Abstract
Activating KRAS mutations and p16Ink4a inactivation are near universal events in human pancreatic ductal adenocarcinoma (PDAC). In mouse models, KrasG12D initiates formation of premalignant pancreatic ductal lesions, and loss of either Ink4a/Arf (p16Ink4a/p19Arf) or p53 enables their malignant progression. As recent mouse modeling studies have suggested a less prominent role for p16Ink4a in constraining malignant progression, we sought to assess the pathological and genomic impact of inactivation of p16Ink4a, p19Arf, and/or p53 in the KrasG12D model. Rapidly progressive PDAC was observed in the setting of homozygous deletion of either p53 or p16Ink4a, the latter with intact germ-line p53 and p19Arf sequences. Additionally, KrasG12D in the context of heterozygosity either for p53 plus p16Ink4a or for p16Ink4a/p19Arf produced PDAC with longer latency and greater propensity for distant metastases relative to mice with homozygous deletion of p53 or p16Ink4a/p19Arf. Tumors from the double-heterozygous cohorts showed frequent p16Ink4a inactivation and loss of either p53 or p19Arf. Different genotypes were associated with specific histopathologic characteristics, most notably a trend toward less differentiated features in the homozygous p16Ink4a/p19Arf mutant model. High-resolution genomic analysis revealed that the tumor suppressor genotype influenced the specific genomic patterns of these tumors and showed overlap in regional chromosomal alterations between murine and human PDAC. Collectively, our results establish that disruptions of p16Ink4a and the p19ARF-p53 circuit play critical and cooperative roles in PDAC progression, with specific tumor suppressor genotypes provocatively influencing the tumor biological phenotypes and genomic profiles of the resultant tumors.
Keywords: array comparative genomic hybridization, mouse models, pancreatic cancer, KRAS, tumor suppressor
Pancreatic ductal adenocarcinoma (PDAC) ranks as the fourth leading cause of cancer mortality in the United States and causes >200,000 deaths worldwide annually (1, 2). Histopathological analyses have identified precursor lesions, pancreatic intraepithelial neoplasias (PanIN), which appear to progress through increasingly severe stages of cellular atypia leading to invasive PDAC (3). These lesions show multistep molecular progression that includes early activating KRAS mutations and telomere attrition, and subsequent inactivation of p16Ink4a, p14ARF, p53, and/or SMAD4 tumor suppressors in a high percentage of cases (4–6).
The Ink4a/Arf locus (hereafter denoted p16Ink4a/p19Arf) encodes tumor suppressors p16INK4A and p14ARF (p19Arf in the mouse). p16INK4A is a G1 cyclin-dependent kinase (CDK) inhibitor that binds to CDK4 and CDK6 and prevents their association with D-type cyclins (7), thereby facilitating CDK4/6-cyclin D-mediated phosphorylation and inactivation of retinoblastoma protein (RB) and S-phase entry. p16INK4A-mediated tumor suppression may relate to its induction by activated oncogenes and consequent promotion of oncogene-induced senescence (8, 9). p14ARF inhibits MDM2-mediated degradation of p53 (10, 11); thus, loss of p14ARF results in reduced p53 protein levels (12). Mounting evidence suggests that p14ARF also possesses p53-independent functions including the inhibition of ribosomal RNA processing (13, 14).
The central role of p16INK4A in PDAC is evidenced by its inactivation in 80–95% of sporadic cases (15) and by increased PDAC risk associated with germ-line p16INK4A mutations (16, 17). Whereas mutations exclusively targeting p16INK4A and sparing p14ARF have been identified in human PDAC, p14ARF-specific mutations have not been reported. However, the pathogenetic relevance of p14ARF is suggested by the occurrence of homozygous p16INK4A/p14ARF deletions in a subset of PanIN lesions, as well as in ≈40% of PDAC (6, 18). An important unresolved issue is the extent to which this correlation reflects the functional benefits of eliminating both p16INK4A and p14ARF, or rather a bystander phenomenon whereby p14ARF loss occurs as a consequence of targeting the overlapping p16Ink4a coding sequences.
p53 regulates target genes governing diverse tumor suppressor processes (19). p53 is mutated in 50–75% of human PDAC coupled with loss of the remaining WT allele (6). These mutations typically occur in advanced PanIN lesions that have previously incurred KRAS activation and p16INK4A loss (20, 21). p53 mutations and p14ARF deletions coexist in ≈38% of human PDAC cases (6, 18, 20, 22). Although such data may imply nonoverlapping tumor suppressor roles for these proteins, the distinct requirements for p53 vs. p14ARF inactivation in PDAC development have not been explored by genetic means.
The frequency and temporal occurrence of mutations in KRAS, p16INK4A/p19ARF and p53 in human PanIN and PDAC support the view that activated KRAS cooperates with defects in the RB and p53 tumor suppressor pathways to drive the initiation and progression of the disease. This hypothesis has received additional support from genetically engineered mouse models of PDAC. Endogenous KrasG12D expression in the mouse pancreas promotes development of PanINs (23, 24) that can progress to PDAC after a long latency (23). Furthermore, pancreas-specific expression of KrasG12D in the setting of homozygous deficiency at the p16Ink4a/p19Arf locus results in rapid advancement of PanIN to invasive PDAC (24). These observations indicate that activated Kras initiates PanIN and p16Ink4a/p19Arf-deficiency promotes PanIN-to-PDAC progression. The relative contributions of p16Ink4a and p19Arf to tumorigenesis was not addressed in this model.
A recent model combining KrasG12D and p53R172H alleles produced invasive PDAC, demonstrating that p53 normally functions to suppress the emergence of PDAC (25). Notably, a functional p16Ink4a/p19Arf locus was retained in the KrasG12D p53R172H tumors. Because both p53 and p16INK4A are characteristically lost in human PDAC, these data suggest that there may be cross-species differences in the role of the p16INK4A-RB pathway in cellular transformation (26) or, intriguingly, raise the possibility that antecedent loss of p53 function or the gain-of-function properties of the p53R172H protein may serve to neutralize rate-limiting components of the RB pathway. Taken together, the aforementioned studies establish a critical need to genetically examine the cooperative contribution of p16INK4A inactivation in PDAC progression.
Results
p53 or p16Ink4a Cooperate to Constrain PDAC Progression.
To study the genetic requirements for PDAC progression, we crossed mice with Pdx1-Cre (27) and LSL-KrasG12D alleles (28) and engineered null mutations in p53 and p16Ink4a or in p16Ink4a/p19Arf (24, 29, 30). Each of these tumor suppressor mutations, alone or in combination, cooperated with KrasG12D activation to promote invasive pancreatic cancers, although with differing tumor latencies and histopathological and genetic properties (Table 1). Hereafter, because all tumors analyzed possess Pdx1-Cre and LSL-KrasG12D alleles, specimens are referred to by their tumor suppressor genotypes. p53 nullizygosity (p53lox/lox) caused the most rapid progression, yielding lethal tumors by 8 weeks of age, which is comparable to the latency of homozygous p16Ink4a/p19Arf deletion (23). Notably, in this homozygous-null p53 model, progression kinetics were not appreciably altered by p16Ink4a status (Table 1), suggesting either cross-species differences or that developmental inactivation of p53 may diminish the need for p16Ink4a loss (see below). Thus, it is important to note that p16Ink4a−/− animals, with WT p53 and p19Arf germ-line status, also developed lethal pancreatic tumors (mean 18.3 weeks).
Table 1.
PDAC Incidence, Latency, and Histological Phenotype
| Genotype | No. of tumors | Average latency, weeks | Metastasis, % | Histology | aCGH‡ | |||
|---|---|---|---|---|---|---|---|---|
| Adenocarcinoma | Sarcomatoid | Anaplastic | Kras | Myc | ||||
| p16/p19lox/lox | 27 | 8.5 | 11 | 48* (81)† | 26 (37) | 26 (70) | 13/16 | 1/16 |
| p16/p19lox/+ | 12 | 34.2 | 69 | 57 (57) | 43 (43) | 0 (0) | 5/8 | 1/8 |
| p53lox/lox;p16+/+ | 3 | 6.2 | 0 | 100 (100) | 0 (33) | 0 (33) | 1/3 | 0/3 |
| p53lox/lox;p16+/− | 5 | 6.5 | 0 | 80 (100) | 0 (0) | 20 (100) | 3/4 | 3/4 |
| p53lox/lox;p16−/− | 5 | 7.2 | 20 | 40 (80) | 0 (0) | 60 (100) | 0/3 | 0/3 |
| p53lox/+;p16+/+ | 3 | 21.8 | 33 | 100 (100) | 0 (33) | 0 (33) | ||
| p53lox/+;p16+/− | 16 | 14.7 | 25 | 81 (88) | 19 (19) | 0 (44) | 1/6 | 2/6 |
| p53lox/+;p16−/− | 4 | 13.1 | 25 | 75 (100) | 25 (50) | 0 (50) | — | |
| p53+/+;p16−/− | 3 | 18.3 | 33 | 0 (33) | 100 (100) | 0 (0) | ||
| KrasG12D | 3 | 57 | 67 | 0 (33) | 100 (100) | 0 (0) | ||
*Percentage of tumors in which the particular histology predominates (see Materials and Methods).
†The percentage of tumors in which the particular histology is present in any proportion.
‡Number of tumors with copy number gains of Kras or Myc.
Because dual inactivation of p53 and p16Ink4a occurs in most human PDACs, we assessed Pdx1-Cre LSL-KrasG12D mice with heterozygous mutations in either or both of these tumor suppressor genes. Tumor latency in p53lox/+ p16Ink4a+/− animals was significantly reduced compared with those with p53lox/+ alone (see Table 1; 14.7 vs. 21.8 weeks), suggesting cooperative roles for p53 and p16INK4A in PDAC suppression. Finally, with regard to the relative impact of p53 vs. p19Arf in constraining PDAC progression, we observed that p16Ink4a/p19Arf lox/+ mice developed PDAC as well, but with longer latency relative to p53lox/+ p16Ink4a+/− animals (34.2 vs. 21.8 weeks), suggesting that p53 functions as a more potent barrier to PDAC progression. Overall, the clinical presentation across the various PDAC models was similar and typified by weight loss and jaundice.
Influence of Genotype on Histopathological Presentation.
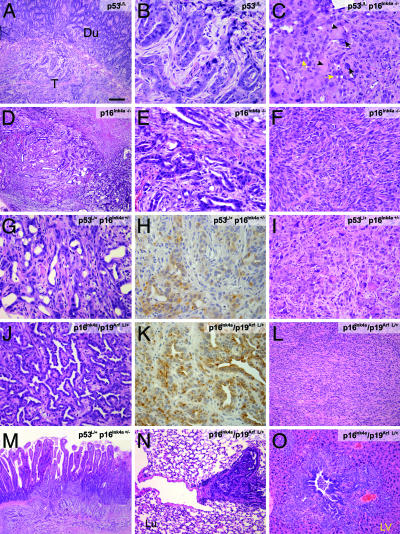
As in other LSL-KrasG12D models (31), histological analysis revealed that the tumors in this study were carcinomas, predominantly ductal adenocarcinomas, defined by the presence of neoplastic glandular (ductal) cells in a dense fibrous stroma or were alternatively, sarcomatoid or anaplastic carcinoma variants, characterized respectively by spindle-cell morphology or by detached tumor cells with marked nuclear and cytoplasmic pleomorphism, occurring in conjunction with ductal differentiation. The distribution of histological phenotypes was different in the various genetic backgrounds. For example, the proportion of ductal adenocarcinomas was highest in the context of p53lox genotypes (Fig. 1A and B). In the p53lox/lox background, p16Ink4a deficiency was associated with a higher frequency of tumors with anaplastic features (Table 1 and Fig. 1C). Nevertheless, mice with a p16Ink4a deficiency but WT p53 germ-line status developed ductal adenocarcinomas with regions of sarcomatoid differentiation (Fig. 1 D–F). In compound heterozygous mice, ductal adenocarcinomas were more common in the p53lox/+ p16Ink4a+/− mice (81% vs. 57% in the p16Ink4a/p19Arf lox/+ cohort) (Table 1 and Fig. 1 G and J), whereas sarcomatoid histology was more common in the p16Ink4a/p19Arf lox/+ tumors (43% compared with 19% in the p53lox/+ p16Ink4a+/− tumors) (Table 1 and Fig. 1L). Finally, the anaplastic features often observed in tumors from homozygous p16Ink4a/p19Arf lox/lox mice (24) were absent in the heterozygous p16Ink4a/p19Arf lox/+ cohort but were often present focally within ductal adenocarcinomas arising in the p53lox/+ p16Ink4a+/− colony (44%)(Table 1 and Fig. 1I). Immunohistochemical analysis confirmed the ductal phenotypes of the well differentiated tumors, with positive staining for ductal markers (CK-19 and DBA lectin) and negative for markers of acinar cell (chymotrypsin) or islet cell (insulin) lineages (Fig. 1 H and K and data not shown). Each model showed invasion and metastasis, although these features were more pronounced in the heterozygous models (Fig. 1 M–O). Overall, these results demonstrate significant roles for p16Ink4a, p19Arf, and p53 in suppressing pancreatic cancer development and suggest specific impact of mutations in these genes in regulating the differentiation state of the ensuing tumors, most prominently indicating that losses of components of the p16Ink4a/p19Arf locus facilitate the development of more poorly differentiated tumors.
Fig. 1.
Deficiency in p53 or p16Ink4a cooperates with oncogenic KrasG12D to produce PDAC. (A) Hematoxylin/eosin stain of a PDAC (T, tumor) arising in a p53lox/lox p16+/+ mouse. Note invasion of duodenum (Du). (Scale bar: A and D, 200 μm; I, N, and O, 100 μm; B, C, E–H, and J–L, 50 μm.) (B) High-magnification view of the tumor in A showing features of ductal adenocarcinoma. (C) Tumor from p53lox/lox p16−/− mouse showing poorly differentiated adenocarcinoma (yellow arrows) admixed with anaplastic epithelioid cells characterized by giant tumor nuclei (black arrows) and eosinophilic inclusions (arrowheads). (D) Invasive PDAC arising in Pdx1-Cre LSL-KrasG12Dp16Ink4a−/− mouse. (E) High-magnification of D showing ductal adenocarcinoma histology. (F) Another region of the tumor in E showing sarcomatoid differentiation. (G) PDAC from p53lox/+;p16Ink4a+/− mouse. (H) Positive staining of the tumor in G for the ductal marker cytokeratin 19. (I) Anaplastic histology in PDAC from the p53lox/+;p16+/−model. (J) Well differentiated p16Ink4a/p19Arf lox/+ PDAC. (K) Positive staining of tumor in J for cytokeratin 19. (L) p16Ink4a/p19Arf lox/+ tumor showing sarcomatoid histology. (M) p53lox/+p16+/− tumor invading the duodenum. (N and O) p16Ink4a/p19Arf lox/+ tumors with metastases to the lung (N) and liver (LV; O).
Somatic Inactivation of PDAC Tumor Suppressor Genes.
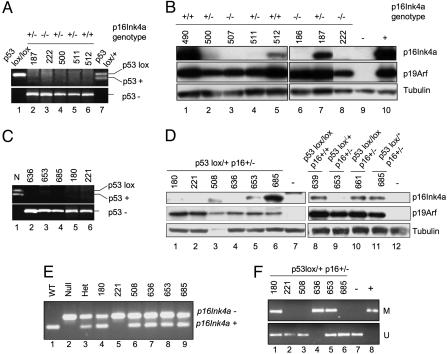
Because our genetic data implicated germ-line lesions in p16Ink4a, p19Arf, and p53 in promoting PDAC progression, we determined the presence of somatic alterations in these genes in primary tumors and early passage tumor cell cultures. In the p53lox/lox colony, elimination of p53 sequences was verified in all samples analyzed, and 10 of 10 tumors showed robust p19Arf expression and WT p19Arf coding sequences (Fig. 2 A and B and data not shown). Examination of p16INK4A status showed that four of four p53lox/lox p16Ink4a+/+ tumor cell lines retained p16INK4A expression, whereas two of four p53lox/lox p16Ink4a+/− tumors showed markedly reduced or absent p16INK4A expression (Fig. 2B and data not shown). These observations, coupled with retention of p53 in all PDAC specimens from p16Ink4a/p19Arf lox/lox mice (24), suggest that p53 and p19Arf serve critical yet largely redundant roles in constraining PDAC progression in the mouse and indicate that p16Ink4a loss is not required for PDAC progression in the context of early p53 inactivation. As noted above, however, a cooperative tumorigenic effect of p53 and p16Ink4a lesions was observed in the p53lox/+ p16Ink4a+/− model. Analysis of derivative tumor cultures from these animals revealed loss of the WT p53 allele in six of six cases (Fig. 2C and data not shown) and low/absent p16INK4A expression in five of six cases (Fig. 2D, lanes 1–6). In contrast, all tumor cell lines expressed p19Arf at comparable levels to the p53lox/lox model (Fig. 2D). Consistent with the reduced/absent p16INK4A expression, there was deletion of WT p16Ink4a sequences in one of six tumor cultures (Fig. 2E, lane 5) and hypermethylation of the p16Ink4a promoter in three of six cases, as determined by methylation-specific PCR (Fig. 2F, lanes 1, 4, and 5). Similar findings were obtained in a larger group of tumors that were not exposed to cell culture (see the supporting information, which is published on the PNAS web site).
Fig. 2.
Molecular analyses of PDAC cell lines and primary tumor specimens from mice with p53 and p16 mutant animals. (A) PCR reactions to detect the p53-WT (+) and p53lox alleles (Upper) and p53-null (−) allele (Lower) in normal tissue from p53lox/lox (lane 1) and p53lox/+ (lane 7) mice and tumor cell lines (lanes 2–6). All tumor cell lines show only the p53-null allele. Germ-line p16Ink4a status is indicated at the top. (B) Western blot for p16Ink4a and p19Arf expression in cell lines derived from p53lox/lox mice with various p16Ink4a genotypes. The negative and positive controls are in lanes 9 and 10, respectively. α-Tubulin is shown as a loading control. (C) PCR analysis of the p53+ and p53lox alleles (Upper) and the p53− (/) allele (Lower) demonstrates loss of the p53+ allele in all tumors from p53lox/+ p16+/− mice (lanes 2–6). Lane 1 shows the WT control specimen. (D) Western blot analysis shows low or absent p16Ink4a expression in five of six tumor cell lines from p53lox/+ p16+/− mice (Right, lanes 1–6), whereas all retain p19Arf expression. Negative controls are in lanes 7 and 12. p16Ink4a expression in PDAC cell lines from p53lox/lox p16+/+ (lane 8) and p53lox/lox p16+/− (lane 10) mice is shown as a reference (Right). (E) PCR analysis of the p16Ink4a− and p16Ink4a+ alleles demonstrates LOH in one of six samples (lane 5). Normal tissue specimens are shown as controls for the p16Ink4a+/+, p16Ink4a−/−, and p16Ink4a+/+ alleles (lanes 1–3). (F) Methylation-specific PCR assay to detect methylated CpG islands in the p16Ink4a promoter region reveals hypermethylation in three tumor lines (lanes 1, 4, and 5). Upper and Lower show the methylated (M) and unmethylated (U) alleles, respectively. Negative and positive controls are in lanes 7 and 8.
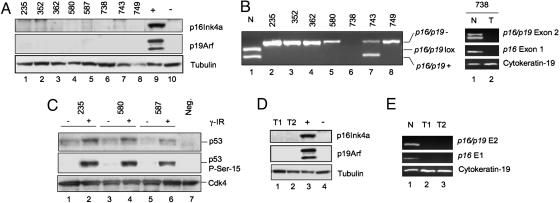
Comparable molecular analysis was performed in the p16Ink4a/p19Arf lox/+ model. As expected, low-passage p16INK4A/p19Arf lox/+ tumor cultures uniformly lacked p16INK4A and p19Arf protein expression (Fig. 3A; n = 8), and seven of eight samples sustained loss of the WT p16Ink4a/p19Arf allele (Fig. 3B). p53 showed low baseline expression, whereas γ-IR led to increased p53 protein levels and Ser-15 phosphorylation consistent with a physically intact p53 allele (Fig. 3C). Additional molecular analysis was performed on two PDACs arising after 1 year in Pdx1-Cre LSL-KrasG12D animals without predisposing tumor suppressor mutations. Early passage tumor cultures and primary tumor lysates showed lack of both p16INK4A and p19Arf expression by Western blot analysis (Fig. 3D and data not shown). Furthermore, extinction of expression was associated with homozygous loss of the p16Ink4a/p19Arf locus as detected by PCR analysis (Fig. 4E). Taken together, our results are in keeping with a model of PDAC tumor suppression in which specific barriers to KrasG12D-directed tumorigenesis are provided by the p19Arf-p53 and p16Ink4 pathways.
Fig. 3.
Molecular analyses of PDAC cell lines from p16Ink4a/p19Arf lox/+ and p16INK4A/p19Arf+/+ mice. (A) Western blot analysis p16Ink4a and p19Arf expression in lysates from tumor cell lines from p16INK4A/p19Arf lox/+ mice. (B) PCR analysis of p16INK4A/p19Arf alleles in tumor cell lines from p16Ink4a/p19Arf lox/+ mice shows only the recombined p16INK4A/p19Arf allele (Left, lanes 2–8). Normal tissue from p16Ink4a/p19Arf lox/+ mice shows WT (+) and unrecombined (lox) alleles. Tumor no. 738 (Left, lane 6; Right, lane 2) shows a biallelic deletion of the entire p16Ink4a/p19Arf locus. (C) Western blot analyses of γ-irradiated tumor cell lines (+) shows induction of total p53 protein levels and phosphorylation of p53 on Ser-15, consistent with present and functional p53 protein. −, untreated. Cdk4 protein levels are shown as a loading control. (D) Western blot analysis shows absence of p16Ink4a and p19Arf expression in Pdx1-Cre LSL-KrasG12D cell lines (lanes 1 and 2). Lanes 3 and 4 show positive and negative controls. (E) PCR analysis for the presence of p16Ink4a/p19Arf exon 2, p16Ink4a exon 1, and cytokeratin 19 sequences in cell lines from Pdx1-Cre LSL-KrasG12D mice (lanes 2 and 3) demonstrates biallelic deletion of p16Ink4a/p19Arf. Lane 1 shows normal control DNA.
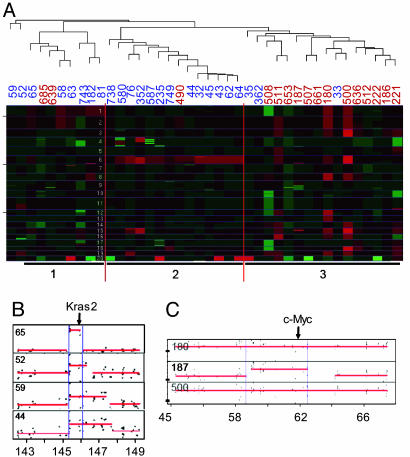
Fig. 4.
aCGH analysis of mouse PDAC. (A) Hierarchical clustering of aCGH profiles reveals three distinct clusters. Sample names are given above the profile followed by an underscore and the genotype class (i.e., “106_6” indicates the profile for tumor no. 106, which is in genotype class 6. The genotype classes are: 1, p53lox/lox;p16Ink4a +/+; 2, p53lox/lox;p16Ink4a +/−; 3, p53lox/lox;p16Ink4a −/−; 4, p53lox/+;p16Ink4a +/−; 5, p16Ink4a/p19Arf lox/+; 6, p16INK4A/p19Arf lox/lox. (B and C) Focal amplifications of Kras2 in p16Ink4a/p19Arf-deficient (B) and Myc in p53-deficient (C) tumors are shown. Minimal common regions of amplification are shown by blue lines.
Genomic Aberrations in Mouse Models of PDAC.
The availability of classes of tumors that either retained or lost expression of p53, p16INK4A, and p19Arf enabled an assessment the impact of these lesions on the level of genomic instability and on the acquisition of specific regional gains or losses of chromosomes. To investigate the possible existence of clonal cooperating genetic lesions, we performed array-comparative genomic hybridization (aCGH) using a high-resolution oligonucleotide microarray on early passage PDAC cells lines from the p16Ink4a/p19Arf lox/lox (n = 16), p16Ink4a/p19Arf lox/+ (n = 8), p53lox/+ p16Ink4a+/− (n = 6), and p53lox/lox (n = 10) models. Intergenotype comparisons of overall genomic instability revealed subtle differences among the various PDAC models, with p53 mutant tumors trending toward an increased genomic complexity (see the supporting information). It is notable that analysis of eight human PCAC cell lines using the same aCGH platform revealed significantly higher numbers of chromosomal copy number alterations (CNAs) compared with the mouse tumors (P < 0.001; see the supporting information).
The aCGH analyses revealed distinct classes of profiles between the mouse genotypes. Hierarchical clustering of the aCGH profiles by using the Pearson correlation as a distance metric yielded three distinct clusters that largely segregated according to p16Ink4a/p19Arf or p53 status (Fig. 4A). p16Ink4a/p19Arf-deficient tumors mainly fell into two clusters (6/8 tumors in cluster 1 and 13/14 tumors in cluster 2), whereas p53-deficient specimens predominated in the final cluster (13/16 tumors in cluster 3). A notable feature of cluster 2 was copy number increases in chromosome 6 containing the Kras locus. Although gains of the entire chromosome were noted, in a number of cases, the alterations were highly focal, spanning <1 Mb, indicating that Kras was the target of this event (Fig. 4B). Amplification of chromosome 6 occurred with much greater frequency in p16Ink4a/p19Arf mutant tumors (75%) than in p53 mutant tumors (31%) (Table 2). Recurrent and genotype-specific alterations also occurred in the form of focal gains in chromosome 15 spanning the c-Myc locus (Fig. 4 C and Table 2; 31% of p53 mutant tumors vs. 8% of the p16Ink4a/p19Arf mutant tumors) and deletion of chromosome 4 in the vicinity of the p16Ink4a/p19Arf locus (Fig. 4A; 50% of tumors from p16Ink4a/p19Arf lox/+ mice and one of six tumors from p53lox/+ p16Ink4a+/− mice). These genomic distinctions within the data set were validated by using an alternative unsupervised clustering approach (D. Carrasco, unpublished work; see supporting information). These approaches demonstrate that CNA patterns including amplifications of Kras and c-Myc effectively identify classes of tumors that reflect the underlying tumor suppressor genotype. Other CNAs were also documented in these mouse PDAC genomes, including recurrent gains on chromosomes 1q, 4q, 8q, 16q, 17q, and 18q as well as losses on chromosomes 9q, 10q, 14q, and 17q (Fig. 4A and supporting information). Importantly, several focal and recurrent CNAs, such as 16q amplification and 17q deletion, are syntenic to recurrent CNAs in sporadic human PDAC (32, 33). Thus, these CNAs provide both a measure of molecular validation of murine PDAC models and a potential means of facilitating identification of PDAC progression genes.
Table 2.
Gain/amplification of Kras2 and Myc in mouse PDAC
| Genotype | Kras2, % | Myc, % |
|---|---|---|
| p53lox/lox (n = 10) | 40 | 30 |
| p53lox/+ (n = 6) | 17 | 33 |
| All p53 (n = 16) | 31 | 31 |
| p16Ink4a/p19Arf lox/lox (n = 16) | 81 | 6 |
| p16Ink4a/p19Arf lox/+ (n = 8) | 63 | 13 |
| All p16Ink4a/p19Arf (n = 24) | 75 | 8 |
Discussion
Both p16Ink4a and Arf-p53 Are Barriers to Mouse PDAC Progression.
This study provides several lines of evidence establishing that both components of the p16Ink4a/p19Arf locus provide critical barriers to PDAC progression. Specifically, mice with homozygous mutations in either p53 or p16Ink4a developed tumors with short latency; moreover, mice with combined heterozygous mutations in p53 and p16Ink4a developed PDAC that predominantly lost expression of products of both genes but showed robust expression of p19Arf. Tumors arising in p16Ink4a/p19Arf lox/+ animals lost both products of this locus but retained p53. Two aspects of this analysis are notable. First, although sharp distinctions have been drawn between Arf and p53 function in several other models and p53 has Arf-independent activities and vice versa (12), our PDAC models demonstrate that the roles of these tumor suppressors overlap significantly with regard to PDAC progression in the mouse. A second observation relates to the role of p16INK4A loss in the setting of p53 inactivation. Specifically, whereas heterozygous deletion of p53 and p16Ink4a during Kras initiation begets subsequent inactivation of the WT alleles of both genes during PDAC progression, loss of p53 function in the context of intact p16Ink4 is not associated with subsequent p16INK4A loss (this study and ref. 25). Possible explanations for this discrepancy include: (i) early and/or uniform homozygous loss of p53 coincident with Kras activation enables bypass of p16INK4A -mediated RB pathway tumor suppressor functions that would otherwise be operative; or (ii) there are species-specific differences in the tumor suppressor functions between human and mouse p16INK4A and p53, as suggested by other studies (26). Irrespective, the double-heterozygous models reflect well the human situation in regard to the complimentary benefits of their concomitant loss and, thus, will provide a highly appropriate model system for studying aspects of PDAC progression.
Tumor Suppressor Gene Status Influences Tumor Biology.
Different combinations of tumor suppressor gene mutations, in conjunction with KrasG12D expression, all promoted the progression of PanIN to PDAC but produced tumors with varying spectra of clinical and histological features. Although tumors in all genotypes showed extensive local invasion and micrometastases, gross metastases were only prominent in mice with engineered heterozygous tumor suppressor deletions (p16Ink4a/p19Arf lox/+ or p53lox/+ p16Ink4a lox/+ mice) but not in mice with engineered homozygous deletions. Because the homozygous animals rapidly developed a lethal tumor burden, often with multiple primary tumors, these results raise the possibility that factors relating to tumor latency rather than the specific p16INK4A-RB or p19Arf-p53 pathway lesions are primarily responsible for modulating metastatic behavior.
With respect the effect of genotype on tumor histology, the p53-deficient cohorts showed the higher prevalence of well differentiated ductal adenocarcinoma compared with the p16Ink4a/p19Arf-deficient animals (Table 1). Conversely, undifferentiated sarcomatoid histology was significantly reduced in p53-deficient models. These results are consistent with observations that KrasG12D and p53R172H alleles produce primarily ductal adenocarcinomas (25). Anaplastic carcinoma, although common in the homozygous models, was never the principal histological component in the heterozygous p53lox/+ p16Ink4a+/− tumors and was completely absent in the p16Ink4a/p19Arf lox/+ model. In humans, ductal adenocarcinoma histology predominates, and the sarcomatoid and anaplastic subtypes are considered uncommon variants of PDAC with more aggressive clinical behavior, although they all appear to have comparable spectra of genetic lesions (34, 35). Our mouse models collectively recapitulate these different histologic variants, albeit at different frequencies than seen in spontaneous human tumors. Overall, these observations suggest that the set of tumor suppressor lesions strongly influences the cell differentiation phenotypes of the resulting tumors.
Recurrent Genomic Aberrations Arise During PDAC Progression.
Genomic analysis revealed an effect of tumor suppressor genotype on the profile of tumor-associated chromosomal alterations and only modest trends toward increased genomic instability in p53 mutant tumors on the basis of the number of CNAs per tumor. Notably, the degree of genomic instability occurring in each of these PDAC mouse models is significantly less than that observed in human specimens (this study and refs. 32 and 33). This may reflect a bias imposed through the use of genetically engineered mouse models (i.e., p53 loss and KRAS activation may obviate the need for many further cytogenetic aberrations) or cross-species differences in chromosome biology and structure, particularly telomeres (5, 36, 37). The patterns of regional alterations in chromosomal copy number differed between tumors of different genotypes, suggesting that chromosomal alterations may harbor genes in which altered expression may specifically cooperate with distinct tumor suppressor gene mutations. Notable copy number gains at chromosome 6 encompassing the Kras locus were more commonly observed in the p16Ink4a/p19Arf mutant tumors. This correlation may reflect the capacity of Kras to activate both the p16INK4A and p19Arf promoters leading to cellular senescence in vivo (8, 38). p16INK4A/p19Arf-deficient tumors may be especially permissive for KrasG12D amplification, facilitating the cellular transformation associated with increased Kras signaling. In contrast, chromosome 15 gains overlapping the Myc locus were much more common in the p53 mutant tumors. The selective amplification of Myc in association with p53 mutation has been noted in genomic analysis of melanomas harboring either p53 or p16Ink4a/p19Arf mutations (39). In these contexts, Myc amplification may serve to bypass the inhibition of CDK4 activity and consequent cell-cycle arrest conferred by elevated p16Ink4a expression (39, 40). It is notable that, in the subset of p53lox/lox and p53lox/+ tumors retaining robust p16INK4A expression, other RB pathway components remained intact as evidenced by the absence of p16Ink4a and Cdk4 mutations in full-length sequence analyses and the lack of perturbations in expression of RB and cyclin D1 (data not shown).
Amplifications of KRAS on chromosome 12q12 and MYC at 8q24 as well as deletions of p16INK4A on 9p21 are commonly observed in human pancreatic adenocarcinoma (32, 33). In addition to CNAs harboring validated pancreatic cancer genes, we have also identified additional mouse CNAs that are syntenic to human PDAC loci (see supporting information). Recurrence of these loci in multiple different tumor specimens and their evolutionary conservation indicate that they may harbor cancer genes with prime importance in PDAC pathogenesis. Moreover, the targeting of syntenic loci in these mouse PDACs highlights the potential of mouse models of cancer to serve as effective filters in the identification of cancer genes present in complex human copy number data sets.
Materials and Methods
Mouse Strains, Histopathology, and Establishment of Primary PDAC Cell Lines.
The mouse strains in this study included the following alleles: LSL-KrasG12D (41), Pdx1-Cre (27), conditional p53lox (30), conditional p16Ink4a/p19Arf lox (24), and a germ-line p16Ink4a-specific null allele (that retains WT p19Arf) (29). All experiments were performed on >87.5% FVB/n background. Some mice with the p16Ink4a/p19Arf lox allele developed lymphomas (latency 28–60 weeks) that occurred independently of concurrent expression of Pdx1-Cre or LSL-KrasG12D. The p16Ink4a/p19Arf lox allele shows intact expression and function of the p16Ink4a/p19Arf locus in cell culture-based assays in vitro, and it appears that the lymphomas arising in these mice are attributable to hypomorphic activity of p16Ink4a/p19Arf lox allele. In addition, a subset of mice from both the p16Ink4a/p19Arf lox/+ and p53lox/+ p16Ink4a+/− colonies developed progressive wasting in the absence of evident neoplastic growth, whereas some others developed cutaneous papillomas. These phenotypes are likely due to the extrapancreatic expression of KrasG12D induced by the Pdx1-Cre transgene because they are not observed in the context of the p48-Cre strain in which expression pattern is more tightly restricted to the pancreas (data not shown). Tissue processing, immunohistochemical staining, and establishment of primary PDAC cell lines were performed as described in ref. 24. The histological classification of each tumor reported in Table 1 was determined by evaluation of cross-sectional slides from two axes. The histological type representing the largest cross-sectional area was considered the primary component.
Molecular Analysis.
RNA and DNA isolation, protein extract preparation from PDAC cell lines, and primary tumors was performed as described in ref. 24. For loss of heterozygosity analysis of p16Ink4a, p53, and p16Ink4a/p19Arf, DNA was extracted from the early passage cell lines and amplified by PCR. Methylation-specific PCR analysis was performed as described in ref. 42.
Genomic Analysis.
aCGH and data analysis was performed as described on Agilent mouse development or human 1A oligonucleotide microarrays (see supporting information) (32).
Supplementary Material
Acknowledgments
We thank David Tuveson and Tyler Jacks for the LSL-KrasG12D mice; Anton Berns for the p53lox mice; Doug Melton for the Pdx1-Cre mice; and Alice Yu, Karen Marmon, and Shan Jiang for expert monitoring of the mouse colony. T. R. Devereux generously provided the Sp6c lung cancer cell line. This work was supported by grants from the National Institutes of Health (to R.A.D. and N.B.). R.A.D. is an American Cancer Society Research Professor.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- PanIN
pancreatic intraepithelial neoplasias
- CDK
cyclin-dependent kinase
- RB
retinoblastoma protein
- aCGH
array-comparative genomic hybridization
- CNAs
copy number alterations.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Parkin D. M., Bray F., Ferlay J., Pisani P. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Murray T., Ward E., Samuels A., Tiwari R. C., Ghafoor A., Feuer E. J., Thun M. J. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Hruban R. H., Wilentz R. E., Kern S. E. Am. J. Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiGiuseppe J. A., Hruban R. H., Offerhaus G. J., Clement M. J., van den Berg F. M., Cameron J. L., van Mansfeld A. D. Am. J. Pathol. 1994;144:889–895. [PMC free article] [PubMed] [Google Scholar]
- 5.van Heek N. T., Meeker A. K., Kern S. E., Yeo C. J., Lillemoe K. D., Cameron J. L., Offerhaus G. J., Hicks J. L., Wilentz R. E., Goggins M. G., et al. Am. J. Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozenblum E., Schutte M., Goggins M., Hahn S. A., Panzer S., Zahurak M., Goodman S. N., Sohn T. A., Hruban R. H., Yeo C. J., Kern S. E. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 7.Sharpless N. E. Mutat. Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 9.Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., Peeper D. S. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 10.Pomerantz J., Schreiber-Agus N., Liegeois N. J., Silverman A., Alland L., Chin L., Potes J., Chen K., Orlow I., Lee H. W., et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Xiong Y., Yarbrough W. G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 12.Lowe S. W., Sherr C. J. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto M., Kuo M. L., Roussel M. F., Sherr C. J. Mol. Cell. 2003;11:415–424. doi: 10.1016/s1097-2765(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 14.Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 15.Schutte M., Hruban R. H., Geradts J., Maynard R., Hilgers W., Rabindran S. K., Moskaluk C. A., Hahn S. A., Schwarte-Waldhoff I., Schmiegel W., et al. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 16.Whelan A. J., Bartsch D., Goodfellow P. J. N. Engl. J. Med. 1995;333:975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein A. M., Fraser M. C., Struewing J. P., Hussussian C. J., Ranade K., Zametkin D. P., Fontaine L. S., Organic S. M., Dracopoli N. C., Clark W. H., Jr, et al. N. Engl. J. Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 18.Hustinx S. R., Leoni L. M., Yeo C. J., Brown P. N., Goggins M., Kern S. E., Hruban R. H., Maitra A. Mod. Pathol. 2005;18:959–963. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 19.Vousden K. H., Lu X. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 20.Maitra A., Adsay N. V., Argani P., Iacobuzio-Donahue C., De Marzo A., Cameron J. L., Yeo C. J., Hruban R. H. Mod. Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 21.Boschman C. R., Stryker S., Reddy J. K., Rao M. S. Am. J. Pathol. 1994;145:1291–1295. [PMC free article] [PubMed] [Google Scholar]
- 22.Heinmoller E., Dietmaier W., Zirngibl H., Heinmoller P., Scaringe W., Jauch K. W., Hofstadter F., Ruschoff J. Am. J. Pathol. 2000;157:83–92. doi: 10.1016/S0002-9440(10)64520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingorani S. R., Petricoin E. F., Maitra A., Rajapakse V., King C., Jacobetz M. A., Ross S., Conrads T. P., Veenstra T. D., Hitt B. A., et al. Cancer Cells. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 24.Aguirre A. J., Bardeesy N., Sinha M., Lopez L., Tuveson D. A., Horner J., Redston M. S., DePinho R. A. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hingorani S. R., Wang L., Multani A. S., Combs C., Deramaudt T. B., Hruban R. H., Rustgi A. K., Chang S., Tuveson D. A. Cancer Cells. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Rangarajan A., Hong S. J., Gifford A., Weinberg R. A. Cancer Cells. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Gu G., Dubauskaite J., Melton D. A. Development (Cambridge, U.K.) 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 28.Tuveson D. A., Shaw A. T., Willis N. A., Silver D. P., Jackson E. L., Chang S., Mercer K. L., Grochow R., Hock H., Crowley D., et al. Cancer Cells. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 29.Sharpless N. E., Bardeesy N., Lee K. H., Carrasco D., Castrillon D. H., Aguirre A. J., Wu E. A., Horner J. W., DePinho R. A. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 30.Marino S., Vooijs M., van Der Gulden H., Jonkers J., Berns A. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 31.Solcia E., Capella C., Kloppel G. Tumors of the Pancreas. Washington, DC: Armed Forces Institute for Pathology; 1995. [Google Scholar]
- 32.Aguirre A. J., Brennan C., Bailey G., Sinha R., Feng B., Leo C., Zhang Y., Zhang J., Gans J. D., Bardeesy N., et al. Proc. Natl. Acad. Sci. USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidenblad M., Schoenmakers E. F., Jonson T., Gorunova L., Veltman J. A., van Kessel A. G., Hoglund M. Cancer Res. 2004;64:3052–3059. doi: 10.1158/0008-5472.can-03-3159. [DOI] [PubMed] [Google Scholar]
- 34.Hoorens A., Prenzel K., Lemoine N. R., Kloppel G. J. Pathol. 1998;185:53–60. doi: 10.1002/(SICI)1096-9896(199805)185:1<53::AID-PATH45>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Deckard-Janatpour K., Kragel S., Teplitz R. L., Min B. H., Gumerlock P. H., Frey C. F., Ruebner B. H. Arch. Pathol. Lab. Med. 1998;122:266–272. [PubMed] [Google Scholar]
- 36.Maser R. S., DePinho R. A. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 37.Blasco M. A. EMBO J. 2005;24:1095–1103. doi: 10.1038/sj.emboj.7600598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmero I., Pantoja C., Serrano M. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 39.Bardeesy N., Bastian B. C., Hezel A., Pinkel D., DePinho R. A., Chin L. Mol. Cell. Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alevizopoulos K., Vlach J., Hennecke S., Amati B. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson E. L., Willis N., Mercer K., Bronson R. T., Crowley D., Montoya R., Jacks T., Tuveson D. A. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bardeesy N., Morgan J., Sinha M., Signoretti S., Srivastava S., Loda M., Merlino G., DePinho R. A. Mol. Cell. Biol. 2002;22:635–643. doi: 10.1128/MCB.22.2.635-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.