Abstract
Chaperone-mediated autophagy (CMA) is a selective pathway for the degradation of cytosolic proteins in lysosomes. CMA declines with age because of a decrease in the levels of lysosome-associated membrane protein (LAMP) type 2A, a lysosomal receptor for this pathway. We have selectively blocked the expression of LAMP-2A in mouse fibroblasts in culture and analyzed the cellular consequences of reduced CMA activity. CMA-defective cells maintain normal rates of long-lived protein degradation by up-regulating macroautophagy, the major form of autophagy. Constitutive up-regulation of macroautophagy is unable, however, to compensate for all CMA functions. Thus, CMA-defective cells are more sensitive to stressors, suggesting that, although protein turnover is maintained, the selectivity of CMA is necessary as part of the cellular response to stress. Our results also denote the existence of cross-talk among different forms of autophagy.
Keywords: lysosome membrane proteins, lysosomes, proteases, protein degradation, macroautophagy
In mammalian cells, three different mechanisms contribute to the degradation of intracellular components inside lysosomes (autophagy) (1, 2). Two of these mechanisms, macroautophagy and microautophagy, are high-capacity processes that allow the simultaneous sequestration of multiple cytosolic constituents (soluble proteins and organelles) and their degradation, all at once, in the lysosomal lumen (1–3). In contrast, chaperone-mediated autophagy (CMA) allows the lysosomal degradation of specific cytosolic proteins on a molecule-by-molecule basis (4, 5). The selectivity of this pathway is conferred by means of the recognition of a pentapeptide amino acid motif in the CMA substrates by a cytosolic chaperone [heat shock cognate (hsc) protein of 70 kDa] (6). The substrate–chaperone complex is targeted to the lysosomal surface, where it interacts with the lysosome-associated membrane protein (LAMP) type 2A, a lysosomal membrane receptor for this pathway (7). After unfolding (8), the substrate translocates into the lysosomal lumen, assisted by a luminal chaperone (lys-hsc70), where it is rapidly degraded (4, 5). Binding of substrate proteins to LAMP-2A is a limiting step for CMA. Levels of LAMP-2A at the lysosomal membrane are tightly controlled and constitute a regulatory mechanism for CMA (4, 5). Up-regulation of CMA occurs during prolonged nutritional stress (starvation), exposure to toxic compounds, and mild oxidative stress (9) (reviewed in refs. 4 and 5), suggesting a role for this pathway in the selective removal of abnormal or damaged proteins under these conditions. In addition, a role for CMA in antigen presentation has recently been described (10). CMA activity decreases during aging (11), and a blockage of this pathway by mutant forms of synuclein also occurs in familial forms of Parkinson's disease (PD) (12). However, because of the complexity of the phenotypes associated with both aging and PD, the direct consequences of blockage of CMA in these systems are difficult to infer.
The LAMP-2 gene undergoes alternative splicing that gives rise to at least three different splicing variants (LAMP-2A, LAMP-2B, and LAMP-2C) (13). These LAMP-2 variants are present in different intracellular locations (14, 15) and probably have both common and isoform-specific functions. LAMP-2A is the only isoform known to participate in CMA (7, 16), whereas a role for LAMP-2B in macroautophagy has been proposed (17). As in Danon disease, a human vacuolopathy resulting from mutations in the LAMP-2 gene (17, 18), LAMP-2 knockout mice (lacking all three LAMP-2 variants) present a severe phenotype with major alterations in lysosomal biogenesis and autophagy (19–21).
To directly evaluate cellular consequences related to decline in CMA activity, we have selectively blocked the expression of LAMP-2A in cultured fibroblasts without affecting the other two LAMP-2 isoforms. Our results indicate the existence of cross-talk between different forms of autophagy, because macroautophagy is up-regulated in cells with impaired CMA. Despite this compensatory mechanism, cells with reduced CMA activity are more sensitive to many stressors, supporting an essential role for CMA as part of the cellular response to stress.
Results and Discussion
Selective Blockage of LAMP-2A Expression in Culture Cells.
To analyze the consequences of impaired CMA activity, we have reduced LAMP-2A levels in mouse fibroblasts in culture by using vector-mediated stable RNA interference (RNAi) directed specifically against the LAMP-2A exon [referred to as LAMP-2A(−) cells hereafter]. Expression of LAMP-2A could be efficiently reduced without affecting the mRNA and protein levels or the intracellular distribution of the other two LAMP-2 splicing variants, which was critical to ensure that any effect observed in these cells was directly related to the LAMP-2A isoform [Fig. 1; three stable LAMP-2A(−) clones (c1–c3) with RNAi targeting toward three different regions of the exon coding for the 2A isoform and resulting in different levels of blockage of LAMP-2A expression (45–90%) are shown]. Lack of all three LAMP-2 isoforms leads to accumulation of free cholesterol in late endosomes in embryonic fibroblasts (20) and to alterations in lysosomal biogenesis in hepatocytes due to impaired traffic of enzymes to lysosomes (20). Blockage of the expression of only the LAMP-2A isoform did not modify total levels or distribution of intracellular cholesterol (Fig. 6A, which is published as supporting information on the PNAS web site), nor did it alter the activity of lysosomal glycosidases (Fig. 6B) or the intracellular levels and maturation of two of the cathepsins shown to be altered in cells lacking all three LAMP-2 isoforms (Fig. 6C). In addition, we did not find significant morphological changes in the major intracellular compartments (specific markers for Golgi, endoplasmic reticulum, actin cytoskeleton, mitochondria, and caveolae are shown in Fig. 6D).
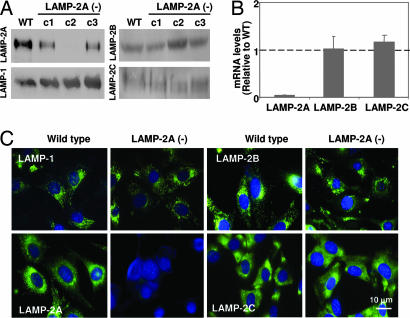
Fig. 1.
Selective blockage of LAMP-2A expression in mouse fibroblasts. Expression of LAMP-2A was reduced in mouse fibroblasts in culture by using a vector-based method for stable RNAi. Three different regions of the LAMP-2 exon 8A were targeted by using three different vectors, and clones stably transfected with each of the vectors [LAMP-2A(−) cells] were isolated by antibiotic selection. (A) Immunoblot for LAMP-2A, -B, and -C and LAMP-1 of total cellular extracts (150 μg of protein) of WT cells or three LAMP-2A(−) clones each carrying RNAi against a different region of LAMP-2A (c1, c2, and c3). (B) Comparison of total levels of LAMP-2A, -B, and -C mRNA in the same cells. Values are expressed as mRNA amount relative to the values in the control cells (arbitrary value of 1) and are the mean + SE of three different clones containing the construct against the intermediate region of exon 8a (c2). (C) Immunofluorescence for LAMP-2A, -B, and -C and LAMP-1 (green). Nuclei were labeled with DAPl (blue).
Impaired CMA Activity in LAMP-2A RNA-Interfered Cells.
CMA activity directly correlates with levels of LAMP-2A at the lysosomal membrane (16), but the consequences of blockage of LAMP-2A expression in CMA have never been analyzed before. The most direct way to assay CMA activity is by measuring the translocation of well characterized CMA substrates by isolated lysosomes in a standardized in vitro system (7, 8, 11, 12, 16). In contrast to fibroblasts lacking all three LAMP-2 isoforms, where changes in the density of lysosomes occur (20), we did not find differences in the density of the lysosomes normally active for CMA (enriched in the luminal chaperone) or in the recovery and enrichment of different lysosomal enzymes in this fraction (β-hexosaminidase and β-N-acetylglucosaminidase activities; data not shown). Levels of other lysosomal membrane proteins (LAMP-1, shown in Fig. 2A) and of cathepsins (cathepsin D, shown in Fig. 2A) in lysosomes from different LAMP-2A(−) cell clones were also comparable to controls. The integrity of the lysosomal membrane (measured as β-hexosaminidase latency; Fig. 2B) right after isolation and after different incubation times was similar to that of the lysosomes isolated from the control cells. Preserved lysosomal membrane integrity is an essential requirement to accurately assay CMA, because leakage of enzymes from the lumen would result in the extralysosomal degradation of the substrate. Degradation of two different well characterized CMA substrates (6, 11) by intact lysosomes (which recapitulates binding, uptake, and proteolysis of the substrate) was significantly lower in lysosomes isolated from different LAMP-2A(−) clones than in controls (Fig. 2C Left), revealing LAMP-2A as an essential component for this pathway. The decrease in CMA in each clone correlated well with the LAMP-2A levels remaining in lysosomes (Fig. 2A) and was due to diminished binding/uptake rather than altered lysosomal protease activity, because when the lysosomal membrane was disrupted (to facilitate free access of the proteases to the substrate), the differences between clones and control cells were no longer observed (Fig. 2C Right). Thus, stably blocking LAMP-2A expression in these cells was an effective method to reduce CMA activity and further analyze the cellular consequences of a failure in this form of autophagy.
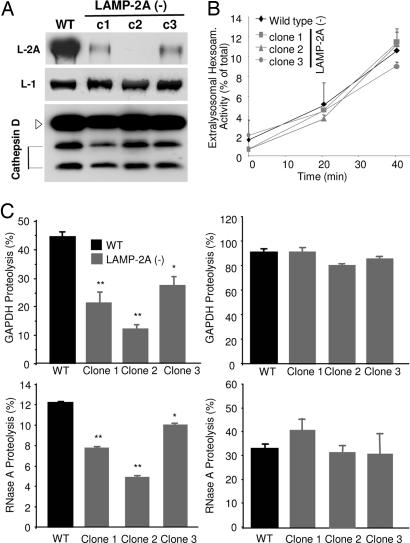
Fig. 2.
Impaired CMA in cells stably expressing RNAi for LAMP-2A. Lysosomes were isolated from WT or three LAMP-2A(−) clones of mouse fibroblasts (RNAi against three different regions of exon 8A). (A) Immunoblot of isolated lysosomes (15 μg of protein) for LAMP-2A, LAMP-1, and cathepsin D. (B) Hexosaminidase release from the isolated lysosomes. Values are expressed as percentage of the total lysosomal activity present in the extralysosomal medium and are the mean + SE of three different experiments. (C) Intact (Left) or disrupted (Right) lysosomes were incubated with radiolabeled GAPDH (Upper) or RNase A (Lower) for 30 min at 37°C. Proteolysis was calculated as the amount of initial acid-precipitable radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and peptides) at the end of the incubation. Values are the mean + SE of five different experiments with triplicate samples. ∗, P < 0.005; ∗∗, P < 0.001.
Compensatory Activation of Macroautophagy in CMA-Impaired Cells.
Intracellular protein turnover is the result of a fine balance between protein synthesis and degradation by different proteolytic pathways (1–3). Rates of protein synthesis (Fig. 3A) and of degradation of short-lived proteins (Fig. 3B) (main substrate for the ubiquitin/proteasome system) were similar in the LAMP-2A(−) cells and in control cells [although in cells subjected to LAMP-2A blockage for >4 months, we have observed a significant decrease in the degradation of short-lived proteins attributable to a reduction in two of the three proteolytic activities of the 26S proteasome and to changes in the subunit composition of this protease (unpublished data)]. Because CMA contributes to the degradation of long-lived proteins, mainly during nutrient deprivation (4, 5), we expected LAMP-2A(−) cells to display reduced protein degradation under those conditions. Surprisingly, the degradation of long-lived proteins in LAMP-2A(−) cells after serum removal was not only not impaired but was even higher than in control cells (Fig. 3C). In fact, the increase in degradation after serum removal was inversely proportional to the levels of LAMP-2A remaining in each clone (Fig. 3D), and it contrasted with the decreased protein degradation (in hepatocytes and HeLa cells) (19–21) or the absence of changes (in mouse embryonic fibroblasts) (22) described in cells lacking all LAMP-2 isoforms. The increased protein degradation detected in LAMP-2A(−) cells was still taking place in lysosomes, because it was inhibited by ammonium chloride, a classical lysosomal inhibitor (Fig. 3C). Treatment with 3-methyladenine (3MA) under conditions known to inhibit macroautophagy reduced protein degradation in LAMP-2A(−) cells after serum removal, whereas, as reported in ref. 23, it did not have any effect in control cells (Fig. 3C). In fact, in agreement with previous reports (24), in control fibroblasts, activation of macroautophagy (3MA-dependent degradation) occurred only during the first 4–6 h of serum removal, decreasing progressively as starvation persists (Fig. 3E). In contrast, in the LAMP-2A(−) cells, macroautophagy activity was detected even in cells maintained in the presence of serum, and it persisted after prolonged serum removal (Fig. 3E). In the three different LAMP-2A(−) clones, 3MA-dependent degradation increased as their LAMP-2A content decreased (Fig. 3D).
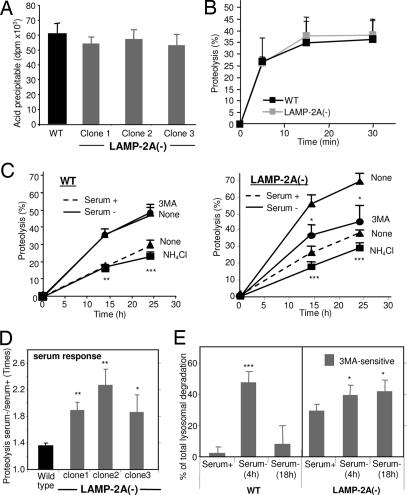
Fig. 3.
Altered protein turnover in LAMP-2A(−) cells. (A) Protein synthesis was measured in WT or three LAMP-2A(−) clones of mouse fibroblasts (c1, c2, and c3) as the incorporation of [3H]leucine (acid-soluble radioactivity) into proteins (acid-precipitable radioactivity) after 10 min of adding the radiolabeled amino acid into the culture medium. Values are expressed in dpm and are the mean + SE of three different experiments. (B) Degradation of short-lived proteins was measured in the same cells after labeling with [3H]leucine for 20 min. Proteolysis was calculated as described in Fig. 2C. Values are the mean + SE of three different experiments. (C) Degradation of long-lived proteins in the same cells was measured after labeling with [3H]leucine for 48 h and then plating the cells in medium supplemented (serum +) or not (serum −) with newborn calf serum. Where indicated, 15 mM NH4Cl or 10 mM 3-methyladenine (3MA) was added into the incubation medium 6 h after removing the serum. Proteolysis was calculated as in Fig. 2C. Values are the mean + SE of four different experiments with triplicate samples. (D) The effect of removal of serum on the degradation of long-lived proteins was calculated in experiments carried out as described in C. Values are expressed as the ratio of proteolysis in serum − cells to that in serum + cells and are the mean + SE of five different experiments with triplicate samples. (E) The contribution of macroautophagy to total protein degradation was calculated in experiments similar to the ones described in C by adding 3MA 2 h before the measuring times. Values are expressed as percentage of lysosomal proteolysis sensitive to 3MA (NH4Cl-sensitive) and are the mean + SE of four different experiments with triplicate samples. ∗, P < 0.01; ∗∗, P < 0.005; ∗∗∗, P < 0.001.
This constitutive activation of macroautophagy in cells with impaired CMA was further confirmed by directly analyzing different macroautophagy markers (Fig. 4). Conversion of the microtubule-associated protein 1 light chain 3 (LC3), a cytosolic protein that conjugates to phosphatidylethanolamine in the membrane of autophagosomes, from an 18-kDa form (LC3-I) to a faster-migrating form (16 kDa; LC3-II), is a well accepted measurement of the amount of autophagic vacuoles inside cells (25). Consistent with the constitutive activation of macroautophagy, we found higher levels of LC3-II in LAMP-2A(−) cells (Fig. 4A shows immunoblots for LC3 with an antibody that recognizes the two LC3 forms and an antibody with higher affinity for the LC3-II form). As in control cells, the removal of serum in LAMP-2A(−) cells reduced the amount of LC3-II (Fig. 4A, lane 8), reflecting increased clearance of autophagic vacuoles by lysosomes under those conditions. In fact, part of the LC3-II associated to autophagosomes is normally degraded in lysosomes after these two compartments fuse, and only when this degradation is inhibited can an accurate measurement of autophagic rates be obtained (26). In the presence of protease inhibitors, levels of LC3-II in LAMP-2A(−) cells were comparable in the presence and absence of serum (lanes 11 and 12) but were always higher than in control cells (lanes 9 and 10), even when starvation was prolonged up to 24 h. These results, together with the proteolysis data (Fig. 3E), are compatible with similar rates of autophagosome formation in LAMP-2A(−) cells maintained in the presence or absence of serum but with more efficient clearance during nutrient deprivation. Changes in the intracellular distribution of LC3 also supported constitutive activation of macroautophagy in LAMP-2A(−) cells (Fig. 4B). In contrast to the diffuse cytosolic localization of LC3 in serum-supplemented control cells, LC3 was detected in LAMP-2A(−) cells as bright puncta (presumably, autophagic vacuoles). This LC3 vesicular pattern was observed in both groups of cells during the first hours of serum removal but persisted after prolonged starvation up to 24 h in LAMP-2A(−) cells only (18-h starvation is shown; Fig. 4B Bottom).
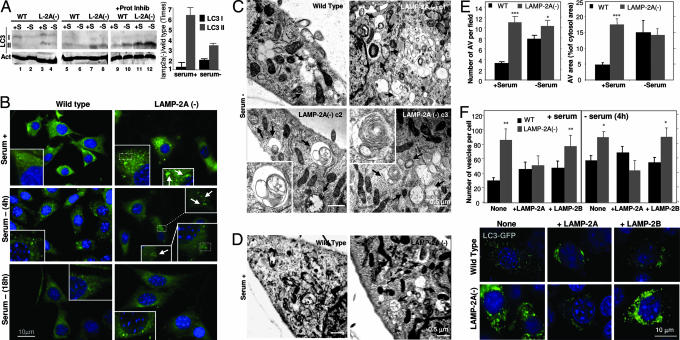
Fig. 4.
Up-regulation of macroautophagy in cells with defective CMA. (A) (Left) Cellular extracts (150 μg of protein) of WT or LAMP-2A(−) mouse fibroblasts were immunoblotted for LC3 with a selective antibody that recognizes both LC3-I and LC3-II proteins (lanes 1–4) and an antibody that recognizes only LC3-II (lanes 5–12). Cells were maintained for 24 h in medium supplemented (+S) or not (−S) with serum. In lanes 9–12, cells were treated with pepstatin A and E-64 as described in ref. 26. Immunoblot for actin is shown at the bottom as a loading control. (Right) The increase in the levels of LC3-I and LC3-II in LAMP-2A(−) cells compared with control was calculated after densitometric quantification of four immunoblots such as those shown here. (B) Immunostaining for endogenous LC3 in WT or LAMP-2A(−) mouse fibroblasts maintained in serum-supplemented medium (serum +) or after 4 or 18 h of removing the serum from the culture medium (serum −) (similar patterns were observed up to 24 h after serum removal). Insets show particular areas at higher magnification to better visualize the ring-shaped vesicles that correspond to autophagic vacuoles. (C and D) Electron microscopy micrographs of WT and three LAMP-2A(−) clones of mouse fibroblasts (c1–c3) maintained in the absence (C) or presence (clone c2) (D) of serum for 12 h. Arrows point to autophagic vacuoles. Insets show at higher magnification structures with morphological characteristics compatible with autophagic vacuoles. (E) The number of autophagic vacuoles per field (Left) and the cytosolic area occupied by those vesicles (Right) was calculated in micrographs such as those in C and D. (F Upper) WT and LAMP-2A(−) cells were transiently transfected with the cDNA for GFP-LC3 alone or plus the cDNA for human LAMP-2A or mouse LAMP-2B, as labeled. The mean number of GFP-positive vesicles (shown as puncta) per cell was quantified in cells maintained in the presence or absence of serum. Fifteen to 30 transfected cells were counted per experiment. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. (F Lower) A representative picture of cells maintained with serum. Nuclei were labeled with DAPl (blue) in B and F.
We also confirmed the presence of a higher content of autophagic vacuoles in LAMP-2A(−) cells by electron microscopy (Fig. 4 C–E). The morphology of most of the autophagic vesicles observed was compatible with that of autophagolysosomes (single-membrane vesicles with partially digested material in their lumen and positive for cathepsin D immunostaining) (Fig. 4 C and D and data not shown). Removal of serum further accelerated the fusion/degradation of autophagic vacuole contents by lysosomes as their size became smaller (for the same number of vesicles, the cytosolic area occupied by the vacuoles was smaller) (Fig. 4E Right). Supporting an increased macroautophagy activity in LAMP-2A(−) cells, we also found expansion of the intracellular acid compartment (stained with monodansylcadaverine) (Fig. 7A, which is published as supporting information on the PNAS web site) and increased levels of other autophagy-related proteins such as beclin (human homologue of Atg6) and Atg12/5 conjugate complex (Fig. 7B). The constitutive activation of productive macroautophagy (involving continuous formation and clearance of autophagic vacuoles) observed in the LAMP-2A(−) cells clearly differs from the impairment in macroautophagy described when all three LAMP-2 isoforms are absent (19, 21) (autophagic vacuoles form but are not properly eliminated).
The decrease in LAMP-2A levels seems to affect autophagy but not other lysosomal functions. In LAMP-2A(−) cells, we found normal rates of internalization by both fluid-phase and receptor-mediated endocytosis (Fig. 8, which is published as supporting information on the PNAS web site) and proper lysosomal degradation of the internalized products (Fig. 8A).
We further confirmed that the observed increase in macroautophagy was directly related to the lack of LAMP-2A (and, consequently, poor CMA activity) and not just to the reduction in total levels of LAMP-2 (independent of the isoform type). Transient transfection of the LAMP-2A(−) cells with human LAMP-2A [which escapes the interference effect but is functionally interchangeable with the mouse protein (7, 16)] significantly reduced the number of LC3-positive vacuoles in the cytosol to levels similar to those observed in control cells, in both the presence and the absence of serum (Fig. 4F). In contrast, the number of autophagic vacuoles was not reduced if LAMP-2A(−) cells were transiently transfected with mouse LAMP-2B (Fig. 4F). Expression levels of both transfected proteins and their proper targeting to lysosomes were verified by immunoblot and immunofluorescence, respectively (data not shown). These results strongly support that activation of macroautophagy was due to the reduced levels of the 2A isoform and that the cytosolic and transmembrane regions of the protein, not the luminal region common to all three isoforms, are important for this function. There is always the possibility of a direct inhibitory role for LAMP-2A over macroautophagy, independent of its role in CMA. However, we consider this direct inhibitory role to be unlikely, because overexpression of LAMP-2A in different cell types does not significantly change macroautophagy activity (7, 16). This compensatory activation of macroautophagy when CMA is impaired could explain the up-regulation of macroautophagy that has been observed by us and others in familial forms of Parkinson's disease (12, 27), where mutant synucleins tightly associate to the lysosomal membrane, interfering with proper CMA function.
Cells with Impaired CMA Have Increased Sensitivity to Different Stressors.
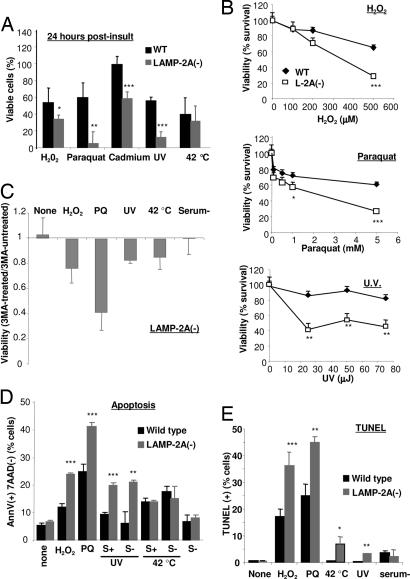
CMA is activated during mild oxidative stress (9) and other conditions that result in protein damage (5), suggesting a possible role for this pathway as part of the response to stress. Although cells with CMA blockage displayed slower rates of growth, their viability was similar to that of control cells under normal conditions (Fig. 9A, which is published as supporting information on the PNAS web site). However, LAMP-2A(−) cells were considerably more susceptible to different damaging stimuli. We observed significantly lower viability in a dose-dependent manner when LAMP-2A(−) cells were exposed to three different types of oxidative stresses (H2O2, paraquat, and cadmium) or to UV light (Fig. 5A and B). Interestingly, their sensitivity to other stressors, such as heat or serum removal, was comparable to that of control cells. Our results support that the compensatory activation of macroautophagy in cells with CMA blockage may be enough to maintain viability under normal conditions but cannot compensate for CMA function under particular stress conditions in which CMA is likely to play an essential role as part of the stress response. Surprisingly, the requirements for active CMA or the possibility of replacing it by macroautophagy differ with the type of insult. Thus, as shown here, cells with impaired CMA but compensatory activation of macroautophagy can accommodate to nutritional stress without changes in their viability, whereas complete blockage of both pathways (i.e., cells lacking all three LAMP-2 isoforms) sensitizes cells to starvation-induced cell death (21). The requirement for one form of autophagy or another may be related to the type of protein damage. It is possible that activation of macroautophagy is preferable to that of CMA if protein damage leads to aggregation, because aggregates would be more efficiently removed by the former. However, if protein damage results mostly in protein unfolding and not aggregation, the selectivity offered by CMA may have some advantages over the less selective macroautophagy.
Fig. 5.
Reduced cell viability in cells with defective CMA after exposure to various stressors. (A) WT and LAMP-2A(−) mouse fibroblasts were subjected to the indicated stresses [100 μM H2O2 (6 h), 50 μM paraquat (PQ) (6 h), 75 μJ of UV light, or incubation at 42°C (30 min)]. Cell viability was measured by the tetrazolium salt assay 6 h after the treatment. Values are expressed as percentage of initial viable cells remaining after the treatment and are the mean + SE of four different experiments. (B) Dose-dependent effect of the various stressors (H2O2, paraquat, and UV light exposure) on the viability of WT and LAMP-2A(−) mouse fibroblasts. Values are the mean + SE of two different experiments with triplicate samples. (C) Effect of blockage of macroautophagy by 3MA on the viability of LAMP-2A(−) mouse fibroblasts exposed to the indicated stressors. Viability was measured 6 h after the treatments. Values are expressed as fraction of the viability in cells not treated with 3MA (arbitrary value of 1) and are the mean + SE of three different experiments with triplicate samples. (D and E) Cells treated as in A were double labeled for 7-aminoactinomycin D (7AAD) and FITC-labeled annexin V (D) or labeled for TUNEL staining (E) and subjected to FACS analysis. After gating, the percentage of total cells positive for annexin V but negative for 7AAD (D) or positive for GFP-TUNEL (E) staining was calculated. Values are the mean + SE of two experiments with triplicate samples. Where indicated, serum was removed 12 h before treatment. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
Accumulation of autophagic vacuoles is characteristic of cellular death type 2 (autophagic), in contrast to type 1 (apoptosis), where no major changes in cytosolic organelles are observed (28). The higher content of autophagic vacuoles in LAMP-2A(−) cells made us consider whether macroautophagy was the mechanism leading to cell death under stress. However, blockage of macroautophagy in these cells did not prevent cellular death; on the contrary, it decreased cell viability even further (Fig. 5C). Exposure of LAMP-2A(−) cells to prooxidants or UV light did not change their autophagic content or activity [measured as levels of LC3-II in the presence and absence of protease inhibitors (Fig. 9B)] but activated apoptosis [significantly higher number of annexin V-positive cells than in control cells (Fig. 5D) and an increase in the number of TUNEL-positive cells (Fig. 5E)]. Activation of apoptosis progressed toward cellular death [dual-labeled cells with annexin V and 7-aminoactinomycin D (Fig. 9C)]. Although most cellular death occurred during the first 6 h after the stress, higher rates of apoptotic cell death persisted in CMA-defective cells up to 24 h after the insult (Fig. 9C). In agreement with our viability studies, we found similar levels of apoptosis in control and LAMP-2A(−) cells after heat shock or prolonged nutritional stress and even when both stressors were combined (Fig. 5D).
In conclusion, our findings provide evidence that a reduction in CMA activity, similar to the one observed in aging or familial forms of Parkinson's disease, is compensated for by activation of macroautophagy, thus supporting the existence of cross-talk between both autophagic pathways and offering an explanation for the up-regulation of macroautophagy previously described in these conditions. Although further studies are necessary to elucidate the molecular basis for this cross-talk, an appealing possibility is that some of the proteins essential for macroautophagy activity could be CMA substrates. This way, blockage of their degradation by CMA, as in this experimental model, would result in maintained activation of macroautophagy. Supporting this possibility is the fact that several of the already described Atg proteins (Atg 6, 7, 8, and 16) contain in their amino acid sequence KFERQ-related motifs. Although replacement of one type of autophagy by the other allows cells to survive under normal conditions, it makes them more susceptible to particular stressors. Thus, to our knowledge, this report provides the first evidence that CMA is an essential component of the stress response orchestrated in mammalian cells to guarantee cellular survival during cell injury.
Materials and Methods
Cells.
Mouse fibroblasts (NIH 3T3) were from the American Type Culture Collection and were cultured as described in refs. 7, 9, and 11. Mild oxidative stress was induced in cultured cells by adding 100 μM H2O2 or 50 μM paraquat for the indicated periods of time. UV exposure was done in a Stratalinker UV crosslinker (Stratagene) (total energy of 75 μJ), and heat shock was performed by placing cells at 42°C for 30 min. Assays were carried out immediately or 6–24 h after removing the stressors, as indicated.
Chemicals.
Sources of chemicals and antibodies were as described in refs. 7, 9, and 11. We used the following antibodies: anti-cytosolic tail of LAMP-2A, -2B, and -2C [prepared in our laboratory (ref. 7 and C. Zhang and A.M.C., unpublished work)], anti-LAMP-1 (Developmental Studies Hybridoma Bank, Iowa City, IA), anti-cathepsins D and L (Santa Cruz Biotechnology), anti-GM130, anti-BiP and anti-caveolin-1 (BD Transduction Laboratories), anti-actin (Abcam, Cambridge, MA), and anti-beclin (Novus Biologicals, Littleton, CO). The two polyclonal antibodies against LC3, the one against Atg12, and the cDNA that codes LC3 fused to GFP were a generous gift from Noburo Mizushima (University of Tokyo, Tokyo). The cDNAs that code for human LAMP-2A and mouse LAMP-2B were obtained from the American Type Culture Collection expressed sequence tag library and inserted into the PCR3.1 mammalian expression vector (Invitrogen).
RNAi.
Stable RNAi was carried out as described in ref. 29. The sequences of the regions targeted by the siRNA in the exon 8a of the LAMP-2A gene were 5′-GACTGCAGTGCAGATGAAG-3′, 5′-CTGCAATCTGATTGATTA-3′, and 5′-TAAACACTGCTTGACCACC-3′, corresponding to bases 1198–1216, 1331–1359, and 1678–1700. The hairpin (sense–loop–antisense) for these sequences was inserted in the multicloning region of the pSuper vector (Ambion, Austin, TX). Cultured cells were transfected by the calcium phosphate method and selected for stable transfectants by resistance to Geneticine (7).
Isolation of Subcellular Fractions.
Lysosomes from cultured cells were isolated from a light mitochondrial–lysosomal fraction in a discontinuous metrizamide/Percoll density gradient as described in ref. 30. Preparations with >10% broken lysosomes, measured as β-hexosaminidase latency, were discarded.
Uptake and Degradation of Substrate Proteins by Isolated Lysosomes.
[14C]GAPDH was incubated in Mops buffer [10 mM 3-(N-morpholino)propanesulfonic acid, pH 7.3/0.3 M sucrose/1 mM DTT] with lysosomes for 30 min. Degradation of [14C]GAPDH was measured after precipitation with trichloroacetic acid as described in refs. 7, 11, and 12. Proteolysis was expressed as the percentage of the initial acid-insoluble radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at the end of the incubation.
Intracellular Protein Turnover.
Rates of protein synthesis and degradation were measured as described in refs. 7 and 12 (see Supporting Methods, which is published as supporting information on the PNAS web site).
Fluorescence and Immunocytochemical Staining.
Immunofluorescence analysis of 3T3 cells was performed as described in ref. 7 (see Supporting Methods for the specific conditions for each type of staining). Images were acquired with an Axiovert 200 fluorescence microscope (Zeiss), subjected to deconvolution with the manufacturer's software, and prepared by using photoshop 6.0 (Adobe Systems, San Jose, CA). Quantification was carried out by using nih image j in three different Z-stack sections for each picture after thresholding.
mRNA Quantification.
mRNA quantification was carried out by using real-time PCR (see Supporting Methods).
Electron Microscopy.
Cells maintained in the presence or absence of serum were washed and fixed in 2.5% glutaraldehyde in SC (100 mM sodium cacodylate, pH 7.43) at room temperature for 45 min. The pellet was then rinsed in SC, postfixed in 1% osmium tetroxide in SC and then 1% uranyl acetate, dehydrated through a graded series of ethanols, and embedded in LX112 resin (Ladd Research Industries, Burlington, VT). Ultrathin sections were cut on a Reichert Ultracut E, stained with uranyl acetate and then lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kV. Morphometric measurements were carried out at 12 h of serum removal to avoid overestimation of size of the autophagic compartment due to cell shrinkage observed after 18 h of serum removal.
Viability and Apoptosis Measurement.
Cell viability was determined by the tetrazolium salt method. The percentage of apoptotic cells was determined by FACS analysis after staining for annexin V and TUNEL by using commercially available kits. (See Supporting Methods for details.)
General Methods.
Protein was determined by the Lowry method, using BSA as a standard. Lysosomal enzymatic activities were measured as reported in ref. 30. Quantification of intracellular cholesterol was performed by using the Amplex Red Cholesterol Assay kit (Molecular Probes) as described in ref. 22. After SDS/PAGE and immunoblotting, the proteins recognized by the specific antibodies were visualized by chemiluminescence methods (Renaissance; NEN–Life Science). Densitometric quantification of the films of the immunoblotted membranes and stained gels was done with an Image Analyzer System (S-100; Inotech, Wohlen, Switzerland). The Student t test was used for statistical analyses.
Supplementary Material
Acknowledgments
We thank Ms. Irene Puga for help with the FACS analysis, the Analytical Imaging Facility (Albert Einstein College of Medicine) for help with electron microscopy, and Dr. Fernando Macian and the members of our laboratory for their valuable suggestions and comments. This work was supported by National Institutes of Health/National Institute on Aging (NIH/NIA) Grant AG021904 and an Ellison Medical Foundation Research Award (to A.M.C.). A.C.M. is supported by NIH/NIA Training Grant T32AG023475. R.K. is an NIH/NIA Postbachelor Fellow.
Abbreviations
- CMA
chaperone-mediated autophagy
- LAMP
lysosome-associated membrane protein
- LC3
microtubule-associated protein 1 light chain 3
- RNAi
RNA interference
- 3MA
3-methyladenine.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cuervo A. M. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Wang C. W., Klionsky D. J. Mol. Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B., Klionsky D. J. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Majeski A., Dice J. F. Int. J. Biochem. Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Massey A. C., Kiffin R., Cuervo A. M. Int. J. Biochem. Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Dice J. F. Trends Biochem. Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 7.Cuervo A. M., Dice J. F. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 8.Salvador N., Aguado C., Horst M., Knecht E. J. Biol. Chem. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 9.Kiffin R., Christian C., Knecht E., Cuervo A. M. Mol. Biol. Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou D., Li P., Lin Y., Lott J., Hislop A. M., Canaday D., Brutkiewicz R., Blum J. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Cuervo A. M., Dice J. F. J. Biol. Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 12.Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 13.Hatem C. L., Gough N. R., Fambrough D. M. J. Cell Sci. 1995;108:2093–2100. doi: 10.1242/jcs.108.5.2093. [DOI] [PubMed] [Google Scholar]
- 14.Gough N. R., Fambrough D. M. J. Cell Biol. 1997;137:1161–1169. doi: 10.1083/jcb.137.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konecki D. S., Foetisch K., Zimmer K. P., Schlotter M., Lichter-Konecki U. Biochem. Biophys. Res. Commun. 1995;215:757–767. doi: 10.1006/bbrc.1995.2528. [DOI] [PubMed] [Google Scholar]
- 16.Cuervo A. M., Dice J. F. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 17.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J., Oh S., Koga Y., et al. Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 18.Saftig P., Tanaka Y., Lullmann-Rauch R., von Figura K. Trends Mol. Med. 2001;7:37–39. doi: 10.1016/s1471-4914(00)01868-2. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lullmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 20.Eskelinen E. L., Illert A. L., Tanaka Y., Schwarzmann G., Blanz J., Von Figura K., Saftig P. Mol. Biol. Cell. 2002;13:3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Polo R. A., Boya P., Pauleau A. L., Jalil A., Larochette N., Souquere S., Eskelinen E. L., Pierron G., Saftig P., Kroemer G. J. Cell Sci. 2005;118:3091–3102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 22.Eskelinen E. L., Schmidt C., Neu S., Willenborg M., Fuertes G., Salvador N., Tanaka Y., Lullmann-Rauch R., Hartmann D., Heeren J., et al. Mol. Biol. Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn P. F., Mesires N. T., Vine M., Dice J. F. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 24.Fuertes G., Martin De Llano J., Villarroya A., Rivett A. J., Knecht E. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 27.Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 28.Debnath J., Baehrecke E. H., Kroemer G. Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 29.Mailand N., Podtelejnikov A. V., Groth A., Mann M., Bartek J., Lukas J. EMBO J. 2002;21:5911–5920. doi: 10.1093/emboj/cdf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storrie B., Madden E. A. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.