Abstract
The molecular mechanism of processive movement of single myosin molecules from classes V and VI along their actin tracks has recently attracted extraordinary attention. Another member of the myosin superfamily, myosin VII, plays vital roles in the sensory function of Drosophila and mammals. We studied the molecular mechanism of Drosophila myosin VIIa, using transient kinetics and single-molecule motility assays. Myosin VIIa moves along actin filaments as a processive, double-headed single molecule when dimerized by the inclusion of a leucine zipper at the C terminus of the coiled-coil domain. Its motility is ≈8–10 times slower than that of myosin V, and its step size is 30 nm, which is consistent with the presence of five IQ motifs in its neck region. The kinetic basis for the processive motility of myosin VIIa is the relative magnitude of the release rate constants of phosphate (fast) and ADP (slow) as in myosins V and VI. The ATPase pathway is rate-limited by a reversible interconversion between two distinct ADP-bound actomyosin states, which results in high steady-state occupancy of a strongly actin-bound myosin species. The distinctive features of myosin VIIa (long run lengths, slow motility) will be very useful in video-based single-molecule applications. In cells, this kinetic behavior would allow myosin VIIa to exert and hold tension on actin filaments and, if dimerized, to function as a processive cargo transporter.
Keywords: fluorescence imaging with one-nanometer accuracy, kinetics, processivity
Myosins constitute a superfamily of actin-dependent motors (1) that play important roles in diverse processes such as vesicle transport, cytokinesis, cell motility, exocytosis, tension maintenance, and filopodia formation (2). Myosin VIIa is essential for sensory function in both Drosophila and mammals (3–5). Mutations in the mammalian myosin VIIa gene cause severe sensory defects (Usher 1B syndrome in humans; shaker-1 mutation in mice) (6). In shaker-1 mice the highly organized arrangement of stereocilia in auditory hair cells is destroyed, and the melanosomes of shaker-1 mice do not enter the apical processes in retinal pigment epithelial cells (7). Slac2-c/MyRIP links melanosome-bound Rab27A and myosin VIIa in retinal cells and thus mediates the transport of retinal melanosomes in pigment epithelium cells in a manner similar to the myosin Va-driven transport of melanosomes in melanocytes through a melanophilin-Rab27a complex (8–11). In Drosophila, myosin VIIa is widely expressed, and mutation of myosin VIIa (called crinkle or ck) is embryonic lethal. Flies that “escape” lethality because of a maternal load of myosin VIIa progress to adulthood and have defects in hearing and actin-based appendages on epithelia (5, 12).
The mechanochemical mechanism of myosin-based motility has been the focus of considerable effort. Myosins that function as single-molecule vesicle transporters must be able to take a large number of steps on the actin filament per diffusional encounter (i.e., they are processive). The processive stepping mechanism of myosins from classes V and VI has been characterized extensively (13–17). In general, these myosins have a kinetic signature where ADP release is rate limiting and phosphate release is fast. Because myosin·ADP binds strongly to actin, these myosins spend most of their kinetic cycle strongly bound to actin and are termed high-duty ratio myosins (18). When dimerized through coiled coils they move processively on actin. Myosin VIIb from both Drosophila and mice also meets this kinetic prerequisite for processivity (19, 20). Here we show that Drosophila myosin VIIa (DmVIIa) has kinetic features tailored for processivity and moves processively on actin filaments as single molecules using a hand-over-hand mechanism with a step size of 30 nm when dimerized through insertion of a leucine zipper sequence at the C terminus of its coiled-coil forming region.
Results
Construct Design.
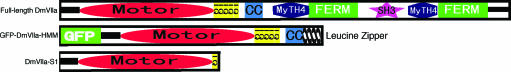
DmVIIa consists of an N-terminal motor domain (containing the actin and ATP binding sites), five light chain-binding IQ motifs, a predicted coiled-coil region that may be responsible for heavy chain dimerization, and various tail domains of effector function (Fig. 1) (5, 12). To investigate the kinetic properties of an isolated head of DmVIIa, we expressed a truncated construct that includes the motor domain and the first predicted IQ motif [DmVIIa–subfragment 1 (S1); Fig. 1]. For in vitro motility assays, we used a two-headed construct, GFP–DmVIIa–heavy meromyosin (HMM), which contained the motor domain, all five predicted IQ motifs, and the entire predicted coiled-coil forming sequence, with an N-terminally fused GFP and a C-terminal GCN4 leucine zipper motif to facilitate dimerization (Fig. 1). In the absence of the GCN4 motif, DmVIIa–HMM failed to dimerize spontaneously.
Fig. 1.
Domain structure of full-length DmVIIa and the constructs used in the present work.
DmVIIa Is a Processive Motor.
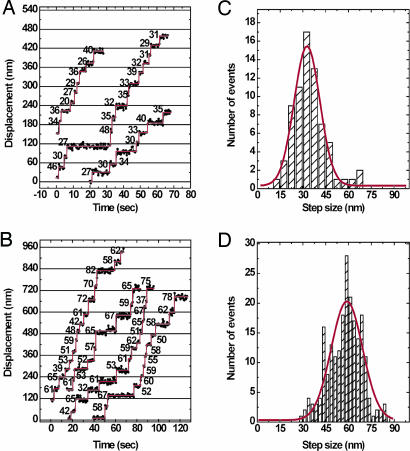
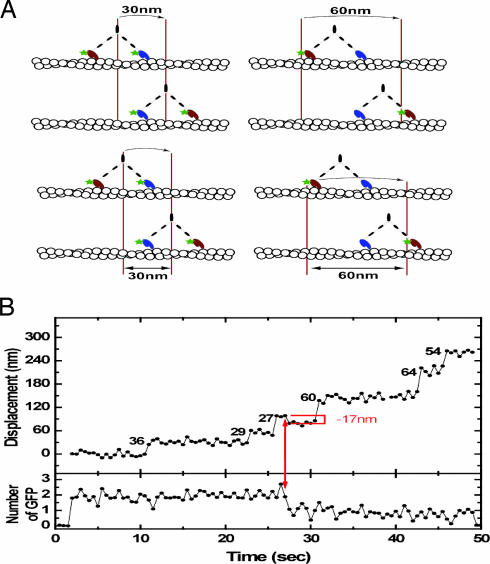
To test directly whether DmVIIa moves processively on actin, we used total internal reflection fluorescence (TIRF) microscopy to record the movement of single GFP–DmVIIa–HMM molecules on actin filaments bound to a coverslip surface. Dimerized GFP–DmVIIa–HMM molecules were observed to move for distances from 0.1 to 3 μm at 72 ± 20 nm/s at saturating ATP concentrations, demonstrating that a double-headed fragment of DmVIIa is processive (see Movie 1, which is published as supporting information on the PNAS web site). The precise run length was not determined because most of the apparent “terminations” would be expected to be photobleaching events given the slow movement rate of this myosin. The fluorescence imaging with one-nanometer accuracy (FIONA) technique, which is capable of determining the position of a single fluorophore to ≈1.5-nm accuracy (16), was used to measure step size. The positions of GFP–DmVIIa–HMM spots were determined every 0.5 s (Fig. 2 A and B). Two populations of step sizes were obtained (Fig. 2). Some molecules showed small steps of ≈30 nm (Fig. 2C), whereas others gave larger steps of 60 nm (Fig. 2D). Our observations are consistent with a “hand-over-hand” stepping mechanism in which heads transit from the trailing to the leading position with a 60-nm step as shown in Fig. 3A. The large steppers (Fig. 3A Right) represent molecules in which one of the GFP labels has photobleached, and we observed the movement of the labeled head as it transitioned from the trailing to leading position, a distance of 60 nm. During this head movement, the center of mass of the molecule would move 30 nm. In contrast, the small step sizes (30 nm) are derived from molecules in which both heads had fluorescing GFP moieties, and the observed movement reflects the change in the position of the center of mass of the two unresolved fluorophores (Fig. 3A Left). Indeed, some stepping traces that initially started out with small steps could be seen to suddenly increase to large steps concomitant with a reduction in intensity of the spot that is associated with photobleaching of one of the two GFP moieties (Fig. 3B; see also Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Stepping trace and step-size histogram of GFP–DmVIIa–HMM with FIONA. (A and B) Stepping traces of GFP–DmVIIa–HMM molecules with double (A) and single (B) GFP signals. Values are the step sizes (in nm) between dwell periods. The experiment was performed at 0.5 μM ATP. (C and D) The horizontal red lines are fit to single Gaussian curves. (C) Step sizes: two GFP molecules; 32.7 ± 16.9 nm (mean ± SD); R = 97; n = 71 steps from 10 GFP–DmVIIa–HMM molecules. (D) Step sizes: one GFP molecule; 59.9 ± 20.5 nm (mean ± SD); R = 0.85; n = 17 steps from 32 GFP–DmVIIa–HMM molecules.
Fig. 3.
DmVIIa moves in a hand-over-hand mechanism. (A) Cartoon showing the proposed stepping mechanism of fluorescently labeled DmVIIa. (Left) Schematic for a molecule with a fluorescing GFP moiety on each head. Here the center of mass of the molecule is followed because the two GFP fluorophores are not resolved. (Right) Schematic for a molecule with only one of the two heads bearing a fluorescing GFP moiety. (B) FIONA stepping trace for a GFP–DmVIIa–HMM molecule that initially has two fluorescing GFP-moieties and, at the arrow, undergoes a photobleaching event of the GFP moiety fused to the lead head.
Kinetic Basis of the Processive Motility of DmVIIa.
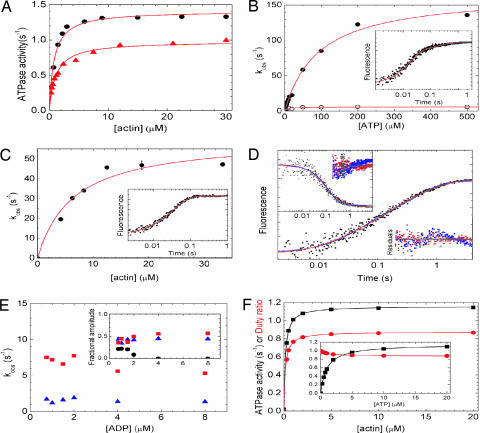
The kinetic properties of DmVIIa were examined to determine the biochemical basis for its processive behavior. DmVIIa–S1 had a basal MgATPase activity of 0.05 ± 0.001 s−1 in the absence of actin, which was activated by actin filaments to a maximum of 1.0 ± 0.02 s−1 with half-saturation at a very low actin concentration (0.98 ± 0.1 μM) in 50 mM KCl (Fig. 4A).
Fig. 4.
Kinetic properties of DmVIIa. (A) Steady-state actin-activated ATPase activity of DmVIIa–S1 and DmVIIa–HMM–Zipper. Hyperbolic fit to the data set yielded a maximal ATPase (Vmax) of 1.0 ± 0.1 s−1 with half-saturation (KATPase) at 1.1 ± 0.10 μM actin for DmVIIa–S1 (triangles) and Vmax = 1.4 ± 0.0 s−1 with a KATPase of 0.8 ± 0.08 μM actin for DmVIIa–HMM–Zipper (circles). (B) Observed rate constants (kobs) of the fast (filled symbols) and slow (open symbols) phases of ATP-induced dissociation of pyrene-acto-DmVIIa–S1 as measured by stopped-flow. The hyperbolic dependence of kobs on [ATP] indicated maximal kobs values of 167 and 5.9 s−1 for the fast and slow phases, respectively, in the experiment shown. Half-saturation occurred at 93 μM ATP for the fast phase. (B Inset) Trace at 50 μM ATP with kobs values of 59 s−1 (fractional amplitude: 80%) and 5.5 s−1 (20%). (C) Observed rate constants (kobs) of Pi release in double-mixing stopped-flow experiments in which mVIIa–S1 was first mixed with substoichiometric amounts of ATP, incubated for 5 s to obtain the DmVIIa–S1–products complex, and then rapidly mixed with various actin concentrations. Pi release was monitored by MDCC-PBP fluorescence (24). The kobs values showed a hyperbolic dependence on actin concentration with a maximum at 61 s−1 and half-saturation occurring at 6.4 μM actin. (C Inset) Trace at 15 μM actin with a single-exponential kobs of 30 s−1. (D) Kinetics of ADP release from acto-DmVIIa–S1. Shown is a pyrene–actin fluorescence trace obtained on mixing 0.5 μM pyrene-acto-DmVIIa–S1 plus 1 μM ADP with 100 μM ATP in the stopped-flow. A triple-exponential fit (red) to the transient shown yielded kobs of 50.1 (22% fractional amplitude), 7.2 s−1 (44%), and 1.2 s−1 (34%). The fastest phase represented ATP-induced dissociation of the nucleotide-free fraction of acto-DmVIIa–S1, and the later two phases originated from two populations of acto-DmVIIa–S1–ADP (compare Fig. 5). (D Inset) A 2′-deoxy-mant-ADP (dmADP) fluorescence transient recorded upon mixing a premixture of 1 μM actin/0.5 μM DmVIIa–S1 plus 5 μM dmADP with 1 mM ATP in the stopped-flow. Similarly to the pyrene–actin fluorescence records, biphasic ADP release was observed, and a double-exponential fit (red) yielded rate constants of 10.6 s−1 (67% fractional amplitude) and 1.3 s−1 (33%). Fits for monophasic ADP release (blue, double-exponential fit in main graph, single exponential in Inset) showed clear systematic deviations from the experimental data (see residual plots; red, biphasic ADP release; blue, monophasic ADP release) in all experiments. Time axes are the same for the data fits and the corresponding residual plots. (E) [ADP] dependence of kobs (main graph) and fractional amplitudes (Inset) of the first (black circles), second (red squares), and third (blue triangles) phase of the transients obtained in experiments described in D. [kobs values of the first phase are not related to ADP release (see above) and are omitted for clarity.] (F) Steady-state ATPase activity (black) and duty ratio (red) of DmVIIa–S1 as obtained in kinetic simulations by using the experimentally determined rate constants of Fig. 5. The duty ratio is defined as the fractional steady-state occupancy of the strongly actin-bound DmVIIa–S1 states (underlined in Fig. 5). DmVIIa–S1 reaches a maximal steady-state ATPase activity of 1.16 s−1 and a maximal duty ratio of 0.88 hyperbolically with half-saturation at submicromolar actin concentrations ([ATP] was 1 mM in the simulations, in the absence of background ADP). (F Inset) DmVIIa–S1 shows a much steeper [ATP]-saturation of its steady-state ATPase activity (half-saturation at 1.0 μM ATP) than other processive myosins ([actin] was 20 μM in the simulations, in the absence of background ADP). At saturating actin and ATP concentrations and in the absence of background ADP, the fractional steady-state occupancy of the various DmVIIa–S1 states (Fig. 5) was the following: AMDclosed, 0.71; AMDopen, 0.15; AMopen, 0.0007; AMclosed, 0.0003; AM-T, 0.008; A-MT, negligible; MT, 0.11; MDP, 0.001; AMDP, 0.02. Conditions were 25°C in 10 mM Mops (pH 7.0)/2 mM MgCl2/0.1 mM EGTA/1 mM ATP/50 mM KCl (10 mM KCl in C).
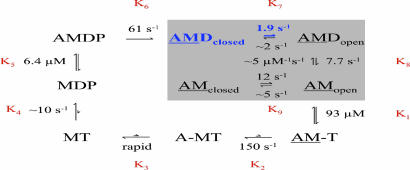
We measured each key step of the acto-DmVIIa ATPase mechanism as summarized and explained in Fig. 5 and also Table 1, which is published as supporting information on the PNAS web site. DmVIIa shares a common ATPase pathway with other myosins as the binding of ATP causes dissociation of the actomyosin complex (K1–K3 in Fig. 5). ATP hydrolysis mainly occurs in an actin-detached state (K4), and rebinding of the myosin head to actin (K5) accelerates Pi release (K6), which is followed by ADP dissociation from actomyosin (K7–K8). Here we will focus on ATP binding to and product (Pi and ADP) release from acto-DmVIIa, the steps that are key determinants of the duty ratio (i.e., steady-state fractional occupancy of strongly actin-bound states) and the processive behavior of DmVIIa.
Fig. 5.
ATPase mechanism of DmVIIa–S1. A, actin; M, DmVIIa-S1; T, ATP; D, ADP; P, phosphate. The rate-limiting step of the cycle and the predominant steady-state intermediate are highlighted in blue. Rate and equilibrium constants are expressed in a clockwise direction. Arrows for associating and dissociating components are omitted for clarity. Strongly bound acto-DmVIIa-S1 complexes are underlined.
To measure the transient kinetics of ATP-induced dissociation of the acto-DmVIIa–S1 complex, we performed stopped-flow experiments using the fluorescence signal of pyrene-labeled actin filaments (21). The transients obtained on rapidly mixing pyrene-acto-DmVIIa–S1 with ATP under pseudo-first-order conditions were biphasic with rate constants saturating around 150 and 10 s−1, respectively (Fig. 4B). The fast phase had a fractional amplitude of ≈70% that was constant at higher ATP concentrations (>50 μM) where the two phases could be well separated (data not shown). This result indicates a multistep ATP-binding model (Fig. 5) in which K1, a rapid and weak equilibrium, is followed by a rather irreversible isomerization (k2). The observed rate constant of the fast phase will thus depend hyperbolically on [ATP] as kfast = K1k2[ATP]/(K1[ATP] + 1). The slow phase originates from an isomerization between two actomyosin states (AMclosed and AMopen in Fig. 5) that interconvert slowly and reversibly (K9). Nucleotide can only bind to the AMopen state. At [ATP] > 50 μM, ATP binding to AMopen will be much faster (kfast > 50 s−1) than the interconversion from AMclosed to AMopen (k−9 = 2 s−1). Thus, in this [ATP] range the reaction consists of two pure phases: kfast determined by K1 and k2 as above, and kslow = k−9. Because the two phases of the reaction were well separated under these conditions, the relative amplitudes of the fast and slow phases were determined solely by the preexisting equilibrium between AMclosed and AMopen (K9), and thus these amplitudes did not depend on ATP concentration. At lower [ATP] (<50 μM), the rate constants of the two phases became similar, and thus their separation was much less accurate, which resulted in uncertain relative amplitudes for the two phases in this [ATP] range. This experimental behavior was also reproducible in kinetic simulations. Similarly to DmVIIa–S1, DmVIIb, brush border myosin I, and rat myosin Ib have been shown to exist in two different nucleotide-free actomyosin states (19, 22, 23).
We used a fluorescently labeled phosphate binding protein (MDCC-PBP) (24) to monitor phosphate release from DmVIIa–S1 and acto-DmVIIa–S1. In double-mixing stopped-flow experiments, DmVIIa–S1 was first mixed with substoichiometric amounts of ATP (single turnover conditions), and then, after a 5-s delay to allow for ATP binding and hydrolysis to occur, the myosin–hydrolysis products complex was rapidly mixed with various actin concentrations to activate Pi release (25). The obtained single-exponential Pi release rate constant was ≈0.02 s−1 in the absence of actin, which corresponds to the basal steady-state ATPase activity of DmVIIa–S1. Pi release was activated by actin to a maximum of 61 s−1 with half-saturation around 6.4 μM actin concentration (Fig. 4C; k6 and K5 in Table 1). Thus, the maximal rate constant of Pi release (k6) is ≈50 times faster than the maximal actin-activated steady-state ATPase activity (Table 1 and Fig. 5).
ADP release from actomyosin is rate-limiting in the processive myosins V and VI (26, 27). Because myosin binds strongly to actin in the presence of ADP (18), the rate-limiting exit from this state causes a high steady-state actin attachment (>50%) of individual myosin heads, which is a kinetic prerequisite for a double-headed processive molecule. We followed ADP release from acto-DmVIIa–S1 by monitoring the effect of ADP on the ATP-induced dissociation of the pyrene-acto-DmVIIa–S1 complex. When pyrene-acto-DmVIIa–S1 was preincubated with increasing concentrations of ADP and then rapidly mixed with a large excess of ATP in the stopped-flow, two phases with rate constants of 7.7 ± 0.8 and 1.9 ± 0.2 s−1, respectively, became predominant with increasing ADP concentration (Fig. 4 D and E). Neither the rate constants nor the relative magnitude of the amplitudes of the two ADP release phases changed systematically with ADP concentration (Fig. 4E). [The fractional occupancy of the AMclosed state in the preincubation mixture is <0.1 at ADP concentrations >3 μM, which indicates that the AMclosed–AMopen transition (k−9) had no major contributions to the 7.7 ± 0.8 s−1 phase in these experiments.] Similar rate constants and amplitudes were observed when the experiment was performed with a constant preincubation ADP concentration and increasing concentrations of ATP after the stopped-flow mix (data not shown). In addition, using a fluorescent ADP analog [2′-deoxy-mant-ADP (dmADP)], we obtained very similar rate constants and amplitudes to those in the pyrene–actin experiments (Fig. 4D Inset). This behavior is consistent with a two-step kinetic model of ADP release from acto-DmVIIa–S1 in which a slower isomerization event (K7: a structural change in the actomyosin–ADP complex, which is the rate-limiting step of the whole ATPase pathway) is followed by a severalfold more rapid ADP dissociation event (K8).
Computational kinetic simulations using the experimentally determined rate and equilibrium constants of the DmVIIa–S1 ATPase cycle highlighted two interesting features of the mechanism. First, although the isomerization between acto-DmVIIa–S1·ADP states is the most prominent rate-limiting step (k7 = 1.9 s−1), the overall steady-state cycling rate will be only ≈60% of this value because of the reversibility of K7 and other steps in the cycle (Fig. 4F). Second, at saturating [actin], the steady-state ATPase activity of DmVIIa–S1 will already be near Vmax at strikingly low ATP concentrations: it reaches half-saturation at 1.0 μM ATP (Fig. 4F Inset), which is 1 and 2 orders of magnitude lower than the corresponding values of myosins V and VI, respectively (26, 27). This property makes DmVIIa uniquely advantageous for single-molecule investigations of the processive stepping mechanism, because its movement can be recorded at saturating ATP concentrations while it remains still within the time resolution of the FIONA technique.
Discussion
We have performed a systematic investigation of the molecular mechanism of the processive motility of DmVIIa including single-molecule motility assays and an in-depth transient-state kinetic characterization. We present evidence that DmVIIa is not only a high-duty ratio motor, but is a processive myosin when dimerized, which makes it functionally capable of vesicle transportation in cells as a single molecule. The kinetic signature of processive myosins is a slow, rate-limiting ADP release and a fast Pi release (18–20, 27), in contrast to nonprocessive myosins where the reciprocal is true (18). DmVIIa also exhibits this signature. All other steps in the ATPase pathway are much faster than the rate-limiting isomerization between two actomyosin–ADP states (K7 in Fig. 5), which as a consequence will be the main determinant of the maximal steady-state actin-activated MgATPase activity (Table 1). Kinetic simulations based on the experimentally determined parameters show that DmVIIa–S1 will exhibit a very high duty ratio (0.88) even at low (a few micromolar) actin concentration and in the absence of ADP (Fig. 4F). This duty ratio is even higher than that of other processive myosins characterized (18–20, 27), which is a kinetic explanation for the long run lengths (and, thus, high processivity) of GFP–DmVIIa–HMM obtained in our single-molecule motility experiments (Fig. 2).
We interpret the biphasic release of ADP from acto-DmVIIa–S1 using a model in which a reversible structural rearrangement (K7 in Fig. 5) precedes the actual ADP release step (K8). Similarly, upon ATP-induced acto-DmVIIa–S1 dissociation, a reversible isomerization (K9) precedes the binding of ATP (K1–K2). Thus, an isomerization occurs both in ADP-bound (K7) and nucleotide-free (K9) forms of the acto-DmVIIa–S1 complex. Nucleotide exchange (release and/or binding) occurs detectably only in one of the end states. We speculate that these isomerizations reflect the same structural state of myosin (28). Therefore, following the nomenclature introduced to describe a similar behavior of rat myosin Ib (29), in Fig. 5 we used the terms open and closed for the states in which nucleotide exchange is allowed or disabled, respectively. This transition is thought to be associated with a swing of the myosin neck region, which may play an important role in the strain dependence of the enzymatic and mechanical performance of myosins (28).
The FIONA measurement of the step size of DmVIIa shows that during double-headed processive stepping, the initial trailing head moves forward by 60 nm as it becomes the lead head, which moves the center of mass by 30 nm. Thus, the step size of DmVIIa is shorter than that of myosin V (36 nm; for review, see ref. 30) but appears to be exactly proportional to the length of its “neck” or lever arm (five IQ motifs in DmVIIa instead of six in vertebrate myosin Va) (31). Because the step size does not correspond to the pseudo repeat distance of an actin filament (36 nm), DmVIIa would have to either spiral around an actin filament or occasionally “wobble” to find an appropriate actin monomer target. The step size of myosin VI when moving processively along actin is also <36 nm, although the distribution of step sizes with this motor is very broad, and it is not thought to act by means of a lever arm mechanism (13, 17). Under saturating ATP conditions, the movement of single myosin VIIa HMM molecules in the total internal reflection fluorescence assay is highly [KCl] dependent, and the motility rates vary from 30 nm/s (10 mM KCl) to 70 nm/s (100 mM KCl) (data not shown).
Several features of DmVIIa make it a very interesting molecule for single-molecule mechanical studies. Its slow, rate-limiting ADP transition (1.9 s−1) is considerably lower than that of myosin V (12 s−1) (32), whereas its ATP binding rate constant (1.3 μM−1·s−1) is equal to that of myosin V. Thus, at ATP concentrations >2 μM, the ADP transition and not ATP binding should be rate limiting, and the stepping rate of DmVIIa should have a half-maximal ATP concentration of ≈1 μM. This finding means that the standard mechanical measurements such as step size measurements, dwell time distributions, etc. can be made at saturating ATP concentrations with high temporal resolution (12).
DmVIIa has only a small segment of predicted coiled-coil forming sequence. We fused a GCN4 leucine zipper at the end of this sequence in the GFP–DmVIIa–HMM construct to ensure that the molecule dimerized efficiently. A construct without the leucine zipper does not spontaneously dimerize or move processively at low protein concentrations. However, preliminary experiments reveal that the unzippered myosin VIIa HMM molecules can be induced to form processive dimers when clustered on actin filaments (Y.Y. and T.S., unpublished data) similarly to myosin VI (33). We speculate that myosins VII and VI, and even myosin X, may have a similar dimerization-regulated mechanism because these myosins contain short and charged-rich putative coiled-coil motifs (34).
DmVIIa plays crucial roles during Drosophila development and hearing (5, 12). The processive stepping mechanism we present here is a strong indication that DmVIIa can function in cells as a single-molecule cargo transporter. Although we know less about which cargoes are implicated in DmVIIa-dependent transport in Drosophila, the studies on melanosome transportation in mammalian retinal pigment epithelial cells suggest that this vesicle transport is indeed driven by myosin VIIa (and not myosin Va). Thus, we speculate that in retinal pigment epithelial cells, mammalian myosin VIIa uses the same motile mechanism to that of DmVIIa. DmVIIa (and also DmVIIb and mammalian myosin VIIa) is a very slow and high-duty-ratio motor, which makes it suitable for tension maintenance (19, 20). The fact that DmVIIa mutations cause marked morphological defects in bristles and hairs and is required for fly hearing suggests that, besides vesicle transport, DmVIIa may also play a mechanical role in the maintenance of structures and perception of stimuli (5, 12).
Materials and Methods
Protein Expression, Purification, and Reagents.
cDNA fragments encoding DmVIIa–S1 (motor domain and the first predicted IQ motif, truncated after codon 757) and DmVIIa–HMM (motor domain, 5 IQ motifs, and predicted coiled-coil motif, truncated after codon 919) were amplified from a DmVIIa clone (12) and subcloned into baculovirus transfer vector pFastBac1 (Invitrogen). A GFP cDNA fragment was inserted into the 5′ end of the DmVIIa–HMM cDNA, and one GCN4 leucine zipper containing 32 aa followed the predicted coiled-coil motif to efficiently dimerize the GFP–DmVIIa–HMM construct. A FLAG tag (sequence: DYKDDDDK) was fused to the C terminus of all constructs to aid purification. Creation and amplification of recombinant baculoviruses was performed according to Invitrogen protocols. Drosophila calmodulin was coexpressed with DmVIIa constructs (35). The expressed proteins were purified as described in ref. 36.
Actin from rabbit skeletal muscle was prepared (37) and pyrene-labeled as described in ref. 38. Phalloidin (Molecular Probes) was added to actin in a 1.5-fold molar excess for stabilization of actin filaments. 2′-deoxy-mant-ADP and fluorescently labeled bacterial phosphate-binding protein (MDCC-PBP) (24) were kindly provided by Howard White (Eastern Virginia Medical School, Norfolk, VA). Other reagents were from Sigma.
Kinetic Measurements.
Steady-state ATPase activities were measured at 25°C (39) in a buffer containing 10 mM Mops (pH 7.0), 2 mM MgCl2, 0.15 mM EGTA, 1 mM ATP, and 10 or 50 mM KCl. Transient kinetic experiments were performed by using an SF-2001 stopped-flow instrument (Kintek, Austin, TX) at 25°C in a buffer containing 20 mM Mops (pH 7.0), 5 mM MgCl2, 0.05 mM EGTA, and 50 mM KCl. Pi release stopped-flow experiments were performed in the above buffer but with 10 mM KCl. Optical setups were as described in ref. 40. Data fitting and analysis was performed by using the Kintek sf-2001 software and originlab 7.0 (Microcal, Amherst, MA). Computational kinetic simulations were performed by using gepasi (Version 3.30; www.gepasi.org).
In Vitro Motility Assays.
Motility assays were performed at 25°C in a buffer containing 20 mM Mops (pH 7.4), 4 mM MgCl2, 0.1 mM EGTA, 100 mM KCl, 50 mM DTT, various concentrations of ATP, and an oxygen scavenging system consisting of 25 μg/ml glucose oxidase, 45 μg/ml catalase, and 2.5 mg/ml glucose. Total internal reflection fluorescence and FIONA assays were performed as described in ref. 31 and quantified with metamorph (Molecular Devices).
Supplementary Material
Acknowledgments
We thank Antoine Smith for technical assistance; Howard White for the gift of MDCC-PBP, dmATP, and 2′-deoxy-mant-ADP; and Lee Sweeney for helpful discussions and sharing unpublished results on the dimerization of myosin VI. M.K. is supported by National Institutes of Health (NIH) Research Grant D43 TW006230 (1 R01 TW007241-01) funded by the Fogarty International Center and the National Heart, Lung, and Blood Institute (NHLBI). D.P.K. is supported by NIH Grant R01 DC007673. T.S. is supported by a fellowship from the Japanese Society for the Promotion of Science Research Fellowships for Japanese Biomedical and Behavioral Researchers at NIH. J.R.S. is supported by the NHLBI Intramural Program.
Abbreviations
- DmVIIa
Drosophila myosin VIIa
- S1
subfragment 1
- HMM
heavy meromyosin
- FIONA
fluorescence imaging with one-nanometer accuracy.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Richards T. A., Cavalier-Smith T. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 2.Kieke M. C., Titus M. A. In: Molecular Motors. Schliwa M., editor. Weinheim, Germany: Wiley–VCH; 2003. pp. 3–44. [Google Scholar]
- 3.Self T., Mahony M., Fleming J., Walsh J., Brown S. D. M., Steel K. P. Development (Cambridge, U.K.) 1998;125:557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 4.El-Amraoui A., Sahly I., Picaud S., Sahel J., Abitbol M., Petit C. Hum. Mol. Genet. 1996;5:1171–1178. doi: 10.1093/hmg/5.8.1171. [DOI] [PubMed] [Google Scholar]
- 5.Todi S. V., Franke J. D., Kiehart D. P., Eberl D. F. Curr. Biol. 2005;15:862–868. doi: 10.1016/j.cub.2005.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman T. B., Sellers J. R., Avraham K. B. Am. J. Med. Genet. 1999;89:147–157. doi: 10.1002/(sici)1096-8628(19990924)89:3<147::aid-ajmg5>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu X. R., Ondek B., Williams D. S. Nat. Genet. 1998;19:117–118. doi: 10.1038/470. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda M., Kuroda T. S. J. Biol. Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- 9.El-Amraoui A., Schonn J. S., Kussel-Andermann P., Blanchard S., Desnos C., Henry J. P., Wolfrum U., Darchen F., Petit C. EMBO Rep. 2002;3:463–470. doi: 10.1093/embo-reports/kvf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda T. S., Fukuda M. J. Biol. Chem. 2005;280:28015–28022. doi: 10.1074/jbc.M501465200. [DOI] [PubMed] [Google Scholar]
- 11.Wu X. S., Rao K., Zhang H., Wang F., Sellers J. R., Matesic L. E., Copeland N. G., Jenkins N. A., Hammer J. A., III Nat. Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 12.Kiehart D. P., Franke J. D., Chee M. K., Montague R. A., Chen T. L., Roote J., Ashburner M. Genetics. 2004;168:1337–1352. doi: 10.1534/genetics.104.026369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rock R. S., Ramamurthy B., Dunn A. R., Beccafico S., Rami B. R., Morris C., Spink B. J., Franzini-Armstrong C., Spudich J. A., Sweeney H. L. Mol. Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Rief M., Rock R. S., Mehta A. D., Mooseker M. S., Cheney R. E., Spudich J. A. Proc. Natl. Acad. Sci. USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veigel C., Schmitz S., Wang F., Sellers J. R. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 16.Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 17.Yildiz A., Park H., Safer D., Yang Z., Chen L. Q., Selvin P. R., Sweeney H. L. J. Biol. Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 18.De La Cruz E. M., Ostap E. M. Curr. Opin. Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., Kovacs M., Xu E., Anderson J. B., Sellers J. R. J. Biol. Chem. 2005;280:32061–32068. doi: 10.1074/jbc.M506765200. [DOI] [PubMed] [Google Scholar]
- 20.Henn A., De La Cruz E. M. J. Biol. Chem. 2005;280:39665–39676. doi: 10.1074/jbc.M507667200. [DOI] [PubMed] [Google Scholar]
- 21.Kouyama T., Mihashi K. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 22.Geeves M. A., Perreault-Micale C., Coluccio L. M. J. Biol. Chem. 2000;275:21624–21630. doi: 10.1074/jbc.M000342200. [DOI] [PubMed] [Google Scholar]
- 23.Jontes J. D., Milligan R. A., Pollard T. D., Ostap E. M. Proc. Natl. Acad. Sci. USA. 1997;94:14332–14337. doi: 10.1073/pnas.94.26.14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brune M., Hunter J. L., Corrie J. E. T., Webb M. R. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 25.White H. D., Belknap B., Webb M. R. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- 26.De La Cruz E. M., Wells A. L., Rosenfeld S. S., Ostap E. M., Sweeney H. L. Proc. Natl. Acad. Sci. USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De La Cruz E. M., Ostap E. M., Sweeney H. L. J. Biol. Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 28.Nyitrai M., Geeves M. A. Philos. Trans. R. Soc. London Ser. B. 2004;359:1867–1877. doi: 10.1098/rstb.2004.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geeves M. A., Chai M., Lehrer S. S. Biochemistry. 2000;39:9345–9350. doi: 10.1021/bi0002232. [DOI] [PubMed] [Google Scholar]
- 30.Sellers J. R., Veigel C. Curr. Opin. Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto T., Yildiz A., Selvin P. R., Sellers J. R. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 32.De La Cruz E. M., Sweeney H. L., Ostap E. M. Biophys. J. 2000;79:1524–1529. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H., Ramamurthy B., Travaglia M., Safer D., Li-Qiong C., Franzini-Armstrong C., Selvin P. R., Sweeney H. L. Mol. Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 34.Knight P. J., Thirumurugan K., Yu Y., Wang F., Kalverda A. P., Stafford W. F., III, Sellers J. R., Peckham M. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 35.Toth J., Kovacs M., Wang F., Nyitray L., Sellers J. R. J. Biol. Chem. 2005;280:30594–30603. doi: 10.1074/jbc.M505209200. [DOI] [PubMed] [Google Scholar]
- 36.Wang F., Harvey E. V., Conti M. A., Wei D., Sellers J. R. Biochemistry. 2000;39:5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- 37.Spudich J. A., Watt S. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 38.Cooper J. A., Walker S. B., Pollard T. D. J. Muscle Res. Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 39.Wang F., Kovacs M., Hu A. H., Limouze J., Harvey E. V., Sellers J. R. J. Biol. Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 40.Kovacs M., Wang F., Sellers J. R. J. Biol. Chem. 2005;280:15071–15083. doi: 10.1074/jbc.M500616200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.