Abstract
Recent human deaths due to infection by highly pathogenic (H5N1) avian influenza A virus have raised the specter of a devastating pandemic like that of 1917–1918, should this avian virus evolve to become readily transmissible among humans. We introduce and use a large-scale stochastic simulation model to investigate the spread of a pandemic strain of influenza virus through the U.S. population of 281 million individuals for R0 (the basic reproductive number) from 1.6 to 2.4. We model the impact that a variety of levels and combinations of influenza antiviral agents, vaccines, and modified social mobility (including school closure and travel restrictions) have on the timing and magnitude of this spread. Our simulations demonstrate that, in a highly mobile population, restricting travel after an outbreak is detected is likely to delay slightly the time course of the outbreak without impacting the eventual number ill. For R0 < 1.9, our model suggests that the rapid production and distribution of vaccines, even if poorly matched to circulating strains, could significantly slow disease spread and limit the number ill to <10% of the population, particularly if children are preferentially vaccinated. Alternatively, the aggressive deployment of several million courses of influenza antiviral agents in a targeted prophylaxis strategy may contain a nascent outbreak with low R0, provided adequate contact tracing and distribution capacities exist. For higher R0, we predict that multiple strategies in combination (involving both social and medical interventions) will be required to achieve similar limits on illness rates.
Keywords: antiviral agents, infectious diseases, simulation modeling, social network dynamics, vaccines
It is inevitable that another influenza pandemic will occur, and recent events suggest that this might happen sooner rather than later (1). A highly pathogenic H5N1 influenza A virus appears to have become endemic in avian hosts in Asia, and it is now spreading in migratory birds westward across eastern Europe. Human infections caused by this virus have a high case fatality rate; together with recent genetic data that implicate direct transmission of avian-adapted influenza virus to humans as the cause of the 1918 influenza pandemic (2), these conditions raise the specter of another devastating pandemic. To date, H5N1 viruses cannot transmit readily from human to human, thus providing a window to plan for the pandemic that will occur should the virus evolve to be readily transmissible among humans. If the nascent pandemic is not contained by timely intervention at its source (3, 4), international travel could carry pandemic viruses around the globe within weeks to months of the initiation of the outbreak, causing a worldwide public health emergency.
Intensive pandemic planning is occurring at the national [U.S. Department of Health and Human Services (HHS) Pandemic Influenza Plan, www.hhs.gov/pandemicflu/plan) and international [World Health Organization (WHO) Global Influenza Preparedness Plan, www.who.int/csr/resources/publications/influenza/WHO_CDS_CSR_GIP_2005_5/en/index.html" levels. The most pressing public health questions are: what might be the time course and geographic spread of the outbreak, and what is the most effective utilization of available therapeutic and social resources to minimize the impact of the outbreak? Precise planning is hampered by several unknowns, most critically the eventual human-to-human transmissibility of the human-adapted avian strain (characterized by the basic reproductive numberR0, the average number of secondary infections caused by a single typical infected individual among a completely susceptible population), and the supply of therapeutic agents. Manufacturers of neuraminidase inhibitors, such as oseltamivir, have committed to considerable increases in production over the next 3–4 years. However, the production of vaccine, the traditional first line of defense against influenza virus infections, is hampered by the inability to predict the antigenic details of the evolved virus at the time that it becomes a pandemic strain and the consequent inability to prepare a highly effective vaccine in advance of a pandemic outbreak. Given these uncertainties, it is important to develop multiple mitigation strategies, involving vaccination, prophylaxis with antiviral drugs, and both voluntary and imposed changes in social patterns such as school closures and travel restrictions.
The course of an influenza outbreak is sensitive to many factors, particularly population mobility and the susceptibility of individuals to the virus. Traditional mathematical models of epidemics often take the form of deterministic SIR differential equations for the population dynamics of susceptible (S), infectious (I), and removed/recovered (R) individuals (5, 6). Such models have also been extended to model the geographic spread of infectious diseases (7, 8). However, the population-based nature of this class of models best describes the dynamics of an epidemic when large numbers of individuals are infected, rather than the initial or final stages of an outbreak, when small numbers of individuals are involved and stochastic person-to-person transmission processes dominate. To satisfactorily model the initial seeding and final quenching of small community-level outbreaks requires a fundamentally different approach. To capture this crucial effect of uncertainty in transmission on epidemic predictions, we develop and use a stochastic agent-based discrete-time simulation model. This class of model has been used to assess vaccination and antiviral prophylaxis strategies on a local level (9–11); larger-scale versions have recently been used to investigate strategies at a regional level for containing an emerging pandemic influenza strain at its source (3, 4). Our national-level model combines an individual-level description of influenza viral infection and transmission dynamics with high-fidelity U.S. Census Bureau and Department of Transportation data on population demographics and mobility, yielding a massive-scale simulation model of the spatiotemporal dynamics of spread of a pandemic strain of influenza virus among an artificial U.S. population of 281 million people. Such an endeavor is only now practical with modern parallel supercomputing platforms and programming techniques.
Results and Discussion
Simulation Model Design.
The model population of 281 million individuals is distributed among 65,334 census tracts to closely represent the actual population distribution according to publicly available 2000 U.S. Census data (www.census.gov/main/www/cen2000.html). Each tract is in turn organized into 2,000-person communities. The model runs in cycles of two 12-hour periods (“day” and “night”), during which we identify seven contexts (“mixing groups”) within which individuals can associate. In five of these contexts (households, household clusters, preschools, playgroups, schools, and work groups), relatively close person-to-person association regularly occurs. Additionally, “neighborhoods” and “communities” provide unspecified contexts (e.g., shopping malls) within which occasional casual person-to-person association occurs. Because each individual may interact with any member of his or her mixing group, the group sizes determine the numbers of people who would be considered for antiviral prophylaxis in our socially targeted strategy of mitigation (below). Daytime contacts occur in neighborhoods and communities as well as in the age-appropriate setting, and nighttime contacts occur only in households, household clusters, neighborhoods, and communities. U.S. Census data on tract-to-tract worker flow is used to model the commute of working adults to their workplace, thus accurately capturing the short- to medium-distance population mobility important for disease spread. In addition, each individual takes occasional long-distance trips (three per year on average), lasting between 1 day and 3 weeks (4.1 days on average), matching Bureau of Transportation Statistics data (www.bts.gov/publications/national_transportation_statistics). Our simple model of long-range travel could be extended to account for different types of travel (e.g., business or leisure) or groups of travelers (such as a family) or to explicitly incorporate the airline network structure, as in ref.8.
The disease transmission and natural history models are briefly described in Materials and Methods, with further details provided in Supporting Text, Figs. 3–5, and Tables 3–5, which are published as supporting information on the PNAS web site. To model the introduction of pandemic influenza into the U.S., we assume that impenetrable borders are either prohibitively expensive or impossible to create, and that international air travel is the dominant mode of influenza introduction from outside the U.S. Consequently, a small random number of incubating individuals, equivalent to 0.04% of arriving international passengers, is introduced each day at each of 14 major international airports in the continental U.S. (see Table 6, which is published as supporting information on the PNAS web site). The simulation covers 180 days, roughly the length of a U.S. influenza season. We assume that, because of the uncertainty in diagnosis of influenza infections and the sporadic nature of the early stages of an outbreak, a cumulative number of 10,000 symptomatic individuals nationwide is required to trigger a nationwide pandemic alert (see Supporting Text for a sensitivity analysis of various response delays, for selected intervention strategies).
Intervention Strategies.
A variety of intervention strategies composed of one or more of the following four actions is considered: (i) socially targeted antiviral prophylaxis (TAP), in which symptomatic individuals and most of their close contacts receive treatment or prophylaxis, respectively, with antiviral drugs; (ii) dynamic mass vaccination, either of a random selection of individuals from the entire population or with preference for children, and with various production and distribution rates and starting dates; (iii) closure of schools, including preschools and play groups; and (iv) social distancing, as a result of legally mandated travel restriction or quarantine programs, or voluntary changes in social behavior.
TAP (11) is triggered by the first symptomatic person to be ascertained within a household (the index case). Because symptomatic diagnosis of influenza viral infection is inaccurate, leading either to delays in accurate diagnosis by biological assay or to excessive use of antivirals due to false positives, we simulate several scenarios. For the majority of our simulations, we assume that there are 0% false positives, and 60% of index cases are ascertained (the rest omitted because of, for example, misdiagnosis or lack of access to health care). When an index case is ascertained, he or she is treated, and all remaining people in this person’s household and household cluster are offered prophylaxis. If an ascertained index case belongs to a daycare, preschool, school, or workplace, then 100% of the people in that daycare or preschool are offered prophylaxis, or 60% of the people in that school or workplace. (Results for other ascertainment percentages, diagnosis delays, false positives, or prophylaxis strategies are presented in Supporting Text.) Due to its labor-intensive nature, TAP is likely to be feasible only during the earliest stages of an outbreak in any particular community, before the community health system is overwhelmed.
The dose and duration for effective treatment and prophylaxis using neuraminidase inhibitors (NAIs) against currently circulating strains of human influenza virus are well known (12), although a recent study suggests that an increased dose and duration of treatment may be needed to counteract H5N1 viral infections (13). On the other hand, these viruses may not retain their current unusually high growth rates if they evolve to be readily human-to-human transmissible. In light of this uncertainty, we use the current manufacturer’s recommended dose (10 tablets of oseltamivir for 5 days of treatment or 10 days of prophylaxis) and current estimates of oseltamivir efficacy in reducing infectiousness and susceptibility (see Supporting Text) (3, 14). Administration of a single course of NAI is initiated the day after the index case is ascertained, providing therapy for the index case and prophylaxis for others. A susceptible individual may receive subsequent courses of NAIs if another index case occurs later in a mixing group of which he or she is a member. We assume that 5% of people who start taking influenza antiviral agents will stop taking them after 1 day of treatment or prophylaxis.
Interventions involving vaccination suffer from uncertainty about the future identity of a pandemic strain, making it impossible to stockpile well matched pandemic vaccines. However, prevaccination based on a killed avian virus precursor to the pandemic strain is possible, providing a perhaps poorly matched but potentially efficacious vaccine. Vaccination can also be based on killed or live attenuated emergent pandemic virus, providing a close match to the subsequent circulating strains but available with a lag of a few months from emergence (in nonpandemic years, vaccine manufacture takes between 6 and 9 months). A “dynamic vaccination” scenario, in which vaccine becomes available incrementally, starting from as early as 2 months before, to as late as 2 months after, the first individual in the U.S. is infected, is investigated, with different production rates, total production amounts, and distribution policies (either uniformly throughout the population or preferentially to children). We compare the administration of the recommended two doses conferring best protection levels to a strategy in which twice as many people are given a single dose, assuming that a single dose of vaccine confers about half the protection of two doses (15). §
Much uncertainty exists about the societal acceptability of options for creating social distance and thereby reduction in transmission. Given the importance of children in the transmission of influenza (16), school closure is likely to be an effective (albeit burdensome) social distancing policy. Although formally imposed quarantine or travel restriction policies are possible, voluntary changes in hygienic and social behavior (including travel plans) will undoubtedly occur. Indeed, the spontaneous public response to news of an approaching pandemic will affect social behavior in unpredictable ways, so the social distancing strategies explored here are hopefully realistic approximations to voluntary or imposed distancing at three different scales: at the levels of schools, local communities, and nationwide travel. At the local scale, this social distancing is assumed to manifest itself in a concentration of interactions within households and household clusters, and at longer scales we consider uniform reductions in the amount of long-range travel to as little as 1% of the normal frequency. (See Supporting Text for details of implementation.) Although the social distancing measures studied here form a necessary first step in modeling such effects on disease transmission, further investigation is needed into variations in contact structure that are not considered in our model (e.g., classroom size variations with geographic region and grade level, parents staying at home with sick children, and other venues and mechanisms for transmission).
Simulation Results.
Independent realizations of our simulations for a given set of parameter values lead to very similar epidemic curves (see Supporting Text, Table 7, and, which are published as supporting information on the PNAS web site, for details including the estimation of R0). In the absence of intervention, for R0 = 1.9, our simulated pandemic begins with sporadic outbreaks occurring across the country in areas of dense population for 24 days before the outbreak is recognized (Table 1and Movie 1, which is published as supporting information on the PNAS web site). The pandemic peaks after 85 days, with a final illness attack rate of 43% (Table 2). The greatest nationwide activity is concentrated in a 2-month period when >100,000 people become ill each day, although local areas differ in the timing and duration of their highly active periods. This coincides quite well with waves of past pandemics; the 1957–1958 influenza A (H2N2) “Asian” virus initially appeared in June and July 1957, as sporadic cases in Iowa, Louisiana, and the West Coast, developing into local outbreaks during August 1957 before peaking in a 60-day period covering September and October 1957 (17). Similarly, the 1968–1969 influenza A (H3N2) “Hong Kong” virus first appeared as sporadic cases along the West Coast in July 1968, developing into local outbreaks 3 months later in October and peaking in December 1968 and January 1969, before finally ending in March 1969 (7).
Table 1.
Characteristics of simulated pandemic influenza in the U.S. in the absence of interventions
| Basic reproductive number, R0 | 1.6 | 1.9 | 2.1 | 2.4 |
| Rate of spread: 1,000th ill person* | 14 | 13 | 12 | 11 |
| 10,000th ill person* | 29 | 24 | 22 | 19 |
| 100,000th ill person* | 48 | 37 | 34 | 29 |
| 1,000,000th ill person* | 70 | 52 | 46 | 39 |
| Peak of epidemic* | 117 | 85 | 75 | 64 |
| Daily number of new cases at peak activity | 2.3 M | 4.5 M | 6.0 M | 7.9 M |
| Number of days with >100,000 new cases | 86 | 68 | 60 | 52 |
| Cumulative number of ill persons | 92 M | 122 M | 136 M | 151 M |
M, million.
*Days after initial introduction.
Table 2.
Simulated mean number of ill people (cumulative incidence per 100) and for TAP, the number of antiviral courses required for various interventions and R0
| Intervention | R0 = 1.6 | R0 = 1.9 | R0 = 2.1 | R0 = 2.4 |
|---|---|---|---|---|
| Baseline (no intervention) | 32.6 | 43.5 | 48.5 | 53.7 |
| Unlimited TAP (no. of courses)* | 0.06 (2.8 M) | 4.3 (182 M) | 12.2 (418 M) | 19.3 (530 M) |
| Dynamic vaccination (one-dose regimen)†‡ | 0.7 | 17.7 | 30.1 | 41.1 |
| Dynamic child-first vaccination†‡ | 0.04 | 2.8 | 16.3 | 35.3 |
| Dynamic vaccination (two-dose regimen)ठ| 3.2 | 33.8 | 41.1 | 48.5 |
| Dynamic child-first vaccinationठ| 0.9 | 25.1 | 37.2 | 47.3 |
| School closure¶ | 1.0 | 29.3 | 37.9 | 46.4 |
| Local social distancing¶ | 25.1 | 39.2 | 44.6 | 50.3 |
| Travel restrictions during entire simulation‖ | 32.8 | 44.0 | 48.9 | 54.1 |
| Local social distancing and travel restictions¶‖ | 19.6 | 39.3 | 44.7 | 50.5 |
| TAP,* school closure,** and social distancing** | 0.02 (0.6 M) | 0.07 (1.6 M) | 0.14 (3.3 M) | 2.8††(20 M) |
| Dynamic vaccination,†‡ social distancing,¶ travel restrictions,¶‖ and school closure** | 0.04 | 0.2 | 0.6 | 4.5 |
| TAP,* dynamic vaccination,†‡ social distancing,¶ travel restrictions,¶‖ and school closure** | 0.02 (0.3 M) | 0.03 (0.7 M) | 0.06 (1.4 M) | 0.1 (3.0 M) |
| Dynamic child-first vaccination,†‡social distancing,¶s travel restrictions,¶‖ and school closure** | 0.02 | 0.2 | 0.9 | 7.7 |
M, million.
*60% TAP, 7 days after pandemic alert, antiviral supply of 20 M courses unless stated.
†10 million doses of a low-efficacy vaccine (single-dose regimen) per week.
‡Intervention continues for 25 weeks, beginning such that the first individuals treated develop an immune response on the date of the first U.S. introduction.
§10 million doses of a high-efficacy vaccine (two-dose regimen) per week.
¶Intervention starting 7 days after pandemic alert.
‖Reduction in long-distance travel, to 10% of normal frequency.
**Intervention starting 14 days after pandemic alert.
††Exhausted the available supply of 20 M antiviral courses.
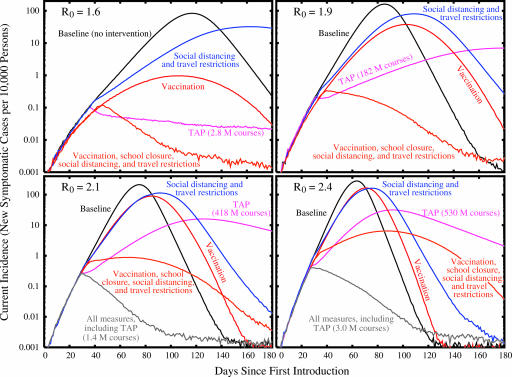
Although the dynamics of the pandemic in the absence of mitigation are clearly sensitive to R0, interestingly, this sensitivity is modest when R0 increases beyond 1.9 compared with the effect of increasing R0 from 1.6 to 1.9 (Table 1). We use as our guideline for adequate mitigation a reduction in the overall rate of illness to no greater than that of a typical influenza epidemic, ≈10%. The results presented in Fig. 2 and Table 2 suggest that as R0 increases from 1.6 to 1.9, a transition occurs from an outbreak that can be mitigated with moderate efforts, to one that can be mitigated only with vigorous application of multiple strategies. For example, several of the single interventions that we simulated are successful for R0 = 1.6, with TAP the most effective single intervention for our model of social mobility and transmission, provided adequate antiviral supplies exist and close contacts can be rapidly identified (see Fig. 1and Movie 2, which is published as supporting information on the PNAS web site). In contrast, for R0 = 1.7, 10.0 million courses are predicted to limit the national illness attack rate to 0.2%, but for R0 = 1.8, a prohibitively large 51 million courses would be required. Fewer courses do not control the pandemic, and an overall attack rate in excess of 10% ensues. An aggressive vaccine production and distribution plan may also be successful for R0 < 1.9 (see Movie 3, which is published as supporting information on the PNAS web site), particularly if initially targeted at children (18). With the exception of school closures for R0 =1.6, social distancing policies alone appear only to slow the pandemic without reducing its impact as measured by morbidity (see Movie 4, which is published as supporting information on the PNAS web site). Regardless of R0, unless drastic travel restrictions are imposed, the extent or duration of the pandemic is insensitive to details of the amount and location(s) of introductions of pandemic influenza virus in our simulations (see, which are published as supporting information on the PNAS web site). Due to the highly mobile U.S. population, the details of the introduction of the pandemic virus only affect the precise geographic spread and timing of the epidemic peak.
Fig. 2.
Epidemic curves (note the logarithmic scale) demonstrating the effectiveness of several different mitigation strategies, as compared to the baseline scenario without any intervention, for different values of R0. See Table 2 for details of each intervention. In the case of vaccination, results shown here are for a uniform coverage of the entire population with a single-dose regimen.
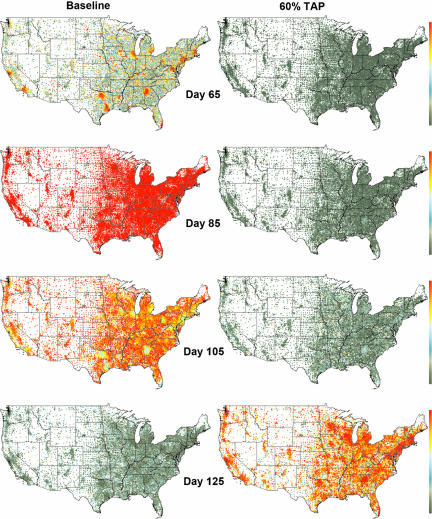
Fig. 1.
Two simulated pandemic influenza outbreaks with R0 = 1.9, initiated by the daily entry of a small number of infected individuals through 14 major international airports in the continental U.S. (beginning on day 0). The tract-level prevalence of symptomatic cases at any point in time is indicated on a logarithmic color scale, from 0.03% (green) to 3% (red) of the population. No mitigation strategies are used in the baseline simulation (Left), resulting in a 43.5% attack rate. (Right) A 60% TAP intervention begins at day 31, or 7 days after the pandemic alert. At day 99, the nationwide supply of 20 million antiviral courses is exhausted, leading to a nationwide pandemic.
For R0> 1.9, no single policy is predicted to be sufficient to mitigate an outbreak. For such highly transmissible strains, a combination of behavioral changes (to slow the spread) and therapeutic and prophylactic measures is essential. Throughout the range of R0 tested, antivirals, provided they are available in sufficient quantities and can be rapidly distributed, are a powerful tool for management. On the other hand, combinations of behavioral changes, together with a steady production of a low-efficacy vaccine throughout the pandemic (dynamic vaccination), can also successfully control pandemics of viruses with all except the highest level of transmissibility (Table 2 and Fig. 2). It is also important to note that the estimated benefits of preferentially vaccinating children are offset by the closing of schools, so that although one measure or the other is highly recommended, both together seem to offer no additional protection. Based on this result, the high societal cost of an extended closing of schools, requiring parents or grandparents to remain home with young children, may be avoidable through such a focused vaccination strategy. Similarly, our model suggests that the combination of TAP, school closure, and social distancing can be successful up to R0 = 2.4, without any vaccination (see Tables 8–10 and Figs. 10 and 11, which are published as supporting information on the PNAS web site, for additional combinations of intervention policies).
These projected major efforts necessary to mitigate pandemic influenza in the U.S. make it obvious that, for the U.S. and other countries, it would be optimal to control a potential pandemic strain of influenza at the source. In the event that a pandemic influenza virus does reach the U.S., according to our results, the U.S. population could begin to experience a nation-wide pandemic within 1 month of the earliest introductions. Our simulations indicate that the rapid imposition of a 90% reduction in domestic travel would slow the virus spread by only a few days to weeks (depending on R0), without reducing the eventual size of the outbreak, unless other behavioral or medical responses are introduced.
Conclusions
In this study, we regard strategies for mitigating pandemic influenza in the U.S. as successful when they limit the national attack rate to that of annual influenza epidemics, ≈10% of the U.S. population. All of our conclusions about the success of mitigation strategies are based on a simplified model of disease transmission and social contacts. Alternative models producing the same R0 may differ in quantitative details, but we expect the following conclusions to hold qualitatively. To achieve the target level of mitigation with antiviral agents alone, a very large stockpile is likely to be required (10 million courses of oseltimavir for R0 = 1.7, or 51 million courses for R0 = 1.8, in our simulations). For larger values of R0, the stockpile would have to be prohibitively large, e.g., 182 million courses for R0 = 1.9. Only for R0 ≤ 1.6 is reasonable control predicted to be achievable with the small currently available stockpile of 5 million courses. Our articulated TAP strategy targets sites of transmission for prophylactic drug use (3, 11), consequently using much less drug than if geographic regions or large groups, such as entire schools, were targeted (3, 4). However, this TAP strategy requires the identification of the effective sizes of the close-contact mixing groups, which is much more difficult in practice than in our assumed contact structure model. Consequently, the implementation of TAP would require considerable up-front preparation or on-the-spot decision making, and its effectiveness may be reduced by unforeseen sites or mechanisms of transmission that are not included in our model. Nevertheless, we believe that, even when antiviral stockpiles are small, the TAP strategy could be quite effective in slowing virus spread until vaccination could be implemented. (Of course, the potential emergence of an antiviral-resistant strain should also be considered in any pandemic planning.)
When vaccine supplies are limited, our simulations indicate that, at a population level, vaccinating n people with the recommended two doses providing maximal protection is less effective at reducing attack rates than vaccinating 2n people with single doses, assuming that a single dose confers roughly half the protection of a two-dose regimen (which may or may not be an option, depending on the particular vaccine). The relative benefits of single-dose vaccination of 2n people and two-dose vaccination of n people are expected to hold for prevaccination using poorly matched avian virus seed stock, although benefits are expected to be less than those presented here. The most effective single mitigation strategy would be a rapid dynamic vaccination of the population, initiated within 2 weeks of the pandemic alert, with a single dose of vaccine from the pandemic virus. Specifically, for R0 ≤ 1.6, spread could potentially be controlled if vaccine could be distributed nationally at the rate of 10 million doses per week for 25 weeks. For 1.9 ≤ R0 ≤ 2.4, single-dose vaccination would likely require augmentation with some combination of TAP, social distancing measures, and travel restrictions to be effective. Assuming that children remain major spreaders during the early stages of a pandemic outbreak, as they are for interpandemic influenza (16), the preferential vaccination of school children should be much more effective than random vaccination unless schools are closed. If vaccination in advance of a pandemic were possible using an avian seed virus, use of this poorly matched vaccine could slow virus spread as much as possible until a well matched vaccine based on the emergent human pandemic virus could be deployed.
Based on the present work, with the assumptions inherent in our model and its parameters, we believe that a large stockpile of avian-based vaccine with potential pandemic influenza antigens, coupled with the capacity to rapidly make a better-matched vaccine based on human strains, would be the best strategy to mitigate pandemic influenza. This effort needs to be coupled with a rapid vaccine distribution system capable of distributing at least 10 million vaccine doses per week to affected regions of the U.S.. For highly transmissible strains (i.e., having R0 ≥ 1.9), social distancing policies, including school closure and/or travel restrictions, may also be required to slow the epidemic spread sufficiently to enable production and distribution of sufficient quantities of vaccine. If antivirals were the preferred therapeutic defense, a stockpile of 20 million courses could be sufficient to effectively reduce national spread of a virus with R0 up to 1.7, provided extensive planning and/or on-the-spot decision making to distribute antivirals in a timely fashion was carried out. If implemented for pandemic planning, such infrastructure for stockpiling and rapid deployment of therapeutics would lead to the more effective use of vaccines (18) and antiviral agents in annual influenza epidemics. On the other hand, travel restrictions alone do not appear to be an effective control strategy, due to the implausibly early and drastic measures required to significantly reduce the large number of local outbreaks that are likely to emerge around the country.
Although our simulation model was specifically designed for the U.S., we believe that the qualitative conclusions reached here will hold for other countries or regions with highly mobile populations. However, for quantitative predictions to hold in settings other than those explicitly studied here, it will be important to demonstrate a robustness to various assumptions inherent in the model and its parameters. (In the event of an actual pandemic, use of a model to make quantitative predictions will require a rapid characterization of the transmission dynamics, disease natural history, and vaccine and antiviral efficacies to estimate these key model parameters.) Then the computational tool introduced here, capturing both the stochastic transmission processes that dominate the initial stages and final extinction of an outbreak and the detailed spatiotemporal dynamics of infectious disease spread, can be applied to public health questions that cannot be effectively addressed with traditional mathematical models (5, 6). In particular, should avian influenza continue to spread throughout the world, it will be important to develop containment strategies, analogous to those proposed for Southeast Asia (3, 4), that anticipate the possibility of a human-to-human transmissible strain of H5N1 influenza emerging first in a highly mobile population such as Europe or the U.S.
Materials and Methods
Disease Transmission Model.
Each class of mixing group is characterized by its own set of age-dependent probabilities of person-to-person contact of sufficient closeness and duration for transmission of normal human influenza virus to plausibly occur within a 12-hour period. Each of these contact probabilities is multiplied by the probability of transmission given contact, a single multiplicative constant that can be varied to model different R0 values.¶ As described in Tables 3 and 4, the contact probabilities were calibrated against total and age-specific illness attack rates of data in past pandemics (3,9,17), although these attack rate data alone do not uniquely determine parameter estimates. Infection of susceptible individuals is modulated by the antiviral and vaccination statuses of both the infectious and susceptible persons. A susceptible individual has a daily probability of becoming infected, accumulated over his/her contacts within each of the mixing groups to which he/she belongs (see Supporting Text for details). Age-dependent distributions are used to determine individual disease progression, whether an infected person becomes ill or remains asymptomatic and, if symptomatic, when (if ever) the person withdraws to household-only contacts.
Disease Natural History Model.
Predictions of the model are sensitive to the assumed disease course, but we can refer to past pandemics for guidance. The disease course for infection with the 1957 and 1968 pandemic influenza viruses and with post-1968 influenza A viruses (17) has been fairly consistent, with an estimated mean latent period of around 1.9 days and mean infectious period of around 4.1 days in several modeling studies (7, 9, 11, 20). The mean serial interval or generation time (i.e., average time between new infection and transmission to another susceptible) is thus ≈4 days. However, a recent reanalysis of incubation period and household transmission data suggests a significantly shorter serial interval of only 2.6 days, also consistent with viral shedding data from experimental infection studies (4). On the other hand, H5N1 virus is quite different from viruses causing past pandemics (including the 1918 pandemic), bearing the distinctive molecular signature of highly pathogenic avian influenza viruses, with possible implications for the resulting disease course in humans. The limited clinical information available to date on the disease course in individuals infected with H5N1 virus suggests a longer time course (13). Because H5N1 has not yet adapted for ready transmission among humans, and disease presentation may change in conjunction with this evolution, we focus on the midrange distributions in our model (see Fig. 3b), with a generation time of 3.5 days (3).
Model Limitations.
No seasonal or environmental effects or viral evolution are modeled (although it would certainly be possible to do so); we assume constant contact, transmission, and disease course parameters throughout the U.S. for the entire duration of an influenza season. Disease-related mortality was also neglected, under the assumption that deaths would occur at the latter stages of the infectious period and thus not significantly affect the spread of disease. It is important to realize that, although we attempt to make realistic estimates of model parameters, model validation in the traditional sense is not possible due to the unpredictability of viral evolution and the impossibility of documenting all cases of influenza in any influenza season.
Supplementary Material
Acknowledgments
We are indebted to Norman Johnson, Peter Lomdahl, and Tim McPherson for several key contributions in the early stages of this work, and to Mike Brown, Neil Ferguson, Brad Holian, Ed MacKerrow, Jeff Newman, Gary Resnick, Tom Wehner, and Shufu Xu for their encouragement and suggestions. We also thank Tony Redondo, Andy White, and the Institutional Computing Program at Los Alamos National Laboratory for providing access to the necessary supercomputing resources. This work was supported by the Department of Homeland Security through program CBLA11MP (to T.C.G., K.K., and C.A.M.) and by National Institute of General Medical Sciences MIDAS Grant U01-GM070749 (to I.M.L.). Los Alamos National Laboratory is operated by the University of California for the U.S. Department of Energy under Contract W-7405-ENG-36.
Abbreviations
- TAP
targeted antiviral prophylaxis
- NAI
neuraminidase inhibitor.
Footnotes
Conflict of interest statement: No conflicts declared.
In fact, efficacy of experimental vaccines against a novel pandemic strain cannot be ascertained in the absence of actual viral challenge; immunogenicity alone can be determined. Experimental vaccines based on avian influenza virus have required much greater amounts of antigen for acceptable levels of immunogenicity than standard human vaccines. This discrepancy does not enter into our calculations of required doses of vaccine. We assume that pandemic vaccines will have the same relationship between efficacy and immunogenicity as that for standard vaccines against human influenza virus.
R0 is a difficult quantity to estimate during an actual epidemic, because it depends critically upon the disease serial interval (or generation time) and to a somewhat lesser extent on the relative durations of the latent and infectious periods (19, 20). Because our model assumes particular values for these quantities, R0 is a useful measure of transmissibility, but care needs to be taken when comparing results for different models or epidemiological data.
References
- 1.Stöhr K., Esveld M. Science. 2004;306:2195–2196. doi: 10.1126/science.1108165. [DOI] [PubMed] [Google Scholar]
- 2.Tumpey T. M., Basler C. F., Aguilar P. V., Zeng H., Solórzano A., Swayne D. E., Cox N. J., Katz J. M., Taubenberger J. K., Palese P., et al. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 3.Longini I. M., Nizam A., Xu S., Ungchusak K., Hanshaoworakul W., Cummings D. A. T., Halloran M. E. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson N. M., Cummings D. A. T., Cauchemez S., Fraser C., Riley S., Meeyai A., Iamsirithaworn S., Burke D. S. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 5.Kermack W. O., McKendrick A. G. Proc. R. Soc. London Ser. A; 1927. pp. 700–721. [Google Scholar]
- 6.Anderson R. M., May R. M. Infectious Diseases of Humans. Oxford, U.K.: Oxford Univ. Press; 1991. [Google Scholar]
- 7.Rvachev L. A., Longini I. M. Math. Biosci. 1985;75:3–22. [Google Scholar]
- 8.Hufnagel L., Brockmann D., Geisel T. Proc. Natl. Acad. Sci. USA. 2004;101:15124–15129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elveback L. R., Fox J. P., Ackerman E., Langworthy A., Boyd M., Gatewood L. Am. J. Epidemiol. 1976;103:152–165. doi: 10.1093/oxfordjournals.aje.a112213. [DOI] [PubMed] [Google Scholar]
- 10.Halloran M. E., Longini I. M., Nizam A., Yang Y. Science. 2002;298:1428–1432. doi: 10.1126/science.1074674. [DOI] [PubMed] [Google Scholar]
- 11.Longini I. M., Halloran M. E., Nizam A., Yang Y. Am. J. Epidemiol. 2004;159:623–633. doi: 10.1093/aje/kwh092. [DOI] [PubMed] [Google Scholar]
- 12.Moscona A. N. Engl. J. Med. 2005;353:1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- 13.Beigel H., Farrar H., Han A. M., Hayden F. G., Hyer R., de Jong M. D., Lochindarat S., Tien N. T. K., Hien N. T., Hien T. T., et al. N. Engl. J. Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y., Longini I. M., Halloran M. E. Appl. Stat. 2006 in press. [Google Scholar]
- 15.Ritzwoller D. P., Bridges C. B., Shetterly S., Yamasaki K., Kolczak M., France E. K. Pediatrics. 2005;116:153–159. doi: 10.1542/peds.2005-0049. [DOI] [PubMed] [Google Scholar]
- 16.Brownstein J. S., Kleinman K. P., Mandl K. D. Am. J. Epidemiol. 2005;162:686–693. doi: 10.1093/aje/kwi257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilbourne E. D. The Influenza Viruses and Influenza. New York: Academic; 1975. [Google Scholar]
- 18.Longini I. M., Halloran M. E. Am. J. Epidemiol. 2005;161:303–306. doi: 10.1093/aje/kwi053. [DOI] [PubMed] [Google Scholar]
- 19.Fraser C., Riley S., Anderson R. M., Ferguson N. M. Proc. Natl. Acad. Sci. USA. 2004;101:6146–6151. doi: 10.1073/pnas.0307506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills C. E., Robins J. M., Lipsitch M. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.