DOI] [PubMed] [Google Scholar]
- 20.Esko J. D., Selleck S. B. Annu. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 21.DeAngelis P. L. Glycobiology. 2002;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- 22.Taylor N., Howells R., Huttly A., Vickers K., Turner S. Proc. Natl. Acad. Sci. USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read S., Bacic A. Science. 2002;295:59–60. doi: 10.1126/science.1068155. [DOI] [PubMed] [Google Scholar]
- 24.Shao M., Zheng H., Hu Y., Liu D., Jang J.-C., Ma H., Huang H. Plant Cell Physiol. 2004;45:1453–1460. doi: 10.1093/pcp/pch168. [DOI] [PubMed] [Google Scholar]
- 25.Orfila C., Sørensen S. O., Harholt J., Geshi N., Crombie H., Truong H.-N., Reid J. S. G., Knox J. P., Scheller H. V. Planta. 2005;222:613–622. doi: 10.1007/s00425-005-0008-z. [DOI] [PubMed] [Google Scholar]
- 26.Weigel P. H. IUBMB Life. 2002;54:201–211. doi: 10.1080/15216540214931. [DOI] [PubMed] [Google Scholar]
- 27.Kusche-Gullberg M., Kjellén L. Curr. Opin. Struct. Biol. 2003;13:605–611. doi: 10.1016/j.sbi.2003.08.002. [DOI] [PubMed] [Google Scholar]