Abstract
Human Wilson protein is a copper-transporting ATPase located in the secretory pathway possessing six N-terminal metal-binding domains. Here we focus on the function of the metal-binding domains closest to the vesicular portion of the copper pump, i.e., domain 4 (WLN4), and a construct of domains 5 and 6 (WLN5–6). For comparison purposes, some experiments were also performed with domain 2 (WLN2). The solution structure of apoWLN5–6 consists of two ferredoxin folds connected by a short linker, and 15N relaxation rate measurements show that it behaves as a unit in solution. An NMR titration of apoWLN5–6 with the metallochaperone Cu(I)HAH1 reveals no complex formation and no copper exchange between the two proteins, whereas titration of Cu(I)HAH1 with WLN4 shows the formation of an adduct that is in fast exchange on the NMR time scale with the isolated protein species as confirmed by 15N relaxation data. A similar interaction is also observed between Cu(I)HAH1 and WLN2; however, the relative amount of the adduct in the protein mixture is lower. An NMR titration of apoWLN5–6 with Cu(I)WLN4 shows copper transfer, first to WLN6 then to WLN5, without the formation of an adduct. Therefore, we suggest that WLN4 and WLN2 are two acceptors of Cu(I) from HAH1, which then somehow route copper to WLN5–6, before the ATP-driven transport of copper across the vesicular membrane.
Keywords: ATPase function, copper transport, metal-binding domain, metallochaperone, Wilson disease protein
Human Wilson protein (WLNP) is a copper-transporting P-type ATPase (1, 2) located in the secretory pathway and plays a crucial role in copper transport and homeostasis (3). Mutations in the gene encoding WLNP are associated with changes in human copper metabolism leading to a severe hepatoneurological disorder, Wilson’s disease (4, 5). In the latter, copper accumulates in a number of tissues, particularly in the liver, brain, and kidneys, causing DNA damage, inactivation of certain enzymes, and lipid peroxidation.
WLNP acquires copper from the cytosolic metallochaperone HAH1 (commonly also called Atox1) (6), and then, after passage of copper into the Golgi, copper is inserted into the multicopper oxidase ceruloplasmin (7, 8). WLNP also translocates from Golgi membrane to the cytosolic membrane for copper detoxification (9). The predicted topological organization of WLNP shows that the protein is composed of four major domains (the N-terminal copper-binding domain, the transmembrane domain, the ATP-binding domain, and the phosphatase domain) and a C-terminal tail. The N-terminal copper-binding domain of WLNP (N-WLNP) is ≈650 aa long, contains six repetitive sequences, each bearing the conserved sequence motif GMT/HCxxCxxxIE (10), and is capable of binding up to one equivalent of Cu(I) per metal-binding domain (11, 12). The structure of N-WLNP has yet to be determined; however, the NMR structures of several single metal-binding domains from the highly homologous human copper-transporting ATPase ATP7A, Menkes protein, have been solved (13–15). Sequence alignments and homology models on the six metal-binding domains of WLNP demonstrate that the ≈70 aa domains of N-WLNP are likely to be folded very similarly into ferredoxin-like units (16). This same fold is also found in both NMR and x-ray structures of the yeast Atx1 and human HAH1 metallochaperones (17–20), the soluble cytosolic proteins that deliver copper to the copper-transporting P-type ATPases (21).
Human missense mutations found within metal-binding domains 1, 5, and 6 are known to give rise to Wilson disease (22), and these mutants show impaired interaction with HAH1 in a column-based assay (6). Mutational studies performed to address the role of the N-WLNP have found that, generally, the domains closest to the membrane seem to be the most important both for the ultimate incorporation of copper into Fet3 in yeast complementation assays and in the copper-dependent translocation of WLNP from the vesicular to the cytosolic membrane (23–25). However, the mechanism by which these domains acquire copper and then transport it across the vesicular membrane is unknown. Several studies indicate that the first four metal-binding domains are important for copper acquisition from HAH1. For example, yeast two-hybrid assays have demonstrated a copper-dependent interaction between HAH1 and constructs containing metal-binding domains 1–4 but notably not with metal-binding domains 5 and 6 (26, 27). Furthermore, Walker et al. (28) proposed that WLN2 is the preferred site of interaction for Cu(I)-HAH1. Therefore, metal-binding domains 1–4 seem to be the most important for acquisition from the metallochaperone, whereas WLN5 and WLN6 are critical for transport of copper into the vesicle.
The six metal-binding domains are connected by linkers of different length. The longest linker between WLN4 and WLN5 provides natural separation of N-WLNP into two parts: WLN1–4 and WLN5–6. Such a feature is not found in the bacterial and yeast copper-transporting ATPases, which have only one or two metal-binding domains (16). This topological organization suggests that the two parts can be treated independently from a structural point of view and that the different linker lengths connecting these domains is likely to be important for copper trafficking and regulation of WLNP in response to copper binding. The spatial separation and relative orientations of the N-terminal copper-binding domains in WLNP is therefore a key point in determining the mechanism by which copper passes through the membrane for incorporation into the transGolgi network. This finding, indeed, is essential to understand the routing of copper from the copper chaperone HAH1 to the transmembrane metal-binding site of WLNP. To address these issues, we have structurally characterized the domains closest to the transmembrane domain, WLN5–6. The properties of the interaction between WLN5–6 and the HAH1 chaperone and two domains belonging to the first WLN1–4 part of N-WLNP, i.e., WLN4 and WLN2, allow us to propose a route for copper transfer. This work, therefore, reports structural evidence for copper acquisition by WLNP.
Results
Solution Structure of Metal-Binding Domains 5 and 6.
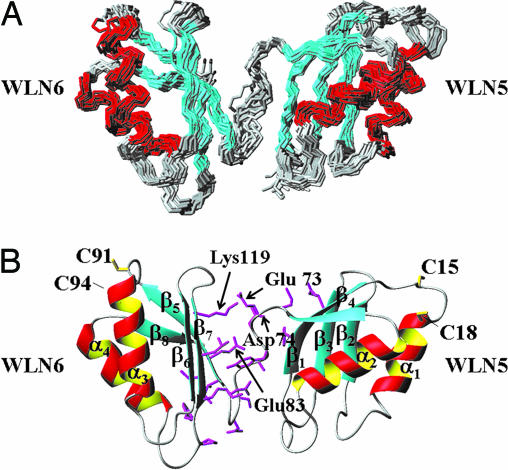
The structural determination of apoWLN5–6 shows that the protein is organized in two domains with the same ferredoxin-like fold (Fig. 1A). The fold of each domain and their copper-binding site structural properties are similar to those found in the single metal-binding domains of the highly homologous human copper-transporting ATPase ATP7A (13–15, 29). The two domains are found with fixed reciprocal orientation, as determined by the presence of nuclear Overhauser effects (NOEs) between each domain and a set of residues of the linker, thus sandwiching the linker between the two domains (see Fig. 1B). The relative orientation of the two WLN5 and WLN6 domains is further refined by measuring the hydrodynamic properties of the molecule using the method of Fushman and coworkers (30) described in the supporting information, which is published on the PNAS web site. Because the two domains tumble as a single molecule (see below), the relaxation data and the hydrodynamic properties can be used to define the relative orientation of the two domains. Both electrostatic interactions and a hydrogen bond network likely hold the two domains in a defined and rigid orientation (Fig. 1B). These interactions involve the negatively charged resides in the linker and strand β5, Glu-73, Glu-83 (which are fully conserved in eukaryotic organisms), and Asp-74 (semiconserved, substituted only by Asn or Gln residues), which constitute a negative patch in close contact with the fully conserved, positively charged residue Lys-119 of WLN6. The structure shows that the two copper-binding sites are distant from each other (Fig. 1), disallowing intramolecular copper transfer. A similar structural arrangement of the metal-binding sites, which are indeed far from each other, was found in the CopA bacterial homologue (31), even if the linker is shorter than in the present case (2 versus 9 aa).
Fig. 1.
Solution structure of apoWLN5–6. (A) Backbone superimposition of the 20 lowest-energy conformers of apoWLN5–6. The secondary structure elements are indicated: β-strands are cyan and α-helices are red. (B) Ribbon diagram of a representative structure of apoWLN5–6 in the same orientation as shown in A. Copper-binding residues (Cys-15, Cys-18, Cys-91, and Cys-94) are yellow. Residues belonging to the linker Met-72–Gly-80, which display NOEs with residues of the two domains, are magenta. The conserved negatively and positively charged residues, likely determining a preferential orientation between the two domains, also are labeled.
Relaxation rate measurements (15N R1, R2, and 1H–15N NOEs are reported in the supporting information), which can provide a picture of the dynamics of the protein from its overall tumbling rate to specific protein internal motions, indicate that WLN5–6 reorients in solution as a single molecule (i.e., as a dumbbell) rather than as independent beads on a string. The overall rotational correlation time (τc) of apoWLN5–6 (9.1 ± 0.6 ns) is indeed two times greater than that (≈4.5 ns) for WLN6 alone and is the value expected for a monomeric protein of a molecular mass of 16 kDa (32). The parameters of the rotational diffusion tensor of apoWLN5–6 are reported in the supporting information. Furthermore, the residues linking the two domains of apoWLN5–6 do not experience conformational exchange processes on the millisecond-to-microsecond time scale (see the supporting information), consistent with the unique relative orientation of the two domains.
Copper Interaction and Transfer Studies.
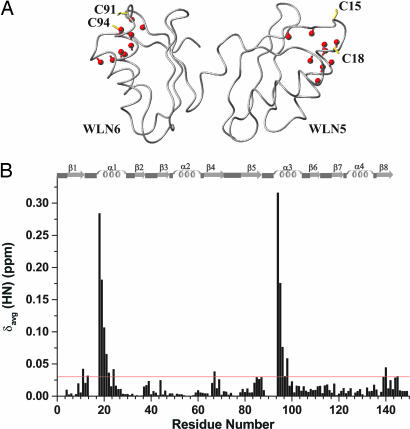
The apoWLN5–6 construct can bind two equivalents of copper provided by small Cu(I) complexes. The copper binding produces spectral changes limited to the two copper-binding regions and does not affect the overall structural properties; in particular, it does not induce changes in the relative orientation of the two domains (Fig. 2). Furthermore, relaxation measurements ruled out the possibility of aggregation phenomena. However, the behavior is dramatically different when apoWLN5–6 is titrated with the physiological partner of WLNP, the Cu(I)HAH1 protein. When apoWLN5–6 and Cu(I)HAH1 are mixed together, no copper transfer occurs from Cu(I)HAH1 to apoWLN5–6, even when more that one equivalent of Cu(I)HAH1 for each metal-binding site is added (HAH1/WLN5–6 ratio of 2.5:1.0) and monitored for 2 weeks. Moreover, the 1H–15N heteronuclear single quantum correlation (HSQC) spectrum of WLN5–6 remains unchanged as apoWLN5–6 is titrated with Cu(I)HAH1, indicating that no adduct in quantities detectable by NMR is formed between HAH1 and WNL5–6. This finding is consistent with the two-hybrid assay data showing that HAH1 does not interact with WLN5–6 (26).
Fig. 2.
Cu(I) interaction studies of apoWLN5–6. (A) The NH crosspeaks, showing chemical shift δavg(HN) differences above the threshold of 0.03 ppm are shown in red on the backbone structure of apoWLN5–6. (B) The weighted average chemical shift differences δavg(HN) (i.e., {[(δH)2 + (δN/5)2]/2}1/2, where δH and δN are chemical shift differences for 1H and 15N, respectively) are shown. Chemical shifts differences are not reported for residues 14–17 and 90–93, because their 1H–15N cross-peaks are not observed for apo and/or Cu(I) forms. The secondary structure elements of apoWLN5–6 are reported at the top of the graph. The threshold is indicated with a horizontal red line.
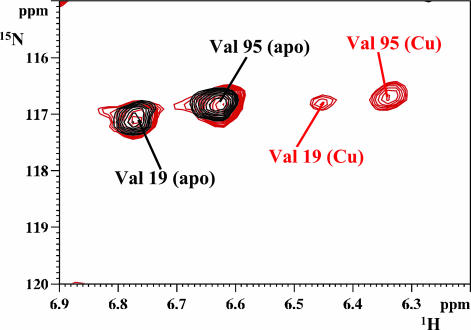
apoWLN5–6 was also titrated with Cu(I)WLN4 to test whether WLN4 could pass the copper ion to the domains closest to the transmembrane copper-binding site. At variance with what occurs with Cu(I)HAH1, partial Cu(I) transfer occurs. By integration of some amide NH signals in the vicinity of the two metal-binding sites, we estimate that, at a 1:1 Cu(I)WLN4/apoWLN5–6 ratio, copper is partially transferred to WLN6 [≈10% of the site is saturated with Cu(I), with no transfer to WLN5]. Further additions of Cu(I)WLN4 up to a ratio of 6:1 cause an increase in the metallation of the WLN6 domain as well as metallation of WLN5 (Fig. 3) [at a 2.5:1 ratio, 25% of Cu(I) is bound to WLN6 and 10% is bound to WLN5; at 6:1 ratio, 45% of Cu(I) is on WLN6 and 35% is on WLN5]. No formation in detectable quantities of a protein–protein adduct is observed: indeed τc of both forms of WLN5–6 [i.e., apo and Cu(I)], in the presence of Cu(I)WLN4, remains unchanged with respect to τc of apoWLN5–6. In addition, the 1H–15N HSQC spectrum of WLN5–6 in the presence of Cu(I)WLN4 is the sum of the spectra of two species, i.e., apoWLN5–6 and Cu(I)WLN5–6.
Fig. 3.
Transfer of Cu(I) from Cu(I)WLN4 to apoWLN5–6. Superposition of the 1H–15N HSQC spectra of apoWLN5–6 (black) and apoWLN5–6 in the presence of unlabeled Cu(I)WLN4 at a 1: 2.5 molar ratio (red), showing the simultaneous presence of the signals of Val-19 and Val-95 in their Cu(I)- and apo-loaded state.
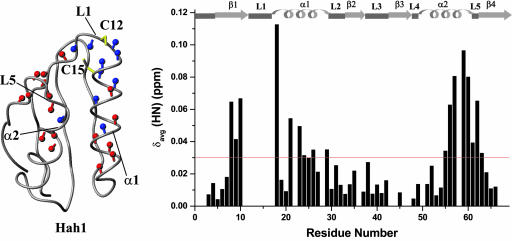
When Cu(I)HAH1 is titrated with apoWLN4, changes in chemical shift in a number of residues are observed (Fig. 4; see also the supporting information), with the shift changes increasing upon addition of apoWLN4. Moreover, a few NH crosspeaks of the residues closer to the metal-binding site of HAH1 broaden beyond detection. The 1:1 Cu(I)HAH1/apoWLN4 mixture shows an increase of τc (7.2 ± 0.7 ns, as obtained from the R2 over R1 ratio; see the supporting information) with respect to isolated Cu(I)HAH1 (4.6 ± 0.4 ns). This value for τc indicates the formation of a complex between WLN4 and HAH1 that is in fast exchange with the free proteins in solution, according to Scheme 1:
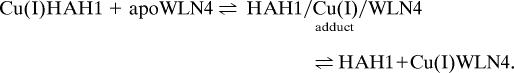 |
At millimolar concentrations and at a 1:1 Cu(I)HAH1/WLN4 ratio, 60% of the proteins are in the adduct form as estimated from the average τc value.§ In the 1H–15N HSQC spectra, it appears that the residues experiencing a δavg(HN) shift variation of >0.03 ppm with respect to both apo and metallated forms are localized in two regions of the protein corresponding to stretches 8–26 and 55–63. These stretches constitute loop 1; helix α1, which contains the metal-binding site; helix α2; and loop 5. The specific chemical shift changes observed upon mixing is a strong indication of the involvement of these regions in the protein–protein interaction. The above equilibrium appears to be very similar to that experienced by Ccc2a and Atx1 and discussed in detail in the literature (33, 34). In the latter case, the formation of the adduct was suggested to be a transient species in the transfer process of copper to the ATPase, which removes copper from one compartment (the cytoplasm) and transfers it to another (the Golgi organelle).
Fig. 4.
Complex formation between Cu(I)HAH1 and WLN4. (Right) The weighted average chemical shift differences δavg(HN) between 15N-Cu(I)HAH1 and the 1:1 15N-Cu(I)HAH1/apoWLN4 mixture. The secondary structure elements of Cu(I)HAH1 are reported at the top of the graph. The threshold is indicated with a horizontal red line. (Left) The NH crosspeaks showing chemical shift δavg(HN) differences above the threshold of 0.03 ppm are shown in red on the backbone structure of Cu(I)HAH1, whereas NH crosspeaks that disappear or significantly broaden are shown in blue. Cys-12 and Cys-15 are shown in yellow.
As it has been recently suggested that the chaperone HAH1 delivers copper specifically to WLN2 of N-WNDP (28), apoWLN2 was titrated with Cu(I)HAH1. Similar to the HAH1/WLN4 titration, changes in chemical shift in a number of residues of WLN2 are observed (see the supporting information), with the shift changes increasing upon addition of Cu(I)HAH1, as well as broadening of a few NH crosspeaks of residues close to the metal-binding site of HAH1. However, the observed chemical shift changes are smaller than those observed in the HAH1/WLN4 titration at the same protein ratio. The chemical shift changes are located in the same regions involved in the WLN4/HAH1 interaction, indicating that similar adducts are formed in both cases. The 1.5:1 Cu(I)HAH1/apoWLN2 mixture shows an increase of τc from a value of 4.6 ± 0.4 ns in the isolated Cu(I)HAH1 to 5.5 ± 0.4 ns for both HAH1 and WLN2 proteins in the mixture (as obtained from the R2 over R1 ratio, see the supporting information for details). The latter value is, however, lower than that observed in the 1:1 Cu(I)HAH1/WLN4 mixture (7.2 ± 0.7 ns), thus indicating that the fraction of formed adduct for WLN2 is lower than WLN4. In conclusion, the data indicate that the equilibrium (Scheme 1) also is present for the HAH1/WLN2 interaction, and, at millimolar concentrations and at a 1.5:1 HAH1/WLN2 ratio, 22% of the proteins are in the adduct state as estimated from the average τc value.
A structural model of the Atx1/Ccc2a complex showed that three negatively charged residues of Ccc2a are involved in electrostatic interactions with four Lys residues of Atx1 (33). These positively charged residues are conserved in HAH1, namely Arg-21, Lys-25, Lys-56, and Lys-57, and show significant chemical shift perturbation in the presence of WLN4 and WLN2 (Fig. 4; see also the supporting information). The three negatively charged residues of Ccc2a are conserved in WLN4, but only two are conserved in WLN2, WLN5, and WLN6 (16). As HAH1 interacts with WLN4 and WLN2 but not with WLN5–6, we may conclude that electrostatic charges are important but not unique factors in the protein–protein interactions.
Discussion
Based on the results of our NMR experiments, we are able to propose a pathway for copper delivery to the N-terminal metal-binding domains of WLNP. First, Cu(I)HAH1 forms a complex with WLN4 and WLN2 that allows transfer of copper between these partners. We did not measure any complex formation and no copper transfer between Cu(I)HAH1 and WLN5–6, indicating that WLN5–6 must acquire copper from one of the other metal-binding domains. Because WLN4 and WLN2 are in close proximity to four other metal-binding domains with similar copper affinity (35), copper may transfer intramolecularly between these sites, and we have shown here the possibility of copper transfer between WLN4 and WLN5–6. The solution structure, hydrodynamic behavior, and relaxation parameters of WLN5–6 reveal that this two-domain construct behaves as a unit, with the metal-binding sites facing away from each other, disallowing interdomain copper transfer between WLN5 and WLN6.
Different Functional Roles of Metal-Binding Domains.
In copper-transporting P-type ATPases, the spatial arrangement of the two-domain WLN5–6 construct is evolutionarily conserved across phylogenetic boundaries from bacteria to yeast to man. A structurally characterized two-domain construct of CopA of Bacillus subtilis (31), denoted CopAab, has a shorter linker than WLN5–6 with a hydrogen-bonding network at the interdomain interface. Like WLN5–6, the metal-binding sites of CopAab are far from each other, and copper binding does not alter protein structure. In contrast to the behavior of WLN5–6, the soluble two-domain constructs of Cu(I)-ATPases in lower organisms typically interact directly with their metallochaperone. For example, CopZ delivers copper to CopAab (36, 37), and yeast Atx1 delivers copper to Ccc2 (38, 39). Study of the biochemistry and function of bacterial and yeast copper trafficking pathways has nonetheless provided crucial information about complex formation in copper donor–target interactions (40, 41).
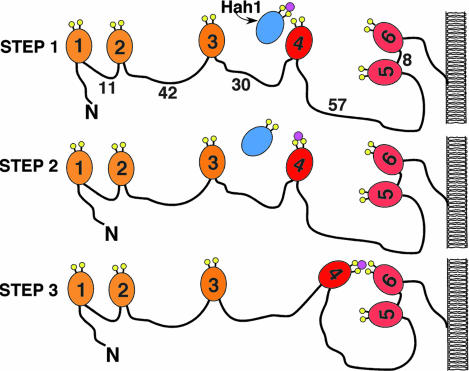
Why is there a need for six seemingly redundant high-affinity copper-binding domains in N-WLNP? Six copper sites rather than one may provide an entropic advantage for copper acquisition; however, we observe inequivalency among the metal-binding domains of WLNP in their surface potentials and interactions with HAH1. Therefore, we propose a model for the function of the N-terminal Wilson metal-binding domains based on our NMR titrations (Fig. 5). The long peptide linker between WLN4 and adjoining domains may allow WLN4 to change orientations and interact with either HAH1 or one of the other metal-binding domains. In our in vitro titration experiments, WLN4 passes copper first to WLN6 and then to WLN5. In vivo, after WLN5–6 acquires copper, it likely pivots and passes copper toward the conserved CIACPC motif in the transmembrane domain. It is possible that other metal-binding domains (i.e., WLN2) also transfer copper to WLN5–6.
Fig. 5.
One of the paths for copper transfer from HAH1 to the N terminus of WLNP. (Step 1) First a complex forms between HAH1 and WLN4 (denoted 4), allowing rapid copper transfer between donor and acceptor. (Step 2) apoHAH1 diffuses away from WLN4, and, then (Step 3), WLN4 transfers copper to WLN5–6. Interdomain residue spacing is noted with numbers. Theoretically, the 57-aa linking region between WLN4 and WLN5–6 could allow the copper-binding site of WLN4 to access both domains of WLN5–6. Other targets of HAH1, i.e., WLN2, may function similarly.
Walker et al. (28) proposed that WLN2 is a primary target for Cu(I)HAH1 based on selective protection of WLN2 (relative to other domains) from labeling with a thiol-specific probe under conditions in which one copper was transferred from HAH1 to N-WLNP. They also showed that a double mutant of WLNP, with the metal-binding Cys of WLN2 mutated to Ala, revealed a decrease in Cu(I)HAH1-dependent catalytic phosphorylation. Similarly, our titration studies between Cu(I)HAH1 and WLN2 indicate the formation of a HAH1/Cu(I)/WLN2 adduct; however, it has lower affinity than that formed between HAH1 and WLN4. The observed lower affinity of WLN2 toward HAH1 with respect to WLN4 is in agreement with the two-hybrid assays that show that the interaction between HAH1 and WLN4 (or WLN1–4) is significantly stronger than the interaction with WLN2 (26, 27). Our observation of a protein–protein adduct of Cu(I)HAH1 with WLN4 or WLN2, which are in a fast exchange regime on the NMR time scale, requires a high dissociative rate constant (42), as observed for the Atx1/Ccc2a adduct with a koff > 103 s−1 (43).
Essential Role of WLN5–6.
A different function for WLN5–6 with respect to the other metal-binding domains has been suggested, because WLN5–6 is found closest to the membrane and therefore may gate copper transfer to the intermembrane CIACPC site. The crucial role that WLN5 and WLN6 play in the function of WLNP are highlighted by two disease-causing missense mutations found in these domains, L492S and G591D (22), located in β1 of WLN5 and turn 2 of WLN6, respectively. In our structure of WLN5–6, Leu 492 (residue 8 of WLN5–6) extends into the hydrophobic core of the protein, and replacement with a hydrophilic residue would disrupt the hydrophobic interactions in the interior of WLN5, thus affecting the protein fold. Gly-591 (residue 107 of WLN5–6) is highly conserved among all of the Wilson domains and is located in turn 2 of WLN6. Alteration of Gly-591 to an Asp likely destabilizes this turn because there is not room for a side chain at that position, consistent with protein expression studies (data not shown) in which the G591D mutant of WLN6 is insoluble, whereas native WLN6 is soluble at millimolar concentrations.
Conclusion.
In summary we have proposed a pathway for copper transfer from the HAH1 metallochaperone to N-WLNP. We have shown that HAH1 does not interact with WLN5–6 but instead forms a protein–protein adduct with WLN4 and WLN2, like that observed between yeast Atx1 and Ccc2a (43), which are known to transfer copper in a reversible manner (38). We also monitored copper transfer between the metal-binding domains WLN4 and WLN5–6. The solution structure of WLN5–6 reveals that these two domains function as a unit but acquire copper differently because of the location of metal-binding motifs on opposite ends of the molecule. Certain metal-binding domains function to accept copper from HAH1, whereas others acquire copper from another ATPase metal-binding domain within the N terminus. The mechanism proposed here is consistent with that recently suggested for the analogous Menkes protein, for which the interaction between a construct containing metal-binding domains 4–6 and HAH1 was studied (44). In addition, in that case, it is proposed that the fourth domain is one of the sites for the initial copper transfer from the partner Cu(I)HAH1.
Materials and Methods
Cloning, Purification, and NMR Sample Preparation of WLN 5–6, WLN2, and WLN4 Domains.
WLN5–6, WLN4, and WLN2 were cloned and expressed in Escherichia coli by using standards methods. 15N and 15N/13C-labeled WLN5–6 were expressed in minimal media. Further details, including metallation, and preparation of NMR samples is described in the supporting information. 15N-labeled and unlabeled HAH1 protein samples were purchased from ProtEra (Florence, Italy).
NMR Experiments and Structure Analysis.
The NMR spectra were acquired on Avance 900, 800, 600, and 500 Bruker spectrometers operating at proton-nominal frequencies of 900.13, 800.13, 600.13, and 500.13 MHz, respectively, using a triple-resonance (TXI 5-mm) probe equipped with pulsed field gradients along the x, y, and z axes. The 900, 800, and 500 MHz have a triple-resonance cryoprobe.
The NMR experiments recorded on 13C/15N and 15N enriched samples of apoWLN5–6 are summarized in the supporting information. All spectra were collected at 298 K, processed with the standard Bruker software (xwinnmr), and analyzed through the cara program. Resonance assignments are reported in the supporting information.
Structure calculations were performed through iterative cycles of dyana (45) followed by restrained energy minimization with amber 5.0 (46) applied to each member of the family. An automated candid (47) approach combined with the fast dyana torsion angle dynamics algorithm was used to assign the ambiguous NOE cross-peaks and to have a preliminary protein structure. The structure of apoWLN5–6 displays the following secondary structure elements: 4–11 (β1), 18–27 (α1), 34–38 (β2), 43–48 (β3), 55–65 (α2), 68–71 (β4), 81–87(β5), 92–104 (α3), 110–114 (β6), 118–124 (β7), 131–141 (α4), and 144–146 (β4). The NMR and refinement statistics for apoWLN5–6 are reported in the supporting information. The 30 structures with the lowest energy were analyzed with procheck-nmr (48). Analysis of the Ramachandran plot showed that 80% of residues were in the most favored regions, 14.7% were in the allowed regions, 3.7% were in the generously allowed regions, and 1.6% were in the disallowed regions. The relative orientation of the two domains was determined by aligning the principal axes of the rotational diffusion tensor of the whole molecule “seen” by each of the domains, using the method of Vardan et al. (30), Ghose et al. (49), and Fushman et al. (50) (for details, see the supporting information). Relaxation measurements and determination of R1 and R2 values are reported in the supporting information.
NMR Titrations of WLN4, WLN2, and WLN5–6.
Titrations of 15N-labeled apoWLN5–6 with unlabeled Cu(I)HAH1 or Cu(I)WLN4 and of 15N-labeled Cu(I)HAH1 with unlabeled WLN4 were performed with NMR spectroscopy by following the 1H–15N spectral changes in 1H–15N HSQC spectra upon addition of increasing amounts of the unlabeled protein partner. Similarly, 15N-labeled apoWLN2 was also titrated with 15N-labeled Cu(I)HAH1. All two-dimensional 1H–15N HSQC spectra were recorded with an INEPT delay of 2.5 ms, a recycle time of 1 s, and with spectral windows of 14 and 40 ppm for the 1H and 15N dimensions, respectively. Two-dimensional total correlated spectroscopy and NOESY and three-dimensional NOESY-15N HSQC experiments were also recorded on Cu(I)-15NHAH1/apoWLN4 samples with protein concentration ratios of 1:0.5 and 1:1 to obtain the resonance assignment of HAH1 in the protein complex. Aliquots were added in a Coy chamber under a nitrogen atmosphere at 298 K using a Hamilton syringe to deliver small amounts of unlabeled proteins to the labeled samples in NMR tubes. The 15N, 1H resonance assignment of apo and Cu(I)HAH1 is available from ref. 20.
The metallation of the WLN5 and WLN6 domains was monitored through a few residues (Val-19, Val-95, Ser-20, and His-96), which are next to the Cu(I)-binding Cys-18 and Cys-94, respectively, and therefore show a large chemical shift difference between the apo- and Cu(I)-loaded forms. In addition, their NH crosspeaks are not overlapped in the 1H–15N HSQC maps of apo and Cu(I)WLN5–6, and, therefore, they are easily integrated during the titration steps. The resonance assignment of backbone NH amides of Cu(I)WLN5–6 was obtained from the analysis of two-dimensional 1H–15N HSQC and three-dimensional NOESY-15N HSQC spectra.
The titrations Cu(I)HAH1 plus apo15NWLN5–6, Cu(I)WLN4 plus apo15NWLN5–6, Cu(I)15NHAH1 plus apoWLN4, and Cu(I)15NHAH1 plus apo15NWLN2 were performed at different protein concentrations (a difference of more than one order of magnitude) to verify that interactions were not sample-condition-driven.
Supplementary Material
Acknowledgments
We thank the staff of the Center for Magnetic Resonance at the University of Florence for assistance, Prof. D. Fushman and Dr. R. Varadan (both at University of Maryland, College Park) for providing the tensor alignment program and for helpful discussions, and Dr. Todd Barkman (Western Michigan University) for DNA sequencing. This work was supported by National Institutes of Health Grant GM073634-01 (to D.L.H.) and European Commission Structural Proteomics in Europe Contract QLG2-CT-2002-00988 through Ente Cassa Risparmio di Firenze and Progetto Promelab.
Abbreviations
- HSQC
heteronuclear single quantum correlation
- NOE
nuclear Overhauser effect
- WLNP
Wilson protein
- N-WLNP
N terminus of WLNP.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structural restraints for apoWLD5–6 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2EW9).
This estimate for percent of adduct is based on the fact that the overall rotational correlation time of the adduct is assumed to be double, because it has a molecular mass approximately double that of the single domain.
References
- 1.Tanzi R. E., Petrukhin K., Chernov I., Pellequer J. L., Wasco W., Ross B., Romano D. M., Parano E., Pavone L., Brzustowicz L. M., et al. Nat. Genet. 1993;5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 2.Bull P. C., Thomas G. R., Rommens J. M., Forbes J. R., Cox D. W. Nat. Genet. 1993;5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 3.Lutsenko S., Petris M. J. J. Membr. Biol. 2003;191:1–12. doi: 10.1007/s00232-002-1040-6. [DOI] [PubMed] [Google Scholar]
- 4.Thomas G. R., Forbes J. R., Roberts E. A., Walshe J. M., Cox D. W. Nat. Genet. 1995;9:210–217. doi: 10.1038/ng0295-210. [DOI] [PubMed] [Google Scholar]
- 5.Mercer J. F. Trends Mol. Med. 2001;7:64–69. doi: 10.1016/s1471-4914(01)01920-7. [DOI] [PubMed] [Google Scholar]
- 6.Hamza I., Schaefer M., Klomp L. W., Gitlin J. D. Proc. Natl. Acad. Sci. USA. 1999;96:13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan D. S., Stearman R., Dancis A., Dunn T., Beeler T., Klausner R. D. Proc. Natl. Acad. Sci. USA. 1995;92:2632–2636. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terada K., Nakako T., Yang X. L., Iida M., Aiba N., Minamiya Y., Nakai M., Sakaki T., Miura N., Sugiyama T. J. Biol. Chem. 1998;273:1815–1820. doi: 10.1074/jbc.273.3.1815. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer M., Hopkins R. G., Failla M. L., Gitlin J. D. Am. J. Physiol. 1999;276:G639–G646. doi: 10.1152/ajpgi.1999.276.3.G639. [DOI] [PubMed] [Google Scholar]
- 10.Bull P. C., Cox D. W. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 11.DiDonato M., Narindrasorasak S., Forbes J. R., Cox D. W., Sarkar B. J. Biol. Chem. 1997;272:33279–33282. doi: 10.1074/jbc.272.52.33279. [DOI] [PubMed] [Google Scholar]
- 12.Lutsenko S., Petrukhin K., Cooper M. J., Gilliam C. T., Kaplan J. H. J. Biol. Chem. 1997;272:18939–18944. doi: 10.1074/jbc.272.30.18939. [DOI] [PubMed] [Google Scholar]
- 13.Gitschier J., Moffat B., Reilly D., Wood W. I., Fairbrother W. J. Nat. Struct. Biol. 1998;5:47–54. doi: 10.1038/nsb0198-47. [DOI] [PubMed] [Google Scholar]
- 14.Banci L., Bertini I., Del Conte R., D’Onofrio M., Rosato A. Biochemistry. 2004;43:3396–3403. doi: 10.1021/bi036042s. [DOI] [PubMed] [Google Scholar]
- 15.Banci L., Bertini I., Ciofi-Baffoni S., Chasapis C. T., Hadjiliadis N., Rosato A. FEBS J. 2005;272:865–871. doi: 10.1111/j.1742-4658.2004.04526.x. [DOI] [PubMed] [Google Scholar]
- 16.Arnesano F., Banci L., Bertini I., Ciofi-Baffoni S., Molteni E., Huffman D. L., O’Halloran T. V. Genome Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- 17.Arnesano F., Banci L., Bertini I., Huffman D. L., O’Halloran T. V. Biochemistry. 2001;40:1528–1539. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- 18.Rosenzweig A. C., Huffman D. L., Hou M. Y., Wernimont A. K., Pufahl R. A., O’Halloran T. V. Struct. Folding Des. 1999;7:605–617. doi: 10.1016/s0969-2126(99)80082-3. [DOI] [PubMed] [Google Scholar]
- 19.Wernimont A. K., Huffman D. L., Lamb A. L., O’Halloran T. V., Rosenzweig A. C. Nat. Struct. Biol. 2000;7:766–771. doi: 10.1038/78999. [DOI] [PubMed] [Google Scholar]
- 20.Anastassopoulou I., Banci L., Bertini I., Cantini F., Katsari E., Rosato A. Biochemistry. 2004;43:13046–13053. doi: 10.1021/bi0487591. [DOI] [PubMed] [Google Scholar]
- 21.O’Halloran T. V., Culotta V. C. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 22.Loudianos G., Dessi V., Lovicu M., Angius A., Nurchi A., Sturniolo G. C., Marcellini M., Zancan L., Bragetti P., Akar N., et al. Hum. Mutat. 1998;12:89–94. doi: 10.1002/(SICI)1098-1004(1998)12:2<89::AID-HUMU3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Cater M. A., Forbes J., La Fontaine S., Cox D., Mercer J. F. Biochem. J. 2004;380:805–813. doi: 10.1042/BJ20031804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes J. R., Hsi G., Cox D. W. J. Biol. Chem. 1999;274:12408–12413. doi: 10.1074/jbc.274.18.12408. [DOI] [PubMed] [Google Scholar]
- 25.Iida M., Terada K., Sambongi Y., Wakabayashi T., Miura N., Koyama K., Futai M., Sugiyama T. FEBS Lett. 1998;428:281–285. doi: 10.1016/s0014-5793(98)00546-8. [DOI] [PubMed] [Google Scholar]
- 26.Larin D., Mekios C., Das K., Ross B., Yang A. S., Gilliam T. C. J. Biol. Chem. 1999;274:28497–28504. doi: 10.1074/jbc.274.40.28497. [DOI] [PubMed] [Google Scholar]
- 27.van Dongen E. M., Klomp L. W., Merkx M. Biochem. Biophys. Res. Commun. 2004;323:789–795. doi: 10.1016/j.bbrc.2004.08.160. [DOI] [PubMed] [Google Scholar]
- 28.Walker J. M., Huster D., Ralle M., Morgan C. T., Blackburn N. J., Lutsenko S. J. Biol. Chem. 2004;279:15376–15384. doi: 10.1074/jbc.M400053200. [DOI] [PubMed] [Google Scholar]
- 29.Banci L., Bertini I., Cantini F., Migliardi M., Rosato A., Wang S. J. Mol. Biol. 2005;352:409–417. doi: 10.1016/j.jmb.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 30.Varadan R., Walker O., Pickart C., Fushman D. J. Mol. Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 31.Banci L., Bertini I., Ciofi-Baffoni S., Gonnelli L., Su X. C. J. Biol. Chem. 2003;278:50506–50513. doi: 10.1074/jbc.M307389200. [DOI] [PubMed] [Google Scholar]
- 32.Cantor R. C., Schimmel P. R. Biophysical Chemistry. San Francisco: Freeman; 1980. [Google Scholar]
- 33.Arnesano F., Banci L., Bertini I., Bonvin A. M. Structure (London) 2004;12:669–676. doi: 10.1016/j.str.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Arnesano F., Banci L., Bertini I., Capozzi F., Ciurli S., Luchinat C., Mangani S., Ciofi-Baffoni S., Rosato A., Turano P., Viezzoli M. S. Coord. Chem. Rev. 2006 in press. [Google Scholar]
- 35.Wernimont A. K., Yatsunyk L. A., Rosenzweig A. C. J. Biol. Chem. 2004;279:12269–12276. doi: 10.1074/jbc.M311213200. [DOI] [PubMed] [Google Scholar]
- 36.Radford D. S., Kihlken M. A., Borrelly G. P., Harwood C. R., Le Brun N. E., Cavet J. S. FEMS Microbiol. Lett. 2003;220:105–112. doi: 10.1016/S0378-1097(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 37.Multhaup G., Strausak D., Bissig K. D., Solioz M. Biochem. Biophys. Res. Commun. 2001;288:172–177. doi: 10.1006/bbrc.2001.5757. [DOI] [PubMed] [Google Scholar]
- 38.Huffman D. L., O’Halloran T. V. J. Biol. Chem. 2000;275:18611–18614. doi: 10.1074/jbc.C000172200. [DOI] [PubMed] [Google Scholar]
- 39.Pufahl R. A., Singer C. P., Peariso K. L., Lin S.-J., Schimdt P., Culotta V. C., Penner-Hahn J. E., O’Halloran T. V. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 40.Huffman D. L., O’Halloran T. V. Annu. Rev. Biochem. 2001;70:677–701. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 41.Rosenzweig A. C. Acc. Chem. Res. 2001;34:119–128. doi: 10.1021/ar000012p. [DOI] [PubMed] [Google Scholar]
- 42.Crowley P. B., Ubbink M. Acc. Chem. Res. 2003;36:723–730. doi: 10.1021/ar0200955. [DOI] [PubMed] [Google Scholar]
- 43.Arnesano F., Banci L., Bertini I., Cantini F., Ciofi-Baffoni S., Huffman D. L., O’Halloran T. V. J. Biol. Chem. 2001;276:41365–41376. doi: 10.1074/jbc.M104807200. [DOI] [PubMed] [Google Scholar]
- 44.Banci L., Bertini I., Cantini F., Chasapis C. T., Hadjiliadis N., Rosato A. J. Biol. Chem. 2005;280:38259–38263. doi: 10.1074/jbc.M506219200. [DOI] [PubMed] [Google Scholar]
- 45.Guntert P., Mumenthaler C., Wuthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 46.Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E., Ferguson D. M., Seibel G. L., Singh U. C., Weiner P. K., Kollman P. A. amber 5.0. San Francisco: University of California; 1997. [Google Scholar]
- 47.Herrmann T., Guntert P., Wuthrich K. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., Thornton J. M. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 49.Ghose R., Fushman D., Cowburn D. J. Magn. Reson. 2001;149:204–217. doi: 10.1006/jmre.2001.2295. [DOI] [PubMed] [Google Scholar]
- 50.Fushman D., Varadan R., Assfalg M., Walker O. Prog. Nucl. Magn. Reson. Spectrosc. 2004;44:189–214. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.