Abstract
Understanding allostery may serve to both elucidate mechanisms of protein regulation and provide a basis for engineering active mutants. Herein we describe directed evolution applied to the integrin αL inserted domain for studying allostery by using a yeast surface display system. Many hot spots for activation are identified, and some single mutants exhibit remarkable increases of 10,000-fold in affinity for a physiological ligand, intercellular adhesion molecule-1. The location of activating mutations traces out an allosteric interface in the interior of the inserted domain that connects the ligand binding site to the α7-helix, which communicates allostery to neighboring domains in intact integrins. The combination of two activating mutations (F265S/F292G) leads to an increase of 200,000-fold in affinity to intercellular adhesion molecule-1. The F265S/F292G mutant is potent in antagonizing lymphocyte function-associated antigen 1-dependent lymphocyte adhesion, aggregation, and transmigration.
Allostery is important in many signaling proteins (1). In the inserted domain (I domain), present in some integrin α subunits, allosteric activation pathways have been studied structurally and functionally (2–8). The ligand-binding site of the I domain, a metal ion-dependent adhesion site (MIDAS), exists as two distinct conformations allosterically regulated by the C-terminal α7-helix (Fig. 1). Intercellular adhesion molecule-1 (ICAM-1) is the ligand for αLβ2 integrin. The N-terminal domain (D1) of ICAM-1 binds to the αL I domain through an interface that buries 1,250 Å; at the center of this interface ICAM-1 residue Glu-34 coordinates to the MIDAS metal (Fig. 1A).
Fig. 1.
αL I domain structure and detection by flow cytometry. (A) Ribbon diagram of the αL I domain in an open conformation in complex with ICAM-1 domain 1. The β-strands (yellow), α-helices (cyan), and N and C termini are labeled. I-domain MIDAS and ICAM-1 Glu-34 side chains are shown as stick models with oxygen atoms in red. The MIDAS metal ion and the oxygen atoms of two water molecules are shown as gold and red spheres, respectively. All structure diagrams are drawn in pymol (Delano Scientific, San Carlos, CA). (B) Top view of the αL I domain. The Cα traces, key side chains as sticks, and MIDAS metal ions as large spheres are in magenta (closed) and yellow (open); oxygens are in red. Primary coordinations are shown as dashed lines. (C) The Cα trace and ratchet side chains (Leu-289, Phe-292, Leu-295 shown in stick models) for the closed (magenta), intermediate (green), and open (yellow) conformations of the α7-helix are depicted. The structure of the α7-helix of the closed αL I domain was modeled based on the crystal structure of Protein Data Bank ID codes 1LFA and 1ZON; the structure of the α7-helix of the open αL I domain (Protein Data Bank ID code 1MQ9) was modeled as described (7). (D) Detection by immunofluorescence before (thin lines) and after (thick lines) induction of anti-c-myc antibody 9E10, I domain-specific antibody MEM83, I domain activation-dependent antibody AL-57, and ICAM-1-Fcγ. The percentages of the cells within the gated regions are shown.
Allosteric activation in I domains was first suggested from structural studies of I domains that were crystallized in two conformations termed closed and open; subsequent mutational studies and cocrystallization with ligands demonstrated that closed and open conformations have low affinity and high affinity (HA), respectively, for ligand (5, 8, 9). The two conformations differ in the MIDAS (Fig. 1B) and the position of the α7-helix (Fig. 1C), which exhibits axial C-terminal or “downward” displacement of 10 Å in the open conformation. In an intact integrin, interaction of the linker region after the α7-helix with the neighboring I-like domain of the β subunit exerts the force that pulls the α7-helix downward (3, 10).
Experts using structural insight have designed mutations for studying allostery in α I domains. In the αM I domain, two buried hydrophobic residues (Phe-275 and Phe-302) become more exposed to solvent in the open conformation. The mutations F275R and F302W led to a 2-fold increase in cell adhesion (2). The allosteric role of the α7-helix was more directly demonstrated in the αL I domain. Pairs of cysteines introduced to stabilize the α7-helix in intermediate or open positions led, respectively, to 500- and 10,000-fold higher affinity to ICAM-1 (4, 5).
Here, we have asked whether directed evolution, an approach based on random mutagenesis coupled with functional screening, and requiring no a priori knowledge of protein structure, could be used to identify key residues in the transmission of allostery within the I domain. In this study, we have chosen the αL I domain as a model system and demonstrate that directed evolution using a yeast display system (11) can be used to probe protein allostery with great efficiency. Furthermore, we have constructed an allosteric mutant with a 200,000-fold increase in affinity.
Results
Expression and Affinity Selection of I Domain Libraries.
As positive controls, wild-type I domain, and disulfide-bonded, intermediate affinity (IA) and HA mutant I domains were displayed on yeast. Good display on the yeast cell wall was shown by the binding of the anti-hemagglutinin (12CA5) and anti-c-myc (9E10) antibodies and I domain-specific antibodies TS1/22 and MEM83 (Fig. 1D and data not shown). Binding was also measured for ICAM-1-Fcγ chimera and a mAb selective for the open conformation, AL-57 (Fig. 1D). Binding was undetectable to the wild-type I domain, and ICAM-1 and AL-57 bound 56 ± 19% and 65 ± 12%, respectively, as well to the IA as to the HA I domain as measured by adjusted specific fluorescence intensity (ASFI).
Next, the yeast display library was tested for the binding of antibodies and ICAM-1. Binding of 12CA5, 9E10, TS1/22, and MEM83 mAbs was reduced significantly after error-prone mutagenesis (Fig. 1D and data not shown). Only 30% of cells in the mutant library bound to the 9E10 mAb, compared with 80% for the wild-type I domain before mutagenesis. The I-domain mutant library was then selected by magnetic cell sorting for binding to AL-57 or ICAM-1. After sorting once with AL-57, the majority of the cells in the enriched library bound to 9E10, MEM83, and AL-57 mAbs and ICAM-1 (Fig. 1D), suggesting that the vast majority of selected clones contained mutations that stabilized the HA conformation of the αL I domain. When the same mutant library was selected with ICAM-1-Fcγ, multiple cycles were needed to enrich for positive cells. After three cycles, the percentage of cells that bound to 9E10, MEM83, and AL-57 mAbs and ICAM-1 was comparable (Fig. 1D).
Activating Mutations in the Error-Prone PCR Library.
A total of 25 yeast colonies selected with AL-57 and ICAM-1-Fcγ were tested for binding of ICAM-1-Fcγ and sequenced (Table 1). Of these, 15 had unique sequences and were designated as m1–m15, and all bound ICAM-1 better than wild type (Table 1). Five clones contained single mutations, and the remainder contained two to five mutations each (Table 1). From the clones containing multiple mutations, one to three residues were selected and independently tested as a single substitution (designated by a, b, or c suffixes in Table 1) or independently isolated from the ratchet residue library (see below).
Table 1.
ICAM-1 binding to I domains
| Clones | Mutations | Expression, %SFI of HA | ICAM-1, %ASFI of HA |
|---|---|---|---|
| WT | Wild type | 102 | 0 |
| m1 | I258T | 197 | 7 |
| m2 | I258T, T267A, F277S | 76 | 10 |
| m3 | G262E | 110 | 4 |
| m4 | F134L, K252E, F265S | 24 | 31 |
| m5 | L274I, S279T | 138 | 51 |
| m5a | L274I | 81 | 91 |
| m6 | F209Y, L289P | 133 | 33 |
| m7 | F292S | 146 | 47 |
| m8 | F292S, K280E | 131 | 54 |
| m9 | F292S, K178R, L204Q | 155 | 28 |
| m10 | F265L, E293G, K294E, K296E, V308I | 86 | 16 |
| m10a | F265L | 58 | 1 |
| m10b | E293G | 97 | 4 |
| m11 | L295P, Q303R | 77 | 107 |
| m11a | Q303R | 80 | 7 |
| m12 | I150T, G246V, F299L, I309T | 113 | 27 |
| m12a | I150T | 80 | 0 |
| m12b | F299L | 89 | 0 |
| m12c | I309T | 91 | 17 |
| m13 | F265S | 57 | 152 |
| m14 | I288N | 132 | 14 |
| m15 | F277L, I288T | 143 | 30 |
| m15a | F277L | 94 | 135 |
| f1 | L289A | 131 | 1 |
| f2 | L289G | 136 | 59 |
| f3 | L289P | 82 | 24 |
| f4 | L289W | 93 | 51 |
| f5 | F292A | 88 | 29 |
| f6 | F292G | 126 | 63 |
| f7 | F292P | 58 | 5 |
| f8 | L295A | 222 | 25 |
| f9 | L295P | 86 | 6 |
| f10 | L295Q | 129 | 4 |
| f11 | L295W | 99 | 65 |
Clones isolated from the error-prone PCR library are designated as m1 to m15. Clones identified by focused mutagenesis are designated as f1 to f11. Clones tested as single substitutions are designated by a, b, and c after the names of the parental clones. Expression level was quantified from anti-c-myc mAb 9E10 binding.
Some clones containing single mutations (F265S, L274I, and F277L) showed higher binding of ICAM-1 than clones containing these and additional mutations. In contrast, some single mutants that were made showed lower binding to ICAM-1 than their parental multiply mutant clones, suggesting either that the wrong residues were selected for single substitutions or the activating effect of the mutations were interdependent (e.g., L295P and Q303R in m11). The significance of the location of these mutations for understanding I domain allostery is highlighted in Discussion.
Activating Mutations in the Ratchet-Residue Library.
Leu-289, Phe-292, and Leu-295 in the β6–α7 loop of the I domain alternatively occupy the same hydrophobic pocket in the closed, intermediate, and open conformation (5) and are termed “ratchet” residues (Fig. 1C). Mutations in each of these ratchet residues were selected from the error-prone PCR library that increased affinity for ICAM-1 (m6, m7, m8, m9, and m11 in Table 1). To obtain higher saturation of mutagenesis of ratchet residues, oligonucleotides randomized in the first two bases of each ratchet residue were used to construct a focused library, and 11 unique clones designated as f1–f11 were selected with AL-57 (Table 1). The particular residues that were selected suggest that the small amino acids Gly and Ala, Pro, and the large amino acid Trp are particularly activating at the ratchet positions (Table 1).
Kinetics of Binding of Soluble I Domains to ICAM-1.
Several of the most activating mutants were expressed in Escherichia coli and refolded to measure the kinetics of binding to ICAM-1 by surface plasmon resonance (Fig. 2). All of the single mutants, F265S, F292A, and F292G, exhibited an association rate to ICAM-1 in a similar range from 9,500 to 16,000 M−1·s−1 (Table 2). The double-mutant F265S/F292G showed a 2-fold higher kon of 25,000 M−1·s−1. These association rates are lower than observed for the IA and HA mutants of 105,000–133,000 M−1·s−1. In contrast to the association rates, the mutants isolated in this study differ very markedly from one another in their dissociation rates. The F265S and F292G mutants showed a 100-fold slower dissociation rate than the F292A mutant. The double-mutant F265S/F292G exhibited a further 10-fold decrease in dissociation rate compared with the F265S and F292G single mutants. The dissociation rate for the double mutant was slower than the HA I domain by close to 100-fold.
Fig. 2.
Binding of αL I-domain mutants to ICAM-1-Fcγ-coated sensor chip measured with surface plasmon resonance (Biacore). In all sensograms, the signals from the control surface, as described in Materials and Methods, were subtracted. The I-domain mutants were injected in a series of 2-fold dilutions starting from 100 μM for F292A (A) and 1 μM for F265S (B), F292G (C), and F265S/F292G (D).
Table 2.
Kinetics of the binding of the soluble I domains to ICAM-1 measured by Biacore
| I domain | kon, M−1·s−1 × 10−3 | koff, s−1 | KD, μM |
|---|---|---|---|
| Wild type* | 3.1 ± 0.1 | 4.6 ± 0.36 | 1,500 ± 200 |
| IA* | 133 ± 10 | 0.43 ± 0.07 | 3 ± 0.44 |
| HA* | 115 ± 7 | 0.014 ± 0.001 | 0.15 ± 0.016 |
| HA | 105 ± 49 | 0.011 ± 0.0003 | 0.105 ± 0.05 |
| F265S | 11.8 ± 0.9 | 0.0017 ± 0.0003 | 0.145 ± 0.031 |
| F292A | 9.5 ± 1.2 | 0.19 ± 0.005 | 20.0 ± 0.8 |
| F292G | 16.0 ± 0.8 | 0.0019 ± 0.0004 | 0.119 ± 0.017 |
| F265S/F292G | 25.0 ± 0.76 | 0.00015 ± 0.000003 | 0.006 ± 0.0002 |
The values shown are mean ± SD from three independent measurements. The parameters of kon, koff, and KD were calculated by fitting the 1:1 Langmuir binding model to the Biacore sensogram.
*The values were from ref. 5.
The KD of the single mutants F265S and F292G of ≈100 nM is similar to that of the HA I domain. Remarkably, the KD of the double-mutant F265S/F292G is 6 nM, which corresponds to an increase of 200,000-fold in affinity compared with the wild-type I domain. All of the I-domain mutants were monomeric in solution as assessed by size-exclusion chromatography (data not shown), and therefore the slower dissociation rate was not affected by avidity modulation.
Soluble I Domains as Inhibitors of Adhesion, Homotypic Aggregation, and Transmigration.
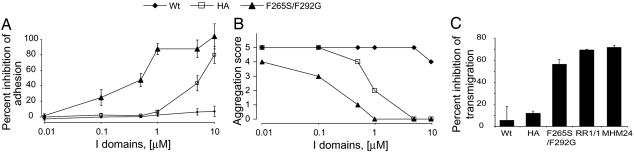
The potential to antagonize lymphocyte function-associated antigen 1 (LFA-1)-dependent lymphocyte adhesion to ICAM-1 with soluble I domains was examined in vitro. Consistent with the differences in affinity measured by surface plasmon resonance, the F265S/F292G mutant (IC50 = 0.35 μM) was much more potent than the disulfide-bonded HA mutant (IC50 = 5.3 μM) in blocking the adhesion of phorbol ester 12-tetradecanoylphorbol-13 acetate (PMA)-activated lymphocytes to ICAM-1-coated surfaces (Fig. 3A). The wild-type I domain gave no inhibition in the same assay.
Fig. 3.
Soluble I domains as inhibitors of LFA-1–ICAM-1 interaction. (A) Inhibition of PMA-stimulated lymphocyte adhesion to ICAM-1-coated surface. Data are mean ± SEM from two experiments, each in triplicate. (B) Inhibition of PMA-stimulated JY lymphoblastoid homotypic aggregation. Scoring scheme for the aggregation assay is as follows (30): 0, no cells are in clusters; 1+, <10% of cells are in aggregates; 2+, <50% of cells are in aggregates; 3+, up to 100% of cells are in small and loose clusters; 4+, up to 100% of cells are aggregated in larger clusters; and 5+, up to 100% of cells are in large, very compact clusters. Data are from two independent experiments, which showed identical results. (C) Lymphocyte migration through endothelial monolayer. I domains were used at 1 μM, and anti-ICAM-1 antibody RR1/1 and anti-LFA-1 antibody MHM24 were at 0.33 μM. Data are mean ± SEM from three experiments.
Homotypic aggregation of lymphocytes stimulated by PMA occurs through the interaction between LFA-1 and ICAMs (12). When the soluble I domains were tested as inhibitors for blocking the aggregation of JY lymphoblastoid cells, the wild-type I domain was slightly effective only at the highest concentration (Fig. 3B). The HA I-domain mutant at as low as 5 μM completely inhibited the aggregation. In contrast, the F265S/F292G mutant at 1 μM completely blocked aggregation, and at 0.5 μM <10% of cells were in aggregates. Even at 0.01 μM, the F265S/F292G was effective in inhibiting the formation of large and compact clusters, which were already observed at 0.1 μM of the HA I domain.
Migration of leukocytes across vascular endothelium is required for emigration from the bloodstream. The interaction between LFA-1 on leukocytes and ICAM-1 and ICAM-2 on endothelium has been identified as important for this process (13). When the I domains were tested at 1 μM, the F265S/F292G mutant was found to be much more efficient than the HA mutant I domain at blocking transmigration (Fig. 3C). The amount of inhibition by the F265S/F292G mutant at 1 μM was comparable to that of the anti-ICAM-1 antibody RR1/1 and the anti-LFA-1 I domain antibody MHM24, which were used at 0.33 μM and are bivalent.
Discussion
In this study, we tested whether directed evolution could be used (i) to engineer HA in a protein in which affinity is allosterically regulated and (ii) to structurally probe allosteric pathways within a protein. Our results show that directed evolution is an efficient method for identifying key residues in the transmission of allostery within the I domain. From 25 clones that were isolated with AL-57 and ICAM-1, all previously speculated or tested hot spots based on expert inspection were found [Phe-265 (2), Leu-289 (4), Phe-292 (2, 4), and Leu-295 (4)], together with many new positions (Ile-258, Gly-262, Leu-274, Phe-277, Ile-288, Glu-293, and Ile-309). Furthermore, using focused mutagenesis of the ratchet positions, we found that the identity of the substituting amino acid was markedly more important than previously suspected. None of the activating mutations were found in the ICAM-1-binding interface, supporting our hypothesis that they are allosteric mutations that shift the conformational equilibrium toward the open, HA conformation.
The mutations found here identify residues that are key in the shape-shifting pathway that connects the ligand-binding site to the α7-helix that communicates allostery to neighboring domains in intact integrins. In the transition of the MIDAS from closed to open conformation, the MIDAS metal moves ≈2 Å toward Thr-206 and away from Asp-239 (Fig. 1B). The α1-helix, particularly near its N-terminal end, and the β1–α1 loop bearing Ser-139 and Ser-141, move inward along with the Mg2+ ion. Furthermore, there is a backbone flip at Gly-240 of the β4–α5 loop, enabling its Asp-239 side chain to alter metal ion coordination and its Glu-241 side chain to form a crucial salt bridge to ICAM-1 (Fig. 1B). These changes all involve MIDAS loops and are directly involved in ligand binding; we will refer to the region containing them as the “active site” region (Fig. 4A). Remarkably, none of the allosteric activating mutations we have selected map to the active site; instead they define another region that we term the region of “switch allostery.” The switch-allostery region corresponds to a single segment of primary structure containing secondary structure elements β5, α6, β6, and α7 (Fig. 4A).
Fig. 4.
Active-site and switch-allostery regions and hot spots for activation in αL I domain. (A) Wild-type I-domain sequence is shown with the secondary-structure assignment. Hot-spot residues with activating mutations are in red. Active-site and switch-allostery regions are underlined in green and orange, respectively. (B) Stereoview of the I domain in the closed conformation. Cα trace is green for the active site, yellow for switch allostery, and gray for the rest of the I domain. MIDAS side chains are green, and hot-spot side chains are cyan. (C and D) En face view of the switch-allostery region Cα trace and the hot-spot side chains in the closed (C) and open (D) structures. Black lines in B–D are drawn by connecting the same Cα atoms in the closed and open structures.
Our findings provide insights into how the switch-allostery and active-site regions are coupled. The switch-allostery region undergoes a shearing motion with respect to the remainder of the I domain, including the active-site region (Fig. 4B). Previously, the importance of movements of key residues and isolated segments of the switch-allostery regions have been discussed (8, 14), but not their shearing motion as a whole. In allostery, the twist of the central β-sheet increases, with a significant increase in twist between β-strands 4 and 5, and a marked increase between β-strands 5 and 6 (Fig. 4 C and D). This twisting/shearing motion normal to the plane of the central β-sheets is accompanied by movements across the entire interface, as shown in the en face view in Fig. 4 C and D. Indeed, the allosteric mutations we have identified are distributed from one side of this interface to the other, demonstrating that residues throughout this extensive interface participate in balancing the energy between the open and closed conformational states.
In the closed conformation, the residues identified by allosteric mutations all are structurally contiguous and linked by hydrophobic packing, backbone hydrogen bonds, and peptide bonds (Fig. 4C). The hydrophobic side chains of Phe-277 and Leu-274 in the α6-helix and Ile-258 in the β5-strand pack against one another on one side of the switch-allostery–active-site interface. This hydrophobic interface extends by contacts with Leu-274 to the side chains of Phe-265 and Ile-288 (Fig. 4C). Furthermore, backbone hydrogen bonds connect Phe-265 to Gly-262, and in turn Gly-262 to Leu-289 (Fig. 4C). Leu-289 is also covalently linked to Ile-288 in the above hydrophobic cluster. In the closed conformation, Leu-289 packs closely with ratchet-residues Phe-292 and Leu-295, and Leu-295 is in the hydrophobic ratchet pocket, which is formed in part by Phe-292 and Leu-289. These residues outline one rim of the interface between the regions of switch allostery and active site. Notably, the shearing motions between these regions are large along this rim and markedly less at the opposite side of the interface where the switch-allostery region connects to the remainder of the I domain at the α5–β5 loop. During shearing of the switch-allostery region to the open conformation, interaction between the hydrophobic residues Phe-277, Leu-274, Ile-258, Phe-265, and Ile-288 is maintained, although swinging away of the Ile-288 side chain lessens its contacts (Fig. 4D). Furthermore, a change in backbone orientations at Gly-262 severs its hydrogen bonds to Phe-265 and Leu-289, as Leu-289 moves into the ratchet pocket (Fig. 4D).
Our mutational findings demonstrate that the region that regulates allostery is larger than previously suspected and extends all of the way to Phe-277 at one end of the rim. Notably, the F277L and L274I mutations, although conservative in nature and far from the ratchet pocket, increased binding of ICAM-1 as much as the F265S and F292G mutations (Table 1). The importance of hydrophobic packing at Phe-277 and Leu-274 had not previously been noted in discussion of I-domain allostery. A role for Gly-262 in αL or the equivalent Gly-272 in αM or Leu-283 in α2 had also not been previously noted to our knowledge. The two major clusters of hydrophobic residues in the switch-allostery region are kept in a fixed position relative to one another in the closed conformation by backbone hydrogen bonds that link Leu-289, Gly-262, and Phe-265. Breakage of these backbone hydrogen bonds in the open conformation is required in the rearrangements at Leu-289 and Phe-265 and their hydrophobic clusters. Equivalent backbone hydrogen bonds are present and broken in the closed and open conformations, respectively, of the αM and α2 I domains. These findings emphasize the ability of directed evolution to shed light on protein allostery.
The directed-evolution approach allowed us to make the unexpected observation that Gly and Ala substitutions for the ratchet-residues Leu-289 and Phe-292 differ markedly in affinity enhancement (Table 1). Phe-292 in the closed position of the α7-helix is buried in a hydrophobic pocket and becomes exposed to solvent as the α7-helix moves downward to the open position (Fig. 4 C and D). The lower energetic penalty for exposure of Ala compared with Phe, estimated from partition data for benzene, is 2.9 kcal/mol (15), corresponding to a 135-fold stabilization of the open conformation. This result is in close agreement with the observed increase in affinity of 75-fold of the F292A mutant (Table 2). By contrast, the 170-fold higher affinity of the F292G mutant compared with the F292A mutant (Table 2) cannot be explained by exposure of the extra methylene group present in Ala compared with Gly (15), which is predicted to cause only a 3-fold higher affinity. The substantially larger increase in affinity for F292G compared with F292A may therefore be derived from the ability of Gly to assume a wider range of φ/ψ backbone angles and backbone change at residue 292.
We also found that Pro and Trp substitutions at ratchet residues were activating. The largest enhancement by Pro substitution at Leu-289 can be explained by the preference for the backbone angles adopted by proline. The φ angle for proline is constrained at −63° ± 15° by the covalent bond to its backbone nitrogen (16), which is compatible with the φ angle at residue 289 of −58° in the open conformation, but not with the φ angle of −116° in the closed conformation. Trp substitutions for Leu activated at residues 289 and 295, which are buried in both the closed and open conformations. Shifting the conformational equilibrium can result from stabilizing the open conformation, destabilizing the closed conformation, or both; very few of the mutants significantly decreased expression on the yeast surface (Table 1).
By combining point mutations of F265S and F292G, each of which led to a 1,000-fold decrease in dissociation rate and a 10,000-fold increase in affinity, we engineered the double-mutant F265S/F292G, with a 200,000-fold increase in affinity to ICAM-1. Compared with the disulfide-bonded HA I-domain mutants, the kinetics of the mutants isolated in this study (F265S, F292G, and F265S/F292G) were markedly different. The slower association rate of these mutants may indicate that they are less efficient than the HA mutant in adopting the ligand-competent conformation before ICAM-1 binding. The significantly slower dissociation rate of the mutants described here may indicate some subtle improvements in the ligand-bound structures, and as a consequence, a lower free energy of the complex.
The remarkable enhancements achieved here by directed evolution bring the affinity of I domains into the range required for effective biotherapeutics. Integrins that contain I domains are important therapeutic targets in inflammation, autoimmunity, and transplantation. We found that in assays of lymphocyte adhesion to ICAM-1, transmigration through endothelium, and homotypic aggregation, the F265S/F292G mutant was 10-fold or more effective than the HA I domain, which has previously been demonstrated to be potent in antagonizing lymphocyte adhesion and homing to peripheral lymph nodes in vivo (4). The higher potency of the F265S/F292G mutant can be attributed to its 20-fold higher binding affinity than the HA mutant.
We have demonstrated that the identification of mutational hot spots provides important information on the interfaces in proteins that transmit allostery and regulate the difference in free energy between conformational states. The method described here can be readily extended to the study of allostery in other proteins and will be particularly useful in studying proteins where no structural information is available, or where the structure of only one conformer is known. Furthermore, HA allosteric mutants can be used directly as biotherapeutics or to isolate therapeutic antibodies specific for a particular conformational state.
Materials and Methods
Yeast Surface Display System.
We used yeast strain EBY100 (11). The αL I-domain cDNA coding for Asn-129 to Thr-318 was subcloned into the display plasmid pCTCON (17) by using NheI and BamHI sites. The resulting fusion protein contains from the N- to C-terminus Aga2, linker, hemagglutinin tag, I domain, and c-myc tag. Previously characterized disulfide-bonded, I-domain mutants (5) were also cloned into pCTCON.
Error-Prone and Focused Mutagenesis Libraries.
Error-prone libraries were generated (17) using a PCR mutagenesis kit (GeneMorph II random mutagenesis kit, Stratagene). In focused mutagenesis, primers spanning codons for amino acids 278–309 of the I domain with a codon designation of NNX (N = any of A, T, G, or C; X = wild type nucleotide) for Leu-289, Phe-292, or Leu-295 were used to amplify cDNA by two-step overlap PCR coding for the I domain. This process allowed substitutions of all 20 amino acids except Asn, Asp, Cys, His, Ile, Phe, and Tyr for Leu-289 and Leu-295, and all except Gln, Glu, Lys, Met, and Trp for Phe-292. Mixtures of 1 μg of the cDNA (≈3 × 1012 copies) and 0.5 μg of the linearized vector (≈1011 copies) in 5 μl of water were transferred to a cuvette containing ≈108 yeast cells in 50 μl of 10 mM Tris (pH 7.5), 270 mM sucrose, and 1 mM MgCl2. Electroporation was performed according to the published protocols (17). Transformation efficiency was ≈2% estimated by the number of colonies on selective dextrose medium with 2% (wt/vol) agar (17), from which the library size was estimated to be 2% of 108 cells or 2 × 106.
After transformation, cells were grown in selective dextrose media at 30°C with shaking for 24 h and induced in selective galactose media (17) for 24 h at room temperature with shaking. The mutation rate in the error-prone I domain library was measured as ≈3 mutations per 1,000 nt, which upon translation corresponded to ≈7 mutations per 1,000 aa.
Immunofluorescence Flow Cytometry.
Induced cells were harvested, washed with 100 μl of labeling buffer (PBS/0.5% BSA/10 mM MgCl2), and then incubated with ligands at 10 μg/ml in 50 μl of the labeling buffer for 20 min with shaking at 30°C. Ligands used in this study were anti-c-myc antibody 9E10 (Santa Cruz Biotechnology) and anti-hemagglutinin antibody 12CA5 (Roche Applied Science), αL I domain-specific antibodies TS1/22 and MEM83 (18), and ICAM-1-Fcγ (R & D Systems). AL-57, which was engineered from a phage display system (unpublished data), was used as an activation-dependent I domain-specific antibody. After incubation, cells were washed in 100 μl of the labeling buffer and incubated with secondary antibodies at 5 μg/ml in 50 μl of the labeling buffer for 20 min at 4°C. Finally, cells were washed once in 100 μl and suspended in 100 μl of the labeling buffer for flow cytometry (FACScan, BD Biosciences).
To calculate SFI, the mean fluorescence intensity (MFI) of uninduced clones was subtracted from the MFI of induced clones. Because the SFI of AL-57 and ICAM-1 is affected both by the affinity to the I domains and the difference in the level of I domain expression, the SFI of AL-57 and ICAM-1 was corrected by dividing them by the SFI of 9E10 mAb to c-myc tag, and these values were defined as ASFI. For example, the ASFI of ICAM-1 is computed as: [MFI (ICAM-1) of induced − MFI (ICAM-1) of uninduced]/[MFI (9E10) of induced-MFI (9E10) of uninduced]. Then the ASFI of AL-57 and ICAM-1 of clones were compared with that of the clone expressing the HA I-domain mutant, e.g., the percentage HA-ASFI of clone m1 = 100 × ASFI (clone m1)/ASFI (HA).
Magnetic Cell Sorting.
Sorting of the I domain library was performed with a magnetic cell sorter (MACS LS Column, Miltenyi Biotec). The I-domain library (≈108 cells) induced in 1 ml of selective galactose media was spun down, washed with 1 ml of the labeling buffer, and incubated with 5 μg/ml of AL-57 or ICAM-1-Fcγ in 200 μl of the labeling buffer for 20 min with shaking at 30°C. After incubation with primary ligands, cells were washed once with 1 ml of the labeling buffer, incubated in 80 μl of the labeling buffer and 20 μl of mouse anti-human IgG microbeads (Miltenyi Biotec) for 20 min at 4°C, and sorted.
Expression of Soluble I Domains and Surface Plasmon Resonance Measurements.
Mutant I domains (F265S, F292A, F292G, F265S/F292G, and HA) with residues of Asn-129 to Tyr-307 were expressed in E. coli BL21 (DE3) (Novagen), refolded, and purified as described (5). ICAM-1-Fcγ-coupled or mock-coupled CM5 sensor chip as control was prepared with the amine coupling kit (Biacore) as described (4). Surface plasmon resonance was measured by using a Biacore 3000 optical biosensor. I domains were injected over the chip in 20 mM Tris·HCl (pH 8.0), 150 mM NaCl, and 10 mM MgCl2, at a flow rate of 10 μl/min at room temperature.
Cell Adhesion Assays.
V-bottom 96-well plates (Corning) were coated with ICAM-1-Fcγ (10 μg/ml in PBS, pH 7.4) or 2% BSA as a control at 4°C overnight, and then blocked with 2% BSA for 1 h at 37°C. I domains were added in 50 μl of L15 medium, 2.5% FBS. Lymphocytes were isolated and cultured as described (13). PMA at 100 ng/ml and 2′,7′-bis-(2-carboxyethyl)-5-(and-6)- carboxyfluorescein acetoxymethyl ester at 2 μg/ml were added to 106 cells/ml in L15 and 2.5% FBS. After 30 min at 37°C, cells were washed and added to the wells in 50 μl of L15 and 2.5% FBS. The plates were immediately centrifuged at 200 × g for 15 min at room temperature (19). Nonadherent cells that accumulated at the center of the V-bottom were quantified by a fluorescence plate reader (Spectra MAX Gemini XS, Molecular Devices). Percent inhibition was calculated from the fluorescence intensity of experimental measurements (F), the positive control coated with ICAM-1-Fcγ without added I domains (FICAM-1), and the negative control coated with BSA alone (FBSA): 100 × (F − FICAM-1)/(FBSA − FICAM-1).
Homotypic Aggregation.
The effect of the soluble I domains on blocking homotypic aggregation of JY PMA-stimulated B lymphoblastoid cells was evaluated by a qualitative aggregation assay (score 0–5) after 1-h incubation as described (12).
Transmigration Assay.
Primary human dermal microvascular endothelial cells (HDMVECs) in 24-well plates were activated for 12 h with TNF-α (100 ng/ml), washed three times in buffer A (Hanks’ balanced salt solution supplemented with 20 mM Hepes, pH 7.2 and 1% human serum albumin) and preincubated for 5 min at 37°C in buffer A alone (control) or buffer A containing the wild-type, HA, and F265S/F292 I domains (1 μM) or RR1/1 (20) or MHM24 (21) mAbs (0.33 μM) (13). IL-2-cultured primary human lymphocytes (105 cells/ml) in 500 μl of buffer A containing I domains or mAbs were added to HDMVECs and incubated at 37°C for 10 min. For each condition complete Z-stacks were obtained in each of 10 randomly selected fields by using the Bio-Rad Radiance 2000 laser-scanning confocal microscope system and then analyzed to determine the number of cells in the process of, or having completed, diapedesis (13). Percent inhibition of transmigration was calculated as: 100 × (1 − fraction of TEM in the presence of I domains or mAbs/fraction of TEM in control).
Acknowledgments
We thank Dr. Travis Waldron for critical comments on the manuscript. This work was supported by National Institutes of Health Grant CA31798.
Abbreviations
- I domain
inserted domain
- MIDAS
metal ion-dependent adhesion site
- ICAM-1
intercellular adhesion molecule-1
- IA
intermediate affinity
- HA
high affinity
- SFI
specific fluorescence intensity
- ASFI
adjusted SFI
- LFA-1
lymphocyte function-associated antigen 1
- PMA
phorbol ester 12-tetradecanoylphorbol-13 acetate
- MFI
mean fluorescence intensity.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Volkman B. F., Lipson D., Wemmer D. E., Kern D. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 2.Li R., Rieu P., Griffith D. L., Scott D., Arnaout M. A. J. Cell Biol. 1998;143:1523–1534. doi: 10.1083/jcb.143.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huth J. R., Olejniczak E. T., Mendoza R., Liang H., Harris E. A., Lupher M. L., Jr, Wilson A. E., Fesik S. W., Staunton D. E. Proc. Natl. Acad. Sci. USA. 2000;97:5231–5236. doi: 10.1073/pnas.97.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimaoka M., Lu C., Palframan R., von Andrian U. H., Takagi J., Springer T. A. Proc. Natl. Acad. Sci. USA. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimaoka M., Xiao T., Liu J.-H., Yang Y., Dong Y., Jun C.-D., McCormack A., Zhang R., Joachimiak A., Takagi J., et al. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCleverty C. J., Liddington R. C. Biochem. J. 2003;372:121–127. doi: 10.1042/BJ20021273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin M., Andricioaei I., Springer T. A. Structure (London) 2004;12:2137–2147. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Lee J.-O., Bankston L. A., Arnaout M. A., Liddington R. C. Structure (London) 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 9.Qu A., Leahy D. J. Proc. Natl. Acad. Sci. USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso J. L., Essafi M., Xiong J. P., Stehle T., Arnaout M. A. Curr. Biol. 2002;12:R340–R342. doi: 10.1016/s0960-9822(02)00852-7. [DOI] [PubMed] [Google Scholar]
- 11.Boder E. T., Wittrup K. D. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 12.Rothlein R., Springer T. A. J. Exp. Med. 1986;163:1132–1149. doi: 10.1084/jem.163.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carman C. V., Springer T. A. J. Cell Biol. 2004;167:377–388. doi: 10.1083/jcb.200404129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 15.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1985. [Google Scholar]
- 16.MacArthur M. W., Thornton J. M. J. Mol. Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 17.Colby D. W., Kellogg B. A., Graff C. P., Yeung Y. A., Swers J. S., Wittrup K. D. Methods Enzymol. 2004;388:348–358. doi: 10.1016/S0076-6879(04)88027-3. [DOI] [PubMed] [Google Scholar]
- 18.Lu C., Shimaoka M., Salas A., Springer T. A. J. Immunol. 2004;173:3972–3978. doi: 10.4049/jimmunol.173.6.3972. [DOI] [PubMed] [Google Scholar]
- 19.Weetall M., Hugo R., Friedman C., Maida S., West S., Wattanasin S., Bouhel R., Weitz-Schmidt G., Lake P. Anal. Biochem. 2001;293:277–287. doi: 10.1006/abio.2001.5140. [DOI] [PubMed] [Google Scholar]
- 20.Rothlein R., Dustin M. L., Marlin S. D., Springer T. A. J. Immunol. 1986;137:1270–1274. [PubMed] [Google Scholar]
- 21.Hildreth J. E. K., Gotch F. M., Hildreth P. D. K., McMichael A. J. Eur. J. Immunol. 1983;13:202–208. doi: 10.1002/eji.1830130305. [DOI] [PubMed] [Google Scholar]