Abstract
In Anopheles gambiae, the vector for the most deadly malaria parasite Plasmodium falciparum, xanthurenic acid (XA) plays a key role in parasite gametogenesis and fertility. In mosquitoes, XA is produced by transamination of 3-hydroxykynurenine (3-HK), a reaction that represents the main route to prevent the accumulation of the potentially toxic 3-HK excess. Interfering with XA metabolism in A. gambiae therefore appears an attractive avenue for the development of malaria transmission-blocking drugs and insecticides. We have determined the crystal structure of A. gambiae 3-HK transaminase in its pyridoxal 5′-phosphate form and in complex with a newly synthesized competitive enzyme inhibitor. Structural inspection of the enzyme active site reveals the key molecular determinants for ligand recognition and catalysis. Major contributions toward inhibitor binding are provided by a salt bridge between the inhibitor carboxylate and Arg-356 and by a remarkable hydrogen bond network involving the anthranilic moiety of the inhibitor and backbone atoms of residues Gly-25 and Asn-44. This study may be useful for the structure-based design of specific enzyme inhibitors of potential interest as antimalarial agents.
Keywords: kynurenine pathway, xanthurenic acid
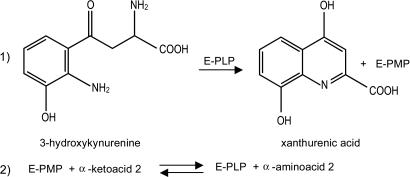
A potentially toxic metabolite of the kynurenine pathway, 3-hydroxykynurenine (3-HK), represents a central intermediate of tryptophan degradation in most living organisms (1, 2). Once formed through the enzymatic hydroxylation at the C3 position of its immediate precursor, l-kynurenine (L-KYN) (3–5), 3-HK can undergo spontaneous oxidation, which in turn can result in free radical generation and apoptotic cell death (6–8). The abnormal levels of 3-HK that have been recorded in the brains of patients with AIDS–dementia complex (9) and Huntington’s disease (10) support a role for this molecule in the etiology and progression of fatal neurodegenerative diseases in humans (11, 12). 3-HK-triggered damage is not restricted to mammalian cells. Indeed, several invertebrate species, which depend on the kynurenine pathway for dietary tryptophan oxidation, are highly sensitive to perturbations in the physiological balance of its intermediate metabolites (13, 14). In particular, the exogenous administration of 3-HK in adult insects profoundly affects locomotive activity and can culminate in irreversible paralysis and death (15, 16). The biochemical description of the kynurenine pathway from a number of vertebrates revealed that 3-HK can undergo two alternative fates: either (i) it is hydrolyzed by a specific kynureninase to alanine and 3-hydroxyanthranilic acid, feeding the de novo synthesis of the essential cofactor NAD (17), or (ii) it can be directly converted into the more chemically stable compound xanthurenic acid (XA) by irreversible transamination. In rodents and humans, this latter function is provided by the same kynurenine aminotransferase (KAT) isozymes that are responsible for the pyridoxal 5′-phosphate (PLP)-dependent transformation of L-KYN into the neuroprotective and anticonvulsant NMDA receptor antagonist kynurenic acid (KA) (18–20). In Anopheles gambiae, the vector of the most deadly malaria parasite Plasmodium falciparum, 3-HK hydrolytic activity has never been documented, even though 3-HK-dependent XA synthesis might represent the only means to prevent the accumulation of this toxic molecule. We recently identified and characterized an A. gambiae 3-HK transaminase (Ag-HKT) (21), the bona fide anopheline ortholog of the previously isolated Aedes aegypti 3-HK transaminase (22, 23). We showed that the recombinant form of Ag-HKT efficiently catalyzes the PLP-dependent transamination of both 3-hydroxykynurenine (3-HK) and kynurenine to XA and KA, respectively, preferentially using the α-ketoacid glyoxylate as the amino group acceptor (Scheme 1). The determination of the kinetic parameters featuring the Ag-HKT-dependent 3-HK transamination revealed that the enzyme is highly active under physiological conditions, thus suggesting a key role of the enzyme in XA biosynthesis in vivo; moreover, the Ag-hkt gene expression profile throughout the lifespan of A. gambiae (21) is in agreement with the proposed 3-HK detoxification activity of the enzyme, confirming earlier observations made in A. aegypti (24). Indeed, the Ag-hkt-specific transcript is abundant in embryos, larvae, and most adult tissues, consistent with the protective role of the enzyme in preventing the 3-HK-triggered oxidative stress response; on the other hand, the observed down-regulation of the Ag-hkt gene transcription in pupae may reflect the physiological 3-HK requirement during compound eye development in the mosquito (25, 26) and the potential role of the molecule in insect tissue remodeling during metamorphosis (27). Analyzed in the context of Plasmodium-infected mosquitoes, XA metabolism represents a metabolic crossroad between the biology of the two species promoting malaria spread throughout human populations. After the initial in vitro studies that disclosed the essential role of XA in Plasmodium gametogenesis and fertility (28–30), the molecular dissection of the signaling cascade triggered by XA and resulting in the sexual maturation of the parasite inside the arthropod host has recently been undertaken (31). In line with these observations, two independent high-throughput microarray screenings identified the 3-HK transaminase coding sequence among a group of genes that are up-regulated upon ingestion of an infected blood meal in distinct Anopheles species (32, 33). Taking also into account that optimal P. falciparum sexual maturation requires the presence of XA in the low micromolar range (30), a value well above the average concentration of the molecule in human plasma (34), the 3-HK transamination in infected mosquitoes could be essential in providing enough XA for sustaining parasite gametogenesis. Therefore, interfering with endogenous XA synthesis appears to be an appealing avenue for the development of innovative malaria transmission-blocking drugs. We report here the crystal structures of the Ag-3-HKT in its PLP form (Ag-HKT:PLP) and in complex with the competitive inhibitor 4-(2-aminophenyl)-4-oxobutyric acid (Ag-HKT:INI) at 2.4- and 2.7-Å resolution, respectively. Our study reports the structural analysis of an enzyme of the tryptophan degradation pathway in A. gambiae and the fourth protein structure of the malaria vector determined so far. By unraveling the molecular determinants responsible for substrate specificity and catalysis, our data may assist the rational design of selective inhibitors of potential interest both as basic tools for investigating the role of the Ag-HKT in vivo and as lead compounds to be developed into innovative antimalarial drugs.
Scheme 1.
Scheme of the two-step reaction catalyzed by the 3-HK transaminase. The first half of the reaction is irreversible.
Results and Discussion
Overall Quality of the Model.
The 3D structure of the recombinant Ag-HKT in its PLP form has been solved by means of the molecular replacement technique, at a resolution of 2.4 Å. Two tight homodimers are present in the asymmetric unit. Three hundred eighty-seven residues of 396, one PLP and one glycerol molecule, were modeled in each of the four subunits, together with a total of 380 solvent molecules. Residues belonging to the four different subunits are numbered A2-A389, B2-B389, C2-C389, and D2-D389, respectively. The A and D subunits build up one of the dimers and the B and C subunits, the other. The final model of the Ag-HKT:INI complex consists of 386 residues (3-389), one PLP and one inhibitor molecule in each of the four subunits, and of a total of 142 solvent molecules. In both structures, residues belonging to the two opposite subunits building a functional homodimer are indicated as 2-389 and 2*-389*. The stereochemistry of both models has been assessed with the program procheck (35). In both forms, 92% of the residues were in the most-favored regions of the Ramachandran plot, and Pro-24 was recognized as the cis conformer in all of the subunits.
Overall Structure.
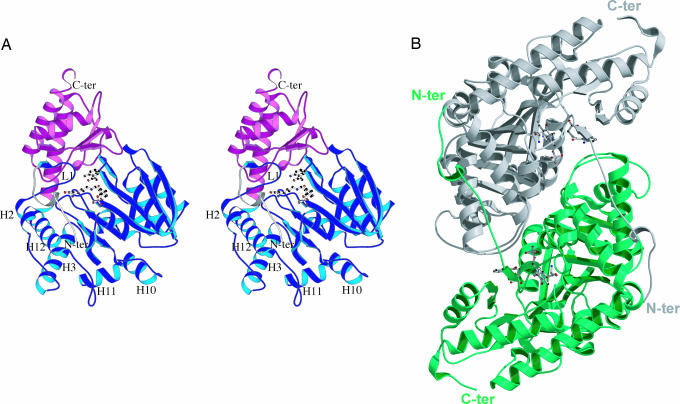
The observed Ag-HKT molecular architecture (Fig. 1) consists of a tight homodimer, with subunits displaying the typical fold of members of the PLP-dependent aminotransferases (36, 37). In each monomer, three distinct regions could be defined: an extended N-terminal arm, a large N-terminal domain, and a small C-terminal domain. The Ag-HKT N-terminal large domain is preceded by a random coiled stretch (residues 3–7 and 11–18), interrupted by an α-helical turn (residues 8–10) and ending in a short β-strand (residues 19–21). This arm runs parallel to the surface of the large domain of the opposite monomer, between the α-helices H3 and H12, where it comes in close contact with the solvent-exposed side chains of several residues, thus contributing to the stability of the functional dimer (Fig. 1B). The N-terminal large domain (residues 22–380), displays a β-α-β architecture organized around an inner core made by a seven-stranded, mainly parallel β-sheet, sandwiched between five conserved α-helices; five additional α-helices and their connecting loops complete the external surface of the large domain, which partially stacks against the corresponding domain of the opposite subunit. The smaller C-terminal domain shows a conserved two-layer α/β fold that consists of five solvent exposed α-helices overlaying onto a four-stranded β-sheet (Fig. 1A). In the Ag-HKT homodimer (Fig. 1B), the two subunits are related by a dyad axis, and the two active sites, one for each monomer, are located at the domain interface in each subunit and at the subunit interface in the dimer. The loops connecting helices H2-H3, H10-H11, and H11-H12 of each monomer (Fig. 1A) form a flat surface that mainly participates in dimer stabilization, stacking against the opposite monomer large domain around the twofold axis. Another contribution toward the stability of the Ag-HKT dimer is provided by the regions encompassing residues 9–19 in the N-terminal arm of one subunit and the helix H3 of the opposite one, where a number of subunit–subunit interactions are observed, including a salt bridge between Lys-19 and Glu-48* (distance of 2.65 Å) and a strong hydrogen bond between Ile-15 main chain oxygen and the Arg-51* side chain (distance of 2.90 Å).
Fig. 1.
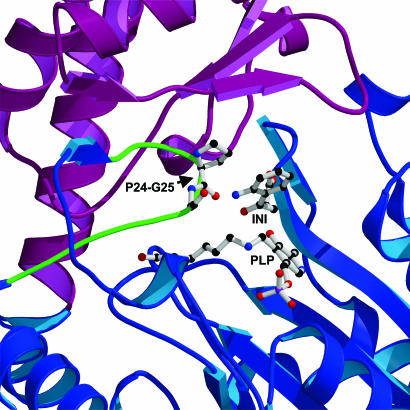
Ag-HKT molecular architecture. (A) Stereo ribbon representation of the Ag-HKT subunit. The N-terminal extension, the large domain, and the small domain are colored in white, blue, and magenta, respectively. The loop protruding into the active site and carrying the cisPro-24-Gly-25 motif is labeled L1. The deep cleft hosting the enzyme active site is placed at the domain interface, where the PLP cofactor and the inhibitor molecules are shown in ball-and-stick representation. The N and the C termini are indicated, as are the helices connected by the loops involved in dimer stabilization. (B) Ribbon representation of the functional Ag-HKT homodimer seen along the dyad axis. The two subunits are colored white and green; PLP and inhibitor molecules are drawn in ball-and-stick representation. The N and the C termini are indicated. The figure was generated with the program molscript (50).
The Ag-HKT Active Site.
Each active site in the functional Ag-HKT homodimer contains one PLP molecule covalently bound by a Schiff-base linkage to the catalytic Lys-205 and is hosted in a deep cleft built up mainly by residues belonging to the large domains of both subunits (Fig. 1). Indeed, the bottom of the PLP-binding pocket is defined by two extended loops encompassing residues 181–190 and 202–211 located at the β-sheet inner edge of the large domain of the corresponding subunit, whereas the lateral walls of the cofactor-binding pocket are formed by distinct arrays of residues contributed by the large domains of both subunits. In particular, the highly conserved residues Ser-154, Asp-179, and Arg-356 reside on the pyridine ring side of the cofactor, whereas the opposite wall consists of residues 77–79 and Gln-204 of one monomer and of residues Tyr-256* and Thr-259* of the other subunit. Moreover, an interesting feature observed in the active site is represented by a loop made by residues 23–27 that protrudes into the active site and points toward the catalytically active PLP C4′ atom. Such a structural element, which is located just after the N-terminal arm playing a relevant role in dimer stabilization (Fig. 1B), is held in place by numerous intrachain contacts as well as by a strong salt bridge between the Lys-19 and the Glu-48* of the opposite monomer.
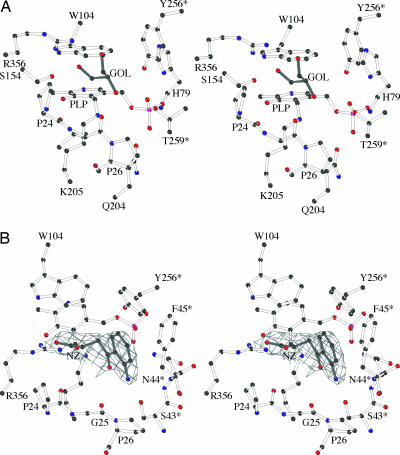
Besides the essential catalytic Lys-205, whose NZ atom is covalently bound to the PLP molecule in the internal aldimine form of the enzyme (Fig. 2A) (38), several other residues contribute to the recognition and anchoring of the cofactor. The PLP pyridine moiety is clamped between the residues Trp-104 and Val-181, forcing the PLP ring plane to form an angle of 115.0° with the Schiff-base linkage. The orientation of the cofactor molecule inside the active site is fixed by the further interactions of the PLP phenolic oxygen with the OG atom of Ser-154 (at a distance of 2.42 Å), whereas the N1 atom in the pyridine ring establishes two hydrogen bonds with the carboxylate group of Asp-179. The oxygen atoms of the PLP phosphate group are involved in a number of interactions strictly conserved throughout the majority of the PLP-dependent enzymes (39). In particular, the OP1 oxygen is in close contact with the residues Tyr-256* (at 2.50 Å from its hydroxyl group) and Thr-259* (at 2.48 Å from its OG1 atom and at 3.0 Å from the backbone nitrogen); the OP2 atom is engaged in two interactions: with Gln-204 (at 2.78 Å from its NE2 atom) and with the backbone nitrogen of Ala-78 (at a distance of 3.26 Å); the OP3 atom makes contact with the OG atom of Ser-77 (at a distance of 2.71 Å) and with the ND1 and main-chain nitrogen atoms of His-79 (at distances of 2.86 and 3.09 Å, respectively); this last residue is also involved in a weak interaction with the OP4 atom. Finally, the N terminus of the α-helix H4 contributes, by its positive α-helical dipole, to keep the PLP phosphate group in electrostatic contact.
Fig. 2.
The Ag-HKT catalytic site. (A) Stereoview of the active site of the Ag-HKT:PLP structure. The PLP cofactor, the glycerol molecule, and protein residues involved in cofactor binding are depicted as ball and stick. (B) Stereo view of the active site in the Ag-HKT:INI structure. Only the 2Fo−Fc electron density map covering the 4-(2-aminophenyl)-4-oxobutyric acid inhibitor (INI) is shown contoured at 1.2σ level. The figure was generated with the program bobscript (51).
Substrate Recognition and Catalysis.
Enzyme kinetics analyses of Ag-HKT revealed that both 3-HK and L-KYN can act efficiently as the aminodonor substrate in the transamination reaction, yielding XA and kynurenic acid, respectively (21). To identify the structural determinants of the Ag-HKT that are involved in substrate recognition and binding, we have determined the structure of the enzyme in complex with 4-(2-aminophenyl)-4-oxobutyric acid (Fig. 2B), a newly synthesized molecule that behaves as a Ag-HKT competitive inhibitor with a Ki in the high micromolar range (Ki = 300 μM). This compound, which chemically closely resembles both 3-HK and L-KYN, cannot be processed by the enzyme due to the absence of a reactive amino group at the C2 position equivalent to the Cα atom of the natural amino acid substrates (see Materials and Methods).
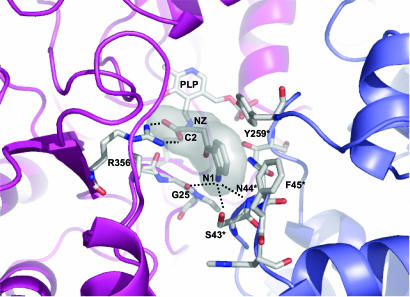
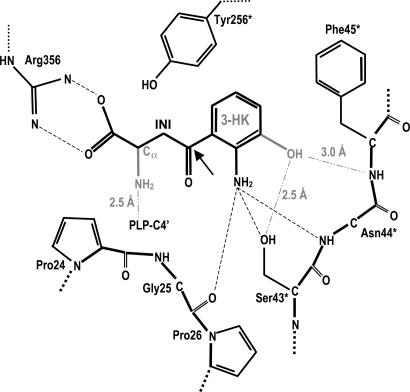
Inspection of the Ag-HKT:INI complex crystal structure reveals that the ligand lies above the pyridine ring plane of the PLP cofactor, which is still in its internal aldimine form (Fig. 2B). Several residues, structurally conserved throughout aminotransferases described so far, define the ligand-binding site and contact the inhibitor. In particular, Arg-356 makes a strong salt bridge with the ligand carboxylate group at a distance of 2.7 Å. A hydrogen-bonding network, involving Ser-154 and Gly-345 backbone oxygen atoms, locks the Arg-356 side chain in the optimal conformation for inhibitor binding, as observed in the Ag-HKT:PLP structure (Fig. 2A) where Arg-356 hydrogen bonds the O1 and O3 atoms of the glycerol molecule invariably occupying the ligand-binding site in all of the subunits. The anchoring of the inhibitor carboxylic moiety by Arg-356 in the Ag-HKT:INI complex (Figs. 2B and 3), results in the positioning of its C2 atom just above the PLP C4′ reactive center at a distance of 3.6 Å. Because the inhibitor C2 atom is structurally equivalent to the Cα atom in the natural substrate, the observed conformation would place the reactive α-amino group in the ideal position for attacking the PLP C4′ atom and forming the external aldimine (38). The inhibitor anthranilic moiety is recognized and stabilized by a number of interactions with the protein milieu (Figs. 2B and 3). In particular, the aromatic ring establishes hydrophobic contacts with residues Tyr-256* and Phe-45* (at minimum distances of 3.2 and 3.0 Å, respectively), and the inhibitor aromatic NH2 amino group is engaged in a bidentate hydrogen bond with the OG atom of Ser-43* (distance of 3.2 Å) and with the Asn-44* backbone nitrogen (distance of 3.1 Å). A further interaction is provided by a hydrogen bond between the inhibitor aromatic NH2 group and the backbone oxygen atom of Gly-25 at a distance of 3.2 Å. Gly-25 is located at the turning point of the loop that dives into and partially plugs the enzyme active site (Fig. 4), thus shielding the ligand-binding pocket from the bulk solvent. The observed Gly-25 backbone oxygen orientation results from Pro-24 being in the cis conformation in both the Ag-HKT:INI and Ag-HKT:PLP structures highlighting the cisPro-24-Gly-25 motif as an important element for ligand binding.
Fig. 3.
Closeup view of the enzyme active site in the Ag-HKT:INI structure. The protein portions building up the active site and contributed by the two subunits of the functional homodimer are shown in a ribbon representation and colored blue and magenta. The PLP cofactor, the inhibitor molecule, and the protein residues involved in ligand binding are shown. The inhibitor C2 atom equivalent to the Cα of the physiological amino acid substrate is indicated. The major interactions established between the inhibitor and residues of Ag-HKT are indicated as dotted lines. The figure was generated by using pymol (www.pymol.org).
Fig. 4.
Closeup view of the protein region surrounding the loop protruding into the active site and carrying the cisPro24-Gly-25 motif participating in ligand binding. The PLP cofactor, the inhibitor molecule, and the cisPro-24-Gly-25 motif are drawn as ball-and-stick representation. The coloring and the orientation are as in Fig. 1A. The figure was generated with the program molscript (50).
As anticipated, both 3-HK and L-KYN, differing only in the presence of a hydroxyl group in the former (Scheme 1), can serve as Ag-HKT substrates. Based on the Ag-HKT:INI structure, the hydroxyl group present in 3-HK can easily be modeled (Scheme 2) and would be placed in an ideal position to make a strong hydrogen bond with the OG of Ser-43*, which therefore appears as a key residue for recognition of both kynurenine and 3-HK.
Scheme 2.
Schematic view of the Ag-HKT active site. The key interactions involved in inhibitor binding are indicated together with the distances observed in the Ag-HKT:INI structure. The chemical groups featuring the physiological substrate 3-HK are shown in light gray, and their interactions with the protein residues, resulting from manual modeling, are indicated. The arrow indicates the C4 position on the 4-(2-aminophenyl)-4-oxobutyric acid that would carry the proposed chemical modification consisting of the replacement of the carbonyl group by an oxy-amino (-O-NH2) moiety.
Implications for Structure-Based Drug Design.
The analysis of the Ag-HKT:INI complex suggests a number of chemical modifications that could be made on the inhibitor structure to improve its efficacy against Ag-HKT. An additional hydroxyl group at position 3 of the anthranilic moiety of our inhibitor (Scheme 2) would establish two further hydrogen bonds with the protein residues Asn-44* and Ser-43* (Fig. 3 and Scheme 2), therefore tightening the inhibitor binding to the enzyme active site. Structural comparison with several other aminotransferases allowed us to propose further chemical manipulation of 4-(2-aminophenyl)-4-oxobutyric acid, aimed at improving its potency and specificity. A dali search (40) identified human alanine-glyoxylate aminotransferase (AGT) (41) as the closest structural ortholog of Ag-HKT. Human AGT and Ag-HKT belong to the same subgroup of PLP-dependent aminotransferases, show 43% sequence identity, and can be superposed with a rms deviation of 1.2 Å for 382 Cα pairs. Human AGT was solved in complex with amino-oxyacetic acid (AOA) (40), a strong competitive inhibitor of the enzyme. Inspection of the active site of Ag-HKT:INI after optimal superimposition to AGT:AOA reveals that the oxy group of AOA almost perfectly superposes the carbonyl atom of our inhibitor, and that the AOA amino group points toward the carbonyl atom of Ag-HKT Gly-25, at an ideal distance for hydrogen bonding. Consequently, the replacement of the carbonyl group in 4-(2-aminophenyl)-4-oxobutyric acid with an amino-oxy group would establish another hydrogen bond with the cisPro-24-Gly-25 motif, which we identified as of key importance for ligand binding in Ag-HKT (Scheme 2). Therefore, the rational design of potent Ag-HKT inhibitors appears feasible. However, a major concern is represented by the selectivity of inhibitor action, because a functional ortholog of the mosquito enzyme exists in humans. Indeed, human kynurenine aminotransferases have been shown to catalyze the irreversible transamination of 3-HK to XA (42). However, in our hands, 4-(2-aminophenyl)-4-oxobutyric acid does not inhibit the human KAT I isozyme in the millimolar range, providing a proof of principle that selective inhibitors toward the mosquito enzyme can be identified. We previously reported the 3D structure of human KAT I in complex with the substrate analogue L-Phe, identifying the structural determinants responsible for ligand binding and catalysis (20). Human KAT I and Ag-HKT belong to different subfamilies of PLP-dependent aminotransferases, show only 14% sequence identity, and can be superposed with a rms deviation of 5.0 Å for 320 Cα pairs. Overall, the two enzyme active sites appear to be poorly structurally conserved, and ligand recognition is achieved by exploiting different structural determinants and different residues in the two enzymes. In keeping with this observation, any attempt we made to manually model 4-(2-aminophenyl)-4-oxobutyric acid into the human KAT I active site failed due to severe sterical collisions with a number of protein residues.
Conclusions
Plasmodium gametogenesis in anophelines requires the presence of XA (28). Making such a molecule unavailable to the mosquito would block the sexual stages of the parasite development inside the insect vector and may therefore represent a promising avenue for the discovery of novel malaria transmission-blocking drugs. Within such a scenario, the inhibition of Ag-HKT, the only known source of endogenous XA, is an important goal. Unfortunately, no inhibitors of Ag-HKT are presently known. The crystal structure of A. gambiae 3-HKT, in complex with a newly synthesized inhibitor reported herein, will be used to guide the structure-based rational design of novel, potent, and highly selective Ag-HKT small molecule inhibitors that may prove useful both as a tool for further investigating the role of Ag-HKT in vivo and as compounds of potential interest as innovative antimalarial agents.
Materials and Methods
Enzyme Expression and Purification.
The procedure adopted for the bacterial overexpression and purification of the recombinant Ag-HKT has been reported (21). The pure and active Ag-HKT used for the crystallization trials was in 20 mM Tris·HCl, pH 8.0/150 mM NaCl/40 μM PLP.
Synthesis of 4-(2-Aminophenyl)-4-oxobutyric Acid.
4-(2-Aminophenyl)-4-oxobutyric acid 3 was synthesized in two steps from commercially available indole-3-propionic acid 1, as shown in Scheme 3.
Scheme 3.
The first step involves oxidative demolition of the five-membered hetero-ring of the indole nucleus (43, 44) by treatment with sodium periodate in a water/methanol mixture to give 4-(2-formylamino-phenyl)-4-oxo-butyric acid 2. Removal of the N-formyl group was performed by acidic hydrolysis (HCl/H2O, room temperature), leading to the target compound 3 (55% overall yield after workup). The structure of compound 3 was confirmed by means of NMR and mass spectrometry analysis.
Crystallization, Data Collection, and Structure Determination.
Crystals of the PLP form of Ag-HKT were obtained by using the vapor diffusion technique in hanging drop. One microliter of a protein solution at a concentration of 8 mg/ml was mixed with an equal volume of a reservoir solution containing 0.2 M tripotassium citrate, 20% wt/vol polyethylene glycol (PEG) 3350, and equilibrated against 500 μl of the reservoir solution, at 20°C. Rod-shaped yellow crystals grew to a maximum length of 0.3 mm in ≈3 days. The crystals of the Ag-HKT:INI were obtained by soaking crystals of the PLP form in their crystallization solution with the addiction of 10 mM inhibitor at 20°C for 12 h. All data collection was performed at 100 K. Crystals were taken from the crystallization droplet; rapidly washed in a cryoprotectant solution containing 0.2 M tripotassium citrate, 20% wt/vol PEG 3350, and 25% glycerol; and flash-frozen under a stream of liquid nitrogen. Diffraction data for the PLP form of the enzyme and for the complex with the inhibitor were collected up to 2.4- and 2.7-Å resolution, respectively, by using synchrotron radiation at the ID14 EH1 beam line, European Synchrotron Radiation Facility, Grenoble, France. Systematic absences and diffraction symmetry allowed us to assign the crystals to the monoclinic P21 space group with the following cell parameters: a = 87.4 Å, b = 84.2 Å, c = 118.8 Å, and β = 99.9°, and containing four molecules in the asymmetric unit with a corresponding solvent content of 55%. Data processing was performed with the programs of the ccp4 suite (45). Statistics for data collection are listed in Table 1. The structure determination of the PLP form of the enzyme was carried out by means of the molecular replacement technique using the coordinates of a monomer of human AGT as the search model (Protein Data Bank ID code 1H0C) (41). The program amore (46) was used to calculate both crossrotation and translation functions in the 10-4-Å resolution range, allowing for the identification of a clear solution of both functions for all four molecules present in the asymmetric unit. The initial model was subjected to iterative cycles of crystallographic refinement with the program refmac (47), alternated with graphic sessions for model building using the program o (48). Tight noncrystallographic symmetry restraints were maintained throughout the different stages of refinement. A random sample containing 1,031 reflections was set apart for the calculation of the free R factor (49). Solvent molecules were manually added at positions with density >4σ in the Fo−Fc map, considering only peaks engaged in at least one hydrogen bond with a protein atom or a solvent atom. The procedure converged to an R factor and free R factor of 0.212 and 0.244, with ideal geometry. For the Ag-HKT:PLP structure, a careful inspection of the electron density in the enzyme active site was performed when the R factor dropped to a value of 0.25 at 2.4 Å and revealed an electron density compatible with one glycerol molecule that was consequently manually modeled based on both the 2Fo−Fc and Fo−Fc electron density maps. The Ag-HKT:INI structure was refined by using as the starting model the final coordinates of the PLP form of the enzyme, from which all solvent and glycerol molecules were removed. The crystallographic refinement was carried out by means of the program refmac (47) alternated with sessions of model building. One thousand twelve randomly chosen reflections were excluded from the refinement of the Ag-HKT:INI structure and used for the free R factor calculation. Solvent molecules were manually added following the same criteria adopted for Ag-HKT:PLP. In the Ag-HKT:INI structure, the ligand molecule was modeled only when the R factor dropped to a value of 0.26- at 2.7-Å resolution. The subsequent crystallographic refinement converged to an R factor and Rfree of 0.212 and 0.266. Refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Ag-HKT:PLP | Ag-HKT:INI | |
|---|---|---|
| Data collection | ||
| Resolution, Å | 2.4 | 2.7 |
| Observations | 206,560 | 125,049 |
| Unique reflections | 65,686 | 45,886 |
| Rmerge | 0.064 | 0.071 |
| Multiplicity | 3.1 | 2.7 |
| Completeness, % | 98.9 | 98.2 |
| Refinement | ||
| No. of protein atoms | 12,180 | 12,143 |
| No. of solvent molecules | 380 | 142 |
| No. of PLP atoms | 60 | 60 |
| No. of INI atoms | — | 56 |
| No. of GOL atoms | 24 | — |
| Rwork, % | 21.2 | 21.2 |
| Rfree, % | 24.4 | 26.6 |
| rms deviation bond lengths, Å | 0.014 | 0.017 |
| rms deviation bond angles, ° | 1.44 | 1.59 |
| Mean B factor main chain, Å2 | 40.0 | 36.5 |
| Mean B factor side chain, Å2 | 41.4 | 38.6 |
| Mean B factor PLP atoms, Å2 | 40.5 | 30.3 |
| Mean B factor INI atoms, Å2 | — | 55.0 |
| Mean B factor GOL atoms, Å2 | 60.0 | — |
| Mean B factor solvent, Å2 | 35.5 | 27.2 |
INI, inhibitor; GOL, glycerol.
Acknowledgments
We thank Dr. Richard Billington (University of Piemonte Orientale) for critical reading of the manuscript. This work was supported by grants from the Regione Piemonte (Ricerca Sanitaria Finalizzata 2004 and Ricerca Scientifica Applicata 2003), Ministero dell’Istruzione, dell’Università, e della Ricerca (FIRB2001), Fondazione Cariplo (project number 2004.1591/11.5437), and National Institutes of Health Grant AI 44399.
Abbreviations
- 3-HK
3-hydroxykynurenine
- XA
xanthurenic acid
- KAT
kynurenine aminotransferase
- PLP
pyridoxal 5′-phosphate
- Ag-HKT
A. gambiae 3-HK transaminase
- AGT
alanine-glyoxylate aminotransferase
- L-KYN
l-kynurenine
- Ag-HKT:INI
Ag-HKT in complex with competitive inhibitor 4-(2-aminophenyl)-4-oxobutyric acid.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2CH1 (Ag-HKT:PLP) and 2CH2 (Ag-HKT:INI)].
References
- 1.Stone T. W. Toxicon. 2001;39:61–73. doi: 10.1016/s0041-0101(00)00156-2. [DOI] [PubMed] [Google Scholar]
- 2.Schwarcz R., Pellicciari R. J. Pharmacol. Exp. Ther. 2002;1:1–10. doi: 10.1124/jpet.102.034439. [DOI] [PubMed] [Google Scholar]
- 3.Breton J., Avanzi N., Magagnin S., Covini N., Magistrelli G., Cozzi L., Isacchi A. Eur. J. Biochem. 2000;267:1092–1099. doi: 10.1046/j.1432-1327.2000.01104.x. [DOI] [PubMed] [Google Scholar]
- 4.Han Q., Calvo E., Marinotti O., Fang J., Rizzi M., James A. A., Li J. Insect Mol. Biol. 2003;12:483–490. doi: 10.1046/j.1365-2583.2003.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirai M., Kiuchi M., Wang J., Ishii A., Matsuoka H. Insect Mol. Biol. 2002;11:497–504. doi: 10.1046/j.1365-2583.2002.00358.x. [DOI] [PubMed] [Google Scholar]
- 6.Okuda S., Nishiyama N., Saito H., Katsuki H. Proc. Natl. Acad. Sci. USA. 1996;93:12553–12558. doi: 10.1073/pnas.93.22.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagami Y., Saito H., Katsuki H. Jpn. J. Pharmacol. 1996;71:183–186. doi: 10.1254/jjp.71.183. [DOI] [PubMed] [Google Scholar]
- 8.Okuda S., Nishiyama N., Saito H., Katsuki H. J. Neurochem. 1998;70:299–307. doi: 10.1046/j.1471-4159.1998.70010299.x. [DOI] [PubMed] [Google Scholar]
- 9.Sardar A. M., Bell J. E., Reynolds G. P. J. Neurochem. 1995;64:932–935. doi: 10.1046/j.1471-4159.1995.64020932.x. [DOI] [PubMed] [Google Scholar]
- 10.Guidetti P., Schwarcz R. Adv. Exp. Med. Biol. 2003;527:137–145. doi: 10.1007/978-1-4615-0135-0_16. [DOI] [PubMed] [Google Scholar]
- 11.Stone T. W., Darlington L. G. Nat. Rev. Drug Discov. 2002;1:609–620. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 12.Schwarcz R. Curr. Opin. Pharmacol. 2004;1:12–17. doi: 10.1016/j.coph.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Savvateeva E. Adv. Exp. Med. Biol. 1991;294:319–328. doi: 10.1007/978-1-4684-5952-4_29. [DOI] [PubMed] [Google Scholar]
- 14.Savvateeva E., Popov A., Kamyshev N., Bragina J., Heisenberg M., Senitz D., Kornhuber J., Riederer P. J. Neural Transm. 2000;107:581–601. doi: 10.1007/s007020070080. [DOI] [PubMed] [Google Scholar]
- 15.Cerstiaens A., Huybrechts J., Kotanen S., Lebeau I., Meylaers K., De Loof A., Schoofs L. Biochem. Biophys. Res. Commun. 2003;312:1171–1177. doi: 10.1016/j.bbrc.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 16.Kotanen S., Huybrechts J., Cerstiaens A., Zoltan K., Daloze D., Baggerman G., Forgo P., De Loof A., Schoofs L. Biochem. Biophys. Res. Commun. 2003;310:64–67. doi: 10.1016/j.bbrc.2003.08.121. [DOI] [PubMed] [Google Scholar]
- 17.Alberati-Giani D., Buchli R., Malherbe P., Broger C., Lang G., Kohler C., Lahm H. W., Cesura A. M. Eur. J. Biochem. 1996;239:460–468. doi: 10.1111/j.1432-1033.1996.0460u.x. [DOI] [PubMed] [Google Scholar]
- 18.Guidetti P., Okuno E., Schwarcz R. J. Neurosci. Res. 1997;50:457–465. doi: 10.1002/(SICI)1097-4547(19971101)50:3<457::AID-JNR12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Okuno E., Nakamura M., Schwarcz R. Brain Res. 1991;542:307–312. doi: 10.1016/0006-8993(91)91583-m. [DOI] [PubMed] [Google Scholar]
- 20.Rossi F., Han Q., Li J., Li J., Rizzi M. J. Biol. Chem. 2004;279:50214–50220. doi: 10.1074/jbc.M409291200. [DOI] [PubMed] [Google Scholar]
- 21.Rossi F., Lombardo F., Paglino A., Cassani C., Miglio G., Arcà B., Rizzi M. FEBS J. 2005;272:5653–5662. doi: 10.1111/j.1742-4658.2005.04961.x. [DOI] [PubMed] [Google Scholar]
- 22.Han Q., Fang J., Li J. J. Biol. Chem. 2002;277:15781–15787. doi: 10.1074/jbc.M201202200. [DOI] [PubMed] [Google Scholar]
- 23.Han Q., Li J. FEBS Lett. 2002;527:199–204. doi: 10.1016/s0014-5793(02)03229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Li J. Insect Biochem. Mol. Biol. 1997;27:859–867. doi: 10.1016/s0965-1748(97)00068-4. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Beerntsen B. T., James A. A. Insect Biochem. Mol. Biol. 1999;29:329–338. doi: 10.1016/s0965-1748(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 26.Cornel A. J., Benedict M. Q., Rafferty C. S., Howells A. J., Collins F. H. Insect Biochem. Mol. Biol. 1997;27:993–997. doi: 10.1016/s0965-1748(97)00084-2. [DOI] [PubMed] [Google Scholar]
- 27.Chiou S. J., Kotanen S., Cerstiaens A., Daloze D., Pasteels J. M., Lesage A., Drijfhout J. W., Verhaert P., Dillen L., Claeys M., et al. Biochem. Biophys. Res. Commun. 1998;246:457–462. doi: 10.1006/bbrc.1998.8644. [DOI] [PubMed] [Google Scholar]
- 28.Billker O., Lindo V., Panico M., Etienne A. E., Paxton T., Dell A., Rogers M., Sinden R. E., Morris H. R. Nature. 1998;392:289–292. doi: 10.1038/32667. [DOI] [PubMed] [Google Scholar]
- 29.Garcia G. E., Wirtz R. A., Barr J. R., Woolfitt A., Rosenberg R. J. Biol. Chem. 1998;273:12003–12005. doi: 10.1074/jbc.273.20.12003. [DOI] [PubMed] [Google Scholar]
- 30.Arai M., Billker O., Morris H. R., Panico M., Delcroix M., Dixon D., Ley S. V., Sinden R. E. Mol. Biochem. Parasitol. 2001;116:17–24. doi: 10.1016/s0166-6851(01)00299-7. [DOI] [PubMed] [Google Scholar]
- 31.Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. Cell. 2004;117:503–514. doi: 10.1016/s0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 32.Dana A. N., Hong Y. S., Kern M. K., Hillenmeyer M. E., Harker B. W., Lobo N. F., Hogan J. R., Romans P., Collins F. H. BMC Genom. 2005;6:5. doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X., Dong Y., Abraham E. G., Kocan A., Srinivasan P., Ghosh A. K., Sinden R. E., Ribeiro J. M., Jacobs-Lorena M., Kafatos F. C., et al. Mol. Biochem. Parasitol. 2005;142:76–87. doi: 10.1016/j.molbiopara.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Truscott R. J., Elderfield A. J. Clin. Sci. 1995;89:591–599. doi: 10.1042/cs0890591. [DOI] [PubMed] [Google Scholar]
- 35.Laskowsky R. A., MacArthur M. W., Moss D. S., Thornton J. M. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 36.Mehta P. K., Hale T. I., Christen P. Eur. J. Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- 37.Christen P., Mehta P. K. Chem. Rec. 2001;1:436–447. doi: 10.1002/tcr.10005. [DOI] [PubMed] [Google Scholar]
- 38.Jansonius J. N. Curr. Opin. Struct. Biol. 1998;8:759–769. doi: 10.1016/s0959-440x(98)80096-1. [DOI] [PubMed] [Google Scholar]
- 39.Denesyuk A. I., Denessiouk K. A., Korpela T., Johnson M. S. J. Mol. Biol. 2002;316:155–172. doi: 10.1006/jmbi.2001.5310. [DOI] [PubMed] [Google Scholar]
- 40.Holm L., Sander C. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X., Roe S. M., Hou Y., Bartlam M., Rao Z., Pearl L. H., Danpure C. J. J. Mol. Biol. 2003;331:643–652. doi: 10.1016/s0022-2836(03)00791-5. [DOI] [PubMed] [Google Scholar]
- 42.Okuno E., Du F., Ishikawa T., Tsujimoto M., Nakamura M., Schwarcz R., Kido R. Brain Res. 1990;534:37–44. doi: 10.1016/0006-8993(90)90109-o. [DOI] [PubMed] [Google Scholar]
- 43.Dolby L. J., Booth D. L. J. Am. Chem. Soc. 1966;88:1049–1051. [Google Scholar]
- 44.Jones G., Tringham G. T. J. Chem. Soc. Perkin Trans. 1975;I:1280–1283. doi: 10.1039/p19750001280. [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computing Project, Number 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 46.Navaza J. Acta Crystallogr. D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Brünger A. T. Nature. 1992;355:472–447. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 50.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 51.Esnouf R. M. J. Mol. Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]