Abstract
The small subpopulation of high-affinity EGF receptors (EGFRs) on living cells revealed by Scatchard analysis of 125I-EGF binding results was discovered nearly three decades ago, yet not much is known about the underlying mechanism. After the determination of the structure of different forms of EGFR extracellular domain it was proposed that the monomeric tethered configuration corresponds to the majority of low-affinity receptors, whereas the extended dimeric configuration corresponds to the minority of the high-affinity class of EGFRs. Mathematical modeling of EGF-binding experiments to different conformational mutants of EGFR has shown that the high-affinity class of EGFR on living cells does not correspond to the extended configuration of EGFR and can only be accounted for by including in the mathematical model an additional binding event that is attributed to the dynamic nature of EGFR on living cells. To circumvent this problem we have performed similar experiments in the background of an EGFR mutant that does not form high-affinity sites. Quantitative analysis and mathematical modeling of these data show that release of the intramolecular tether causes a 2-fold increase in EGF-binding affinity, whereas elimination of the dimerization arm reduces EGF-binding affinity by ≈6-fold. These experiments confirm the salient features of the structural model for EGFR regulation and argue further that the intramolecular tether provides only limited autoinhibitory control of EGFR activity and that the low-affinity class of EGF-binding sites on living cells reflects interconverting, tethered, and extended receptor configurations.
Keywords: cell signaling, growth factors, ligand binding, mathematical modeling, surface receptors
The protein tyrosine kinase (PTK) activities of the EGF receptor (EGFR) family of receptor tyrosine kinases (RTKs) are normally subject to multiple regulatory mechanisms before and after growth factor stimulation (reviewed in refs. 1 and 2). It has been shown that deregulation of the PTK activity or constitutive tyrosine phosphorylation of members of the EGFR family of RTKs play an important role in a great variety of human cancers (reviewed in ref. 3).
One of the oldest unsolved questions related to the mechanism of action of EGFR is the origin of the curvilinear Scatchard plots seen in quantitative analyses of 125I-labeled EGF binding to living cells bearing EGFRs (4). A region of the Scatchard plot with steep slope is thought to represent high-affinity receptors that bind EGF with a KD of ≈10–100 pM, accounting for 2–5% of the receptors. The second, more shallow, slope is thought to correspond to an apparent low-affinity class (KD 1–10 nM), representing the majority (95–98%) of receptors (4, 5). Several studies have proposed that the high-affinity receptors are the primary mediators of EGFR signaling (reviewed in ref. 5).
At first glance, the two affinity states of EGFR detected by Scatchard analysis in living cells seem to be consistent with the occurrence of two distinct conformations of the receptor extracellular domain that were proposed on the basis of structural studies (reviewed in ref. 6). The monomeric, unoccupied EGFR is maintained in an autoinhibited low-affinity state by intramolecular interactions between a specific loop from the cysteine-rich domain II and a binding site on domain IV (designated the “tethered” configuration). It is thought that this tethered monomeric configuration is in equilibrium with an “extended” configuration that exhibits higher ligand-binding affinity and is competent for dimer formation and resulting tyrosine kinase activation. EGF binding promotes dimerization of EGFR that is driven by receptor–receptor interactions involving the same loop in domain II (the “dimerization arm”) that is responsible for forming the intramolecular tether in the monomer. Quantitative EGF-binding experiments using the soluble extracellular domain of EGFR showed that breaking the tether (thus favoring the extended configuration) increases EGF binding affinity by ≈3-fold (7).
Although the tethered and extended forms have different EGF-binding affinities, our mathematical modeling showed that a population of receptors alternating between these two states cannot produce the experimentally observed curvilinear Scatchard plots of EGF-binding experiments to living cells (8). To yield such curved plots, an unspecified “external” site that binds dimerized receptors with high affinity had to be included in the model (8). Furthermore, our attempts to study the effect of extracellular domain mutations on EGF-binding affinities suffered from complications that are likely caused by the dynamic nature of EGFRs in living cells that undergo rapid endocytosis and degradation (9). These processes are mediated by tyrosine phosphorylation and interactions with components of coated pits and other elements of the endocytic machinery.
To remove this complication from our studies, we have taken advantage of an observation that we made two decades ago. Deletion of the EGFR cytoplasmic domain leads to defects in internalization and down-regulation (10) and, most interestingly, abolishes the high-affinity binding sites seen with wild-type EGFR on the cell surface. The mutated EGFR retains the capacity to undergo EGF-induced dimerization and EGF-induced heterodimerization with wild-type EGFR expressed in the same cells (11, 12). Importantly, the internalization-deficient EGFR mutant (EGFR-P) gives linear rather than the typical curved Scatchard plots (10), simplifying analysis of the effects of extracellular mutations on ligand-binding affinity. Interestingly, a similar loss of high-affinity EGF-binding sites is seen upon overexpression of a dynamin mutant that impairs clathrin-dependent endocytosis (13).
To enable quantitative analysis of EGF binding to living cells, we introduced targeted point mutations and deletions that disrupt the autoinhibitory tether or receptor dimerization in the background of EGFR-P and have explored the effects of these conformational alterations on EGF-binding characteristics. Because these EGF-binding experiments are performed with receptors expressed on the surface of living cells, this study is a more physiologically relevant system than studies using only the soluble extracellular domain. Our results argue that the so-called low-affinity class of EGF-binding sites reflects interconverting tethered and extended configurations. Moreover, mutational disruption of the tether increased EGF-binding affinity by only ≈2-fold, indicating that the tether exerts only limited autoinhibitory control.
Results and Discussion
We first analyzed the EGF-binding profile of the EGFR cytoplasmicdomain deletion mutant (EGFR-P) that serves as both a platform and a standard for determining the contribution of different critical amino acid residues in the extracellular domain toward EGF binding affinity. The IC50 obtained from an EGF displacement curve for cells expressing EGFR-P treated with 125I-labeled EGF was found to be 6 nM, and Scatchard analysis of 125I-labeled EGF-binding to the same cells showed a best fit to a single binding constant with an apparent KD of 6.4 ± 1.0 nM (Figs. 1 and 2A), similar to the value published by Livneh et al. (10), and in the expected range for the majority of so-called low-affinity EGF-binding sites seen with the wild-type receptor.
Fig. 1.
Homologous binding inhibition of 125I-EGF to EGFR mutants. Mouse 2.2 cells (3T3 devoid of endogenous EGFR) stably expressing EGFR-P (♦), dimerization mutants [D279A/H280A (□), Del:242–259 (×), and Y251A/R285S (•)], and intramolecular tether mutants [D563A/H566A/K585A (■) and Del:560–590 (▴)] were seeded on 24-well plates and grown to confluence. Cells were treated with a fixed concentration of (3.1 ng/ml) 125I-EGF and an increasing concentration of unlabeled EGF for 1 h at room temperature. After washing with PBS the remaining bound radioactivity was detected by using a liquid scintillation counter LS 6500 (Beckman). (Inset) Expression levels of EGFR mutants in 2.2 3T3 cells. Stably transfected cells harboring various extracellular domain mutations in a background of EGFR-P with a myc tag in the C terminus of the molecule are shown. Monoclonal anti-EGFR antibody (mAb108) was used for immunoprecipitation followed by detection with anti-Myc antibodies. The mutants are EGFR-P (a), Del:242–249 (b), Y251/R285S (c), D279/H280 (d), D563A/H566/A/K585A (e), and Del:560–590 (f).
Fig. 2.
Scatchard plots of EGF-binding data obtained from cells expressing EGFR-P (A) or versions of EGFR-P mutated in the autoinhibitory tether region, Del:560–590 (B) and D563A/H566A/K585A (C). Cells were grown to confluence in 24-well plates and incubated with increasing concentrations of 125I-EGF in triplicate for 60 min at room temperature. A 100-fold excess of unlabeled EGF was simultaneously added to the third well to determine nonspecific binding. Curve fitting to saturation binding data (Insets) was performed by nonlinear regression using prism 3.03 software (GraphPad). EGFR-P cells express ≈600,000 EGFR per cell (A). The EGF-binding results best fit a linear Scatchard plot, although signs of convexity can already be observed. The convexity is much more pronounced in Scatchard plots of EGF-binding results to EGFR-P cells expressing 180,000 EGFRs per cell (see Fig. 7).
The Role of Intramolecular Tether.
The effect of the autoinhibitory tether was determined by analyzing two different mutants: one devoid of the entire region in domain IV (amino acids 560–590) responsible for intramolecular tether formation (Del:560–590), and a second in which three critical residues that participate in domain II/IV interactions were replaced with alanine (D563A/H566A/K580A). The IC50 and apparent KD values were 2.5 nM and 2.7 ± 0.5, respectively, for the Del:560–590 mutant (Figs. 1 and 2B) and 3.0 nM and 3.2 ± 0.5 nM, respectively, for the D563A/H566A/K580A mutant (Figs. 1 and 2C). Thus, both of these mutations, which disrupt all tether interactions seen in the crystal structure (7), cause an ≈2- to 3-fold increase in EGF-binding affinity compared with values obtained for EGF binding to cells expressing the EGFR-P standard. In addition, Scatchard plots of the binding data produced concave-down (convex) curves for both mutations. Computational simulation of ligand binding and dimerization of EGFR (see below) demonstrates that concave-down curves observed in Scatchard plots can be generated in the presence of detectable dimerization. We also measured the dissociation constants for EGFR-P and D563A/H566A/K585A mutants by applying the one-phase exponential decay model to 125I-EGF dissociation data. A decrease in the dissociation rate by a factor of two was detected for the extended EGFR extracellular domain (D563A/H566A/K585A) as compared with the EGFR-P control (Fig. 6, which is published as supporting information on the PNAS web site).
As a control, we performed EGF-binding experiments with cells expressing wild-type EGFR. These experiments revealed typical curvilinear Scatchard plots of two affinity-binding classes with apparent KD values of 130 pM and 7 nM, corresponding to the high- and low-affinity receptor classes, respectively (Fig. 7, which is published as supporting information on the PNAS web site). These affinities are in good agreement with the previously reported affinities for the full-length EGFR (4). Taken together, these experiments show that a mutated EGFR that is defective in internalization does not exhibit the high-affinity class seen with wild-type EGFRs on living cells. Moreover, disruption of the intramolecular tether led to a 2- to 3-fold increase in the strength of EGF binding to these low-affinity sites.
Disruption of the Dimerization Interface.
Three different types of mutations were generated in the EGFR-P background to address the question of how disrupting the dimerization interface influences EGF binding characteristics. In the first mutant, the entire dimerization arm (amino acids 242–259) was deleted to generate a mutant designated Del:242–259. This mutant gave a linear Scatchard plot with an apparent KD of ≈36 nM (Fig. 3B). Deleting the entire dimerization arm thus reduces EGF-binding affinity by 6-fold compared with the EGFR-P control (Table 1). In a second mutant, two amino acids forming hydrogen bonds across the dimer interface were mutated (reviewed in ref. 6); Tyr-251 was replaced with alanine, and Arg-285 was replaced with serine (Y251A/R285S). Nonlinear curve fitting to saturation-binding data for Y251A/R285S produced a concave-down curvature with a best fit to an apparent KD of 16 ± 2 nM (Fig. 3C), which represents a 3-fold decrease in affinity compared with EGFR-P (apparent Kd of ≈6 nM) (Fig. 3A). Furthermore, the measurements of IC50, ≈10 nM for this mutant (Fig. 1) compared with ≈ 3 nM for EGFR-P, is in good agreement with these saturation-binding data. Additional important intermolecular contacts across the dimerization interface are provided by the side chains of Asp-279 and His-280 (14), which both were replaced with alanines in D279A/H280A. Nonlinear curve-fitting to 125I-EGF-binding data for (D279A/H280A) produced a best fit of ≈16 nM (Fig. 3D), similar to the value measured for the Y251A/R285S mutant. IC50 values for these two dimerization-defective mutants are in agreement with the affinity constants obtained from saturation-binding data (Table 1). The fact that the two double-point mutations both reduce EGF-binding affinity to a smaller extent than seen with the mutant lacking the entire dimerization arm suggests that receptor dimerization is incompletely disrupted in the Y251A/R285S and D279A/H280A mutants.
Fig. 3.
Scatchard plots of EGF-binding data obtained from various dimerization-impaired EGFR mutants in an EGFR-P background. 3T3 2.2 cells stably expressing EGFR-P (A) or EGFR-P bearing the mutations Del:242–259 (B), Y251A/R285S (C), or D279A/H280A (D) were grown in 24-well plates as described. Triplicate wells were treated with increasing concentrations of 125I-labeled EGF, including 100-fold excess unlabeled EGF in the third well of each concentration at room temperature. The average of three independent experiments is indicated with the SE. (Insets) Saturation binding curves.
Table 1.
Apparent binding affinities and receptor numbers per cell for different EGFR mutants detected by nonlinear curve fitting to binding data
| EGFR mutants | IC50, nM | KD, nM | No. of receptors per cell | Apparent concentrations, μM |
|---|---|---|---|---|
| EGFR-P | 6 | 6.4 ± 1.0 | 180,000 | 5.45 |
| Del:242–259 | 20 | 34.5 ± 1.2 | 120,000 | 2.97 |
| D279A/H280A | 10.5 | 15.6 ± 2.6 | 140,000 | 3.74 |
| Y251A/R285S | 10 | 16 ± 2 | 210,000 | 6.87 |
| D563A/H566A/K585A | 3.0 | 3.2 ± 0.5 | 120,000 | 2.97 |
| Del:560–590 | 2.5 | 2.7 ± 0.5 | 110,000 | 2.61 |
Apparent KD and Bmax values obtained were average of three independent experiments with the indicated standard errors. Model predictions for the apparent receptor concentrations are indicated.
Description of Model.
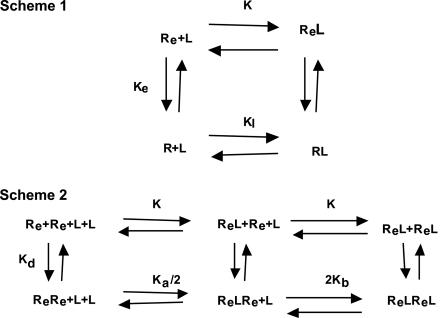
We previously introduced an equilibrium model (8) for ligand-induced dimerization of EGFR that takes into account the autoinhibition of the receptor. Here, we expand this model (Fig. 4) to enable simulation of both the release of the autoinhibitory tether and disruption of receptor dimerization. Both simulations are compared with experimental results. In addition, we incorporate into the model distinct affinity constants for the three distinguishable binding events, namely, binding to receptor monomers, binding to unoccupied receptor dimers, and binding to dimerized receptor that has one ligand molecule bound. This latter extension of the model enables us to account for the convexity displayed in the Scatchard plot of the mutant with complete disruption of the autoinhibitory tether (Figs. 2C and 6). The equilibrium equations (models I, II, and III) used to solve the model are shown below.
Fig. 4.
Schematic depiction of the interactions between EGF and EGFR together with their dissociation constants. In Scheme 1, an EGFR in the conformationally extended state (Re) binds EGF (L) with higher affinity than a receptor in the autoinhibited state (R). In Scheme 2, receptors in the extended state can dimerize in both the absence and presence of ligand.
Rewriting 1–6:
Model I = Eqs. 1′–3′ = disrupted dimerization.
 |
Model II = Eqs. 3′–6′ = disrupted autoinhibition.
 |
Model III = Eqs. 1′–6′ = wild type with deleted cytoplasmic domain.
 |
To compute the apparent affinity in a simulation we used a simple linear regression on the simulated Scatchard plot (15). In the case of convex plots we generally performed the regression on the right-most part of the simulated Scatchard plot to ensure that any systematic bias resulting from the convexity of the plot did not substantially alter the results. Although linear regression is not advisable in connection with experimental data, it can be applied for linear simulations. On the other hand, for convex Scatchard plots the apparent dissociation constants can be highly sensitive to the selection of points. Therefore, one should be careful with quantitative conclusions drawn from convex Scatchard plots. The apparent 3D concentrations of EGFR were calculated from the experimentally determined receptor per cell numbers (Table 1), using the same average-distance-to-nearest-neighbor approach as before (8). We have decreased the values used for K and K1 to ≈1 and 100 nM, respectively, from our previous estimates for binding to soluble forms of EGFR (8) because of the effect of membrane dimensionality on the on-rates and to better fit the experimental data. It is noteworthy that the apparent ligand-binding affinity as measured in this article for the internalization-impaired EGFR-P mutants is in good agreement with the previously determined affinity class corresponding to the shallow (low-affinity) part of the Scatchard plot as measured for full-length EGFRs on living cells. This result argues that the shallow part of the Scatchard plot corresponds to the binding of ligand to a receptor that can exist in the three states of tethered, extended, and dimer, whereas the steeper part of the Scatchard plot corresponds to a binding event involving external factors of as-yet-undetermined nature, for which the intact intracellular domain is required. In Fig. 5IIIa we show a simulated Scatchard plot that is in good agreement with the experimental plot for EGFR-P (Fig. 7B).
Fig. 5.
Parameterization for models I, II, and III. (Ia) Simulated Scatchard plot for model I. In the absence of dimerization the plot is not able to demonstrate the existence of two separate binding affinities. Model inputs were: K = 2 nM, Kl = 100 nM, Ke = 30, and RT = 5 μM. The apparent affinity is 39 nM. (II a–c) Simulated Scatchard plots for model II show that in the absence of autoinhibition dimerization alone is not able to produce positive cooperativity. (IIa) Model inputs were: K = 1 nM, Kd = 1 μM, Ka = 1 nM, Kb = 1 nM, and RT = 5 μM. The apparent affinity is 1 nM. (IIb) Model inputs were: K = 2 nM, Kd = 1 μM, Ka = 1 nM, Kb = 1 nM, and RT = 5 μM. The apparent affinity is 1.15 nM. (IIc) Model inputs were: K = 2 nM, Kd = 1 μM, Ka = 2 nM, Kb = 1 nM, and RT = 5 μM. The apparent affinity is 2.12 nM. (III a–c) Simulated Scatchard plots for model III. Impaired dimerization can still produce concave-down Scatchard plots. (IIIa) Model inputs were: Kl = 100 nM, K = 0.5 nM, Kd = 1 μM, Ka = 0.5 nM, Kb = 0.5 nM, Ke = 30, and RT = 5 μM. The apparent affinity is ≈5–6 nM. (IIIb) Model inputs were: Kl = 100 nM, K = 2 nM, Kd = 10 μM, Ka = 2 nM, Kb = 1 nM, Ke = 30, and RT = 10 μM. The apparent affinity is ≈30–40 nM. (IIIc) Model inputs were: Kl = 100 nM, K = 2 nM, Kd = 5 μM, Ka = 2 nM, Kb = 1 nM, Ke = 30, and RT = 5 μM. The apparent affinity is ≈25–35 nM.
Simulation of Binding Results to Mutants with Impaired Intramolecular Tether.
We used a model that has only one state for receptor (Fig. 4, Scheme 2). If K = Ka = Kb, i.e., if ligand binding is fully independent of dimerization, it is only possible to obtain linear Scatchard plots (Fig. 5IIa). In our experiments with EGFR-P, the Scatchard plots for the mutation Del:560–590 show minor convexity, whereas the plot for D563A/H566A/K585A shows considerable convexity (Fig. 2C). Although we cannot rule out the possibility that the mutations have not completely disrupted the autoinhibition, we also consider the possibility that K, Ka, and Kb are not equal. The primary condition for convexity in our model is that Kb is lower than Ka (Fig. 5 IIb and IIc), i.e., that there is positive cooperativity in EGF binding to EGFR. Because there does not appear to be any direct contact between the two ligands in a fully dimerized complex (6), a potential explanation for such positive cooperativity might be that the first binding event stabilizes a conformation of the second receptor molecule that binds ligand more strongly, so that the cost of binding the second ligand is lower.
Simulation of Binding Results to Mutants with Impaired Dimerization.
To simulate a mutant with impaired dimerization we used a model that has two states for the receptor, namely extended and autoinhibited, but does not allow dimerization (Fig. 4, Scheme 1). This model is only capable of producing linear Scatchard plots (16) (Fig. 5Ia). The simulation of the mutant devoid of the entire dimerization arm (Del:242–259) appears linear. Of the two other mutants studied here, one (D279A/H280A) shows strong convexity (Fig. 3D), whereas another (Y251A/R285S) appears somewhat convex (Fig. 3B). This result suggests that these two mutants are at least capable of limited dimerization, a hypothesis that fits with their apparent EGF-binding affinity. In Fig. 5III b and c we show that it is possible to simulate convex Scatchard plots with model III, using a lower dimerization constant. The apparent affinity obtained in the simulation closely matches that obtained experimentally.
Conclusion
Determination of crystal structures of the EGFR extracellular ligand-binding domain revealed two distinct conformational states: a tethered monomeric autoinhibited configuration and an extended dimeric configuration. Simultaneous engagement of binding sites in both domains I and III by a single ligand molecule causes exposure of a key dimerization arm in domain II and smaller domain II conformational changes that optimize interactions across the dimerization interface (14). Based on the cell surface EGF-binding parameters measured with the approach described here, which removes the complication of a poorly understood high-affinity state, we have refined our previous mathematical model to investigate directly the effects of receptor dimerization and autoinhibition on EGF binding to EGFR in living cells. Using a truncated, EGFR mutant that is impaired in internalization and other parameters, experimental and simulated Scatchard plots are concave-down, indicating positive cooperativity (Fig. 2C). We suggest that this positive cooperativity is normally masked in studies of wild-type EGFR by occurrence of the high-affinity class of receptors, which requires cytoplasmic domain interactions. We analyzed the effects of impairing receptor dimerization on the binding profile of EGF by simulating Scatchard plots at various dimerization affinities and analyzing EGF binding to dimerization site mutants. In both models and experiments, an absence of dimerization converted the concave-down Scatchard plots seen in Fig. 2C, for example, into linear plots (Figs. 3B and 5). Mutants or models that partially disrupt EGFR dimerization retain some concave-down curvature as expected (Figs. 3 C and D and 5). As we showed earlier, however, the concave-up curvature typical of an EGF-binding Scatchard plot for wild-type EGFR can be simulated only by including in the model a saturatable high-affinity external site for receptor dimers (8).
We also show that disruption of the autoinhibitory tether in EGFR-P increases the apparent affinity toward EGF by ≈2- to 3-fold, demonstrating that the intramolecular tether exerts only limited autoinhibitory control on receptor activity (9). Indeed, EGFR mutants devoid of the autoinhibitory tether are not constitutively activated and do not promote deregulated activation of downstream signaling pathways (7, 9, 17). Our experimental findings with EGFR-P, together with mathematical modeling, clearly argue that the so-called low-affinity EGF-binding sites seen in studies of the wild-type EGFR represent the interconverting tethered and extended (dimerizing) form of the receptor extracellular domain inferred from structural studies. Altering the tethered to extended equilibrium directly affects the affinity measured for this average low-affinity state and need not affect the poorly characterized high-affinity state. In addition, these experiments provide further support for the notion that for EGFR to function as a sensitive signaling receptor it must posses multiple layers of not very strong autoinhibitory mechanisms before ligand stimulation (18).
Finally, it is interesting to compare the EGF-binding affinities measured here for cell-surface EGFR-P with those determined by using the isolated extracellular ligand-binding domain, sEGFR (14). The apparent Kd is reduced from 175 nM for wild-type sEGFR to 6.4 nM for wild-type EGFR-P in cell membranes, a 27-fold difference. Considering all mutants studied, anchoring the EGFR extracellular domain to the cell surface appears to increase EGF-binding affinity by 15- to 50-fold, corresponding to ≈2 kcal/mole. Because EGF binding is linked to receptor dimerization, membrane anchoring of the receptor should enhance EGF-binding affinity by increasing the local concentration (and thus dimerization) and reducing the number of degrees of orientational freedom of the receptor. The enhancement in EGF-binding affinity is significantly less than might be predicted based on theoretical considerations (19). However, because experiments with sEGFR (14) use protein concentrations within ≈10-fold of the effective cell-surface EGFR-P concentration (Table 1) the influence of reduced dimensionality in this respect is likely minimal. Enhancement of EGF binding through restricted orientation at the cell surface appears at most to have a modest influence. Thus, our studies of cell-surface EGFR-P argue that the process of EGF binding to two interconverting soluble extracellular domains provides a reasonable physical view of the nature of the low-affinity EGF-binding sites seen with wild-type EGFR in living cells. A major challenge for the future is to determine how this process is altered for a subset of intact receptors and what precisely is the conformation of the minority high-affinity EGF-binding sites on living cells.
Materials and Methods
Preparation of EGFR Mutants and Cell Lines.
Human EGFR was cloned into the retroviral vector pLSV as described (4) and used to prepare a truncated EGFR-P platform mutant lacking most of the cytoplasmic domain. EGFR-P was constructed by fusing together a PCR fragment encoding the N-terminal 630-aa portion of human EGFR with a DNA sequence coding for a myc tag, a stop codon, and a noncoding short stretch of 3′ UTR. This DNA construct was cloned into the mammalian expression vector pBabe-puro by using conventional cloning procedures. All mutations were introduced into the EGFR-P background by using the QuikChange method according to manufacturer specifications (Stratagene) and confirmed by DNA sequencing.
Deletion mutant Del:242–259 lacking the entire dimerization arm of domain II was generated in a single step, whereas deletion mutant Del:560–590 was generated in three steps: removal of residues 560–572, removal of residues 573–581, and finally removal of amino acids 582–590. Lipofectamine 2000 (Invitrogen) was used to introduce viral constructs into GPG cells (a gift from Joan Brugge, Harvard University, Cambridge, MA) according to the manufacturer’s directions. Harvested virus was used to infect 2.2 cells (a derivative of 3T3) lacking endogenous EGFR as described (20).
Immunoprecipitation and Immunoblotting Experiments.
Mouse 3T3 2.2 cells expressing the EGFR mutants were grown in DMEM containing FBS 10%, 100 units/ml streptomycin penicillin, and 1 μg/ml puromycin. Cells were washed twice with cold PBS (1×) and lysed in buffer containing 1% Triton 100-X as described (4, 20, 21). EGFR was immunoprecipitated by incubating cell lysates for 1 h with monoclonal anti-EGFR antibody (mAb108) followed by an additional 1-h incubation with Protein A Sepharose (Zymed). Immunoprecipitates were washed with buffer containing 0.1% Triton X-100, separated by SDS/PAGE, and transferred to nitrocellulose membrane (Bio-Rad). Membrane was blocked for 1 h in 5% BSA/Tris buffered saline and immunoblotted with anti-Myc-tag antibodies (Sigma). Proteins were visualized by incubation with enhanced chemiluminescence (Amersham Pharmacia).
125I-Labeled EGF Binding Experiments.
NIH 3T3 (2.2) cells stably expressing the different EGFR mutants were seeded on 24-well plates and allowed to reach confluence for ≈24 h in DMEM containing 10% FBS. The average cell number per well was obtained from two independent well counts to be used in the calculation of receptor number per cell from the Bmax of nonlinear curve fitting to saturation binding data. Iodination of EGF (Chemicon) was performed by using the Chloramine-T method to 100,000–250,000 cpm per ng (125I from PerkinElmer) as described (9). Briefly, wells in triplicate were treated with concentrations of 125I–EGF ranging from 0.1 to 620 ng/ml at room temperature. Nonspecific binding was determined by including 100-fold excess nonlabeled EGF in the third well of each treated sample. The bound radioactivity was measured by liquid scintillation counter for 10 min. For the ligand replacement experiments the cells were treated with 0.5 nM 125I-EGF in the presence of increasing concentrations of nonlabeled EGF (0–500 ng/ml) for 1 h, and the same procedure described for the saturation binding was followed to quantitate the remaining bound radioactive content. Saturation binding data were analyzed by nonlinear curve fitting by using prism 3.03 software (GraphPad, San Diego).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01-AR051448 (to J.S.), R01-AR051886 (to J.S.), and R01-CA096768 (to M.A.L.) and funds from the Ludwig Institute for Cancer Research (to J.S). F.Ö. is partially supported by a fellowship from the Scientific and Technological Research Council of Turkey.
Abbreviations
- EGFR
EGF receptor
- EGFR-P
internalization-deficient EGFR mutant.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Jorissen R. N., Walker F., Pouliot N., Garrett T. P., Ward C. W., Burgess A. W. Exp. Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P., Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 4.Lax I., Bellot F., Howk R., Ullrich A., Givol D., Schlessinger J. EMBO J. 1989;8:421–427. doi: 10.1002/j.1460-2075.1989.tb03393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ullrich A., Schlessinger J. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 6.Burgess A. W., Cho H. S., Eigenbrot C., Ferguson K. M., Garrett T. P., Leahy D. J., Lemmon M. A., Sliwkowski M. X., Ward C. W., Yokoyama S. Mol. Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson K. M., Berger M. B., Mendrola J. M., Cho H. S., Leahy D. J., Lemmon M. A. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 8.Klein P., Mattoon D., Lemmon M. A., Schlessinger J. Proc. Natl. Acad. Sci. USA. 2004;101:929–934. doi: 10.1073/pnas.0307285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattoon D., Klein P., Lemmon M. A., Lax I., Schlessinger J. Proc. Natl. Acad. Sci. USA. 2004;101:923–928. doi: 10.1073/pnas.0307286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livneh E., Prywes R., Kashles O., Reiss N., Sasson I., Mory Y., Ullrich A., Schlessinger J. J. Biol. Chem. 1986;261:12490–12497. [PubMed] [Google Scholar]
- 11.Kashles O., Yarden Y., Fischer R., Ullrich A., Schlessinger J. Mol. Cell. Biol. 1991;11:1454–1463. doi: 10.1128/mcb.11.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spivak-Kroizman T., Rotin D., Pinchasi D., Ullrich A., Schlessinger J., Lax I. J. Biol. Chem. 1992;267:8056–8063. [PubMed] [Google Scholar]
- 13.Ringerike T., Stang E., Johannessen L. E., Sandnes D., Levy F. O., Madshus I. H. J. Biol. Chem. 1998;273:16639–16642. doi: 10.1074/jbc.273.27.16639. [DOI] [PubMed] [Google Scholar]
- 14.Dawson J. P., Berger M. B., Lin C. C., Schlessinger J., Lemmon M. A., Ferguson K. M. Mol. Cell. Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scatchard G. Ann. N.Y. Acad. Sci. 1949;51:660–672. [Google Scholar]
- 16.Wofsy C., Goldstein B. Math. Biosci. 1992;112:115–154. doi: 10.1016/0025-5564(92)90090-j. [DOI] [PubMed] [Google Scholar]
- 17.Walker F., Orchard S. G., Jorissen R. N., Hall N. E., Zhang H. H., Hoyne P. A., Adams T. E., Johns T. G., Ward C., Garrett T. P., et al. J. Biol. Chem. 2004;279:22387–22398. doi: 10.1074/jbc.M401244200. [DOI] [PubMed] [Google Scholar]
- 18.Schlessinger J. Science. 2003;300:750–752. doi: 10.1126/science.1082024. [DOI] [PubMed] [Google Scholar]
- 19.Grasberger B., Minton A. P., DeLisi C., Metzger H. Proc. Natl. Acad. Sci. USA. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honegger A. M., Szapary D., Schmidt A., Lyall R., Van Obberghen E., Dull T. J., Ullrich A., Schlessinger J. Mol. Cell. Biol. 1987;7:4568–4571. doi: 10.1128/mcb.7.12.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batzer A. G., Rotin D., Urena J. M., Skolnik E. Y., Schlessinger J. Mol. Cell. Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.