Abstract
A single nucleotide polymorphism (SNP) in the sickle β-globin gene (βS) leads to sickle cell anemia. Sickling increases sharply with deoxy sickle Hb concentration and decreases with increasing fetal γ-globin concentration. Measures that decrease sickle Hb concentration should have an antisickling effect. RNA interference (RNAi) uses small interfering (si)RNAs for sequence-specific gene silencing. A βS siRNA with position 10 of the guide strand designed to align with the targeted βS SNP specifically silences βS gene expression without affecting the expression of the γ-globin or normal β-globin (βA) genes. Silencing is increased by altering the 5′ end of the siRNA antisense (guide) strand to enhance its binding to the RNA-induced silencing complex (RISC). Specific βS silencing was demonstrated by using a luciferase reporter and full-length βS cDNA transfected into HeLa cells and mouse erythroleukemia cells, where it was expressed in the context of the endogenous β-globin gene promoter and the locus control region enhancers. When this strategy was used to target βE, silencing was not limited to the mutant gene but also targeted the normal βA gene. siRNAs, mismatched with their target at position 10, guided mRNA cleavage in all cases except when two bulky purines were aligned. The specific silencing of the βS-globin gene, as compared with βE, as well as studies of silencing SNP mutants in other diseases, indicates that siRNAs developed to target a disease-causing SNP will be specific if the mutant residue is a pyrimidine and the normal residue is a purine.
Keywords: sickle cell anemia, specificity of silencing, hemoglobin E, hemoglobin S
Sickle cell anemia (SCA) results from a single nucleotide substitution in codon 6 (GAG→GTG) of the Hb β-chain gene (βS). The Glu→Val substitution produces a Hb variant (HbS) that polymerizes upon deoxygenation to produce long rigid fibers that distort red blood cell (RBC) shape. Sickled RBCs have reduced flexibility that impairs their transit through the microvasculature and leads to vasoocclusion, localized hypoxia, painful crises, and organ damage. The sickled RBCs are also prone to hemolysis with resulting anemia. The intracellular concentration and the polymerization of deoxyHbS are the critical pathogenic determinants of SCA (1, 2). Other Hbs, such as HbA or HbC may permit or promote the polymerization of deoxyHbS, or as with HbF or HbA2, may inhibit the polymerization and have an antisickling effect (2). A determinant of SCA severity is the relative abundance of γ and βS, with elevated levels of fetal HbF (α2γ2) mitigating disease severity. Hydroxyurea (HU) treatment, which increases γ-gene expression, decreases sickle cell crises (3). Similarly, the disease is ameliorated in mouse models of SCA when gene therapy is used to increase the intraerythrocytic concentration of a modified βA mutated in position 87Thr→Gln, a key residue for the antisickling properties of HbF (4).
An alternate therapeutic approach, which we present here, is to decrease the intracellular concentration of HbS by specifically silencing the βS gene without diminishing the expression of the γ or the normal βA gene. In recent years, RNA interference (RNAi) has emerged as a powerful approach for silencing gene expression. RNAi leads to sequence-specific gene silencing in response to small interfering (si)RNAs that guide the degradation of homologous mRNAs (5). One of the siRNA strands (the guide strand) becomes incorporated into the RNA-induced silencing complex (RISC) to direct target mRNA cleavage. The choice of strands that enter RISC is determined by the relative thermodynamic stability of each of the 5′ ends; the strand that is energetically favored for unwinding predominates in RISC (6).
siRNAs with incomplete homology to their target mRNA sequence can also silence gene expression. However, the rules that govern which partially homologous target mRNAs are efficiently silenced by any small interfering (si)RNAs are still uncertain. The central region of the siRNA:mRNA interaction site is thought to be critical for silencing (7). Homology at the site of mRNA cleavage (between positions 10 and 11 from the 5′ end of the siRNA guide strand) is considered essential for efficient silencing by mRNA degradation but may not be required for the less efficient silencing by inhibition of translation (8–10).
The ability to silence specifically an allele with a disease-causing SNP is desirable not only for SCA, but also for other conditions resulting from a mutant SNP. SNP-selective targeting has already been shown to silence an oncogenic ras (K-RASV12) and a spinocerebellar ataxia gene allele (ataxin-3) (11, 12). Here we investigate SNP-selective targeting of disease-causing alleles of human β-globin. To maximize the potential specificity of silencing, β allele-specific siRNAs were designed so that the SNP was aligned with position 10 of the guide strand. These siRNAs were analyzed by using an allele-specific luciferase reporter. The βS siRNA specifically silenced the βS gene, without inhibiting the expression of the normal βA or γ-globin genes. By altering the thermodynamic properties of the 5′ end of the guide strand, silencing could be substantially increased. Despite the high specificity of the βS siRNA, the corresponding βA siRNA not only silenced βA but also silenced the βS reporter and cDNA expression construct, targeting them for degradation. A further examination of different combinations of nucleotides at the mismatched siRNA:mRNA interaction site demonstrated that central mismatches consisting of pyrimidine:pyrimidine or pyrimidine:purine residues maintained a significant level of silencing by mRNA cleavage. However, introducing two bulky mismatched purines at position 10 abrogated silencing. The same observation was made with siRNAs designed to silence another disease-causing allele (βE), which has a codon 26 SNP and produces HbE that is unstable under conditions of oxidative stress and elevated body temperature (13). These findings suggest that mismatches at the mRNA cleavage site involving pyrimidines can be accommodated within the catalytic site of the RISC endonuclease Ago2 and lead to mRNA degradation, despite the lack of a base-paired interaction at the active site. Although earlier studies have examined the position of mismatches on the specificity of silencing, this study shows that the nucleotide composition of the mismatch determines the specificity of the silencing reaction (14).
Results
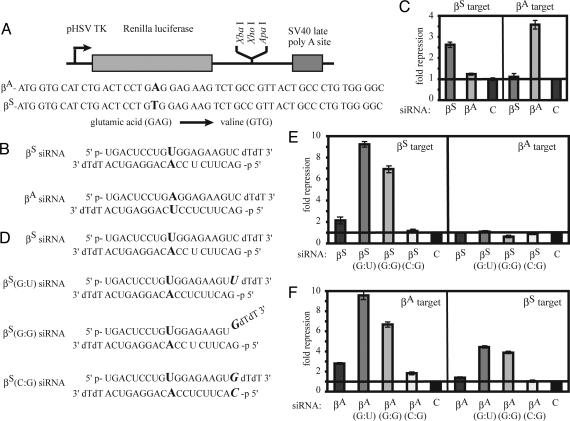
Using RNAi as a potential therapeutic agent for SNP β-hemoglobinopathies requires siRNAs to discriminate between β-globin alleles. For SCA, the siRNA needs to silence βS and have little or no effect on the expression of the γ and βA genes. Because βA and βS differ by only one nucleotide, the most rigorous test of specificity would be to determine whether the βS siRNA would affect βA expression. Specific silencing of βS is particularly important if it is done in conjunction with expression of βA (4). A 50-nt target sequence from either βS or βA containing the region surrounding the SNP was cloned into the 3′ UTR of the Renilla luciferase expression vector pTK-RL (8) (Fig. 1A). These expression plasmids were cotransfected with βS, βA, or control siRNA and the firefly luciferase control vector (Fig. 1B). The SNP was aligned with position 10 of each siRNA guide strand. The βS siRNA silenced the βS reporter target ≈2.5-fold with only minimal effects on βA expression (Fig. 1C). However, the overall silencing was low. To improve silencing, the siRNA was altered at the 5′ terminus of the guide strand to promote the preferential uptake of this strand into the RISC. Replacing the C at position 19 of the sense strand with a U changes the 5′ terminal base pair of the guide strand of the siRNA from the stable G:C pairing to the non-Watson–Crick G:U pairing (Fig. 1D). The βS siRNA containing the G:U base pair [βS(G:U)] dramatically improved silencing of the reporter construct to ≈9-fold compared with the nonspecific control siRNA, without sacrificing specificity (Fig. 1E). Similarly, incorporating a G:G mismatch at the 5′ end of the guide strand by replacing the C residue at position 19 of the target strand with a G, producing a frayed or forked siRNA [βS(G:G)], repressed expression of the βS target construct ≈7-fold and maintained specificity. When the base pairing of the 5′ end of the guide strand was restored by modifying both siRNA strands [βS(C:G)] silencing was minimal (1.2-fold) and reduced compared with the original siRNA. None of the modified βS siRNAs appreciably targeted the βA reporter. Therefore, allele-specific silencing of βS can be achieved, and the efficacy of silencing can be increased by destabilizing base-pairing at the 5′ end of the guide strand.
Fig. 1.
Allele-specific silencing of βS. (A) Renilla luciferase reporter plasmids into which the target βA and βS sequences were cloned. (B) The βA and βS siRNA sequences were designed with the SNP aligned with position 10 of the guide strand. (C) Transfection of βA and βS siRNAs induces allele-specific silencing of the luciferase reporter constructs. The fold repression was determined by the degree of silencing of the Renilla luciferase-targeting construct relative to the firefly luciferase control after background subtraction of the GFP control siRNA-treated samples. (D) Sequences of βS siRNAs destabilized at the 5′ terminus of the guide strand. Similar destabilized siRNAs were made for the βA siRNA. (E) Destabilization of the βS siRNAs increased the efficacy of silencing but retained the specificity of silencing. (F) The destabilized βA siRNA showed enhanced silencing of the βA target but also showed significant silencing of the βS reporter. For both sets of siRNAs, restoring the base pair interaction at the 5′ terminus eliminated silencing.
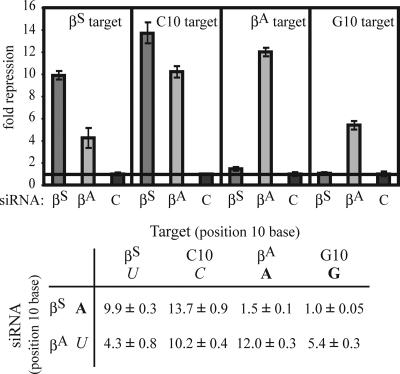
Modifications to the 5′ end of the guide strand of the βA siRNA also enhanced βAsilencing (Fig. 1F). However, in contrast to the specificity of the βS siRNAs, the highly active modified βA siRNA also silenced the βS target by as much as ≈4-fold. These two targeting reactions differ only in the mismatched nucleotides at the site of RISC-mediated endonuclease cleavage, with an A:A mismatch between the βS siRNA and the βA mRNA and a U:U mismatch between the βA siRNA and the βS mRNA. This finding implies that the U:U mismatch is more permissive for silencing than the A:A mismatch. One possible explanation is that juxtaposing two bulky purines (A:A) at the site of mRNA cleavage distorts the expected A-type helix formed between the siRNA and the target mRNA, affecting the positioning of the mRNA in the Ago2 active site and inhibiting cleavage. Smaller mismatched pyrimidines (U:U) might be more readily accommodated and may be permissive for silencing. To address this possibility, the specificity of silencing by the βS and βA siRNAs was tested against reporter constructs modified to contain either a C (pyrimidine) or a G (purine) residue in the mismatched position (Fig. 2). Consistent with the previous experiment, a pyrimidine:pyrimidine (C:U) or pyrimidine:purine (C:A or U:G) mismatch allowed silencing, whereas a purine:purine mismatch (A:G or A:A) strongly inhibited silencing.
Fig. 2.
siRNAs mismatched with their target mRNA at position 10 still retain silencing activity, unless the mismatch contains two purine bases. The βS(G:U) and βA(G:U) siRNAs were tested against reporters which contained either a pyrimidine (U or C) or purine residue (A or G) juxtaposed with position 10 of the siRNAs. Silencing of the Renilla luciferase reporter constructs was determined as in Fig. 1.
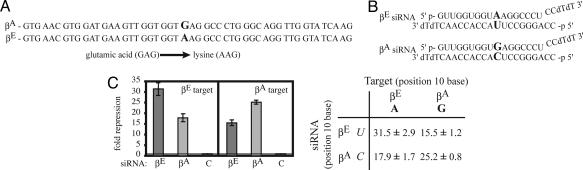
To test further the effect of position 10 base composition on silencing, we designed siRNAs to silence βE, another β-globin disease variant with a SNP in codon 26 (GAG→AAG, Glu→Lys). siRNAs were designed to silence βE, but not βA, and tested by using luciferase reporter constructs containing a 50-nt region of the normal and variant β-globin gene encompassing the βE mutation (Fig. 3A and B). To increase the efficiency of silencing, two mismatched base pairs were incorporated into the siRNA at the 5′ end of the guide strand. The βE siRNA strongly silenced luciferase expression of the βE target reporter (≈32-fold) (Fig. 3C). However, the βE siRNA also significantly silenced the expression of the corresponding βA target (≈16-fold). The βA siRNA effectively silenced the βA target reporter (≈25-fold) but showed almost equivalent silencing of the βE target (≈19-fold). As in the previous analysis, the juxtaposition of a purine and a pyrimidine (A:C or G:U) at the cleavage site was permissive for silencing (Fig. 3D). These findings support the hypothesis that purine:pyrimidine and pyrimidine:pyrimidine mismatches at the siRNA:mRNA cleavage site permit silencing, whereas bulky purine:purine mismatches do not.
Fig. 3.
Lack of allele-specific silencing of βE. (A) Renilla luciferase reporter plasmids into which the target βA and βE sequences were cloned. (B) The sequence of the βA and βE siRNAs (the position 10 mismatch distinguishing each allele is shown in bold). (C) Silencing of the Renilla luciferase reporter is shown relative to the firefly luciferase control, as in Fig. 1. In C, pyrimidine residues are italicized, and purine residues are shown in bold.
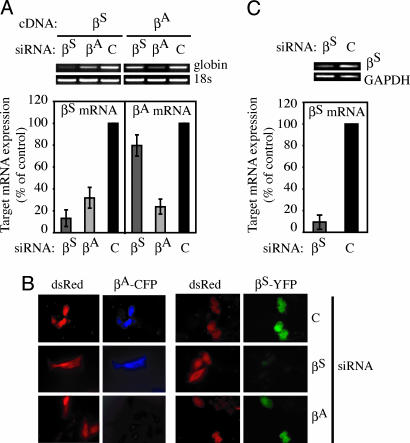
The targeting constructs contain only a small portion of the β-globin genes. To demonstrate that the allele-specific siRNAs can silence full length mRNA, the siRNAs that showed the strongest silencing [βS(G:U) or βA(G:U)] were cotransfected into HeLa cells with a cDNA expression plasmid for either βS or βA. β-globin mRNA was assessed by RT-PCR and quantified by using real-time RT-PCR analysis. βS siRNA substantially reduced βS mRNA without silencing βA mRNA (Fig. 4A). As we observed for the luciferase reporter assay, the βA siRNA silenced both the βA mRNA and the βS mRNA. Because the βA siRNA is not being considered for clinical use, this cross-reactivity is not of therapeutic concern. Despite the lack of base pairing at position 10, gene silencing occurred via mRNA degradation, as demonstrated by decreased βS mRNA in cells treated with βA siRNA (Fig. 4A).
Fig. 4.
Targeted silencing of β-globin mRNA and protein. (A) βS(G:U) siRNA silenced βS mRNA but showed no appreciable targeting of βA mRNA as determined by RT-PCR and quantitative RT-PCR. The percent expression of the target is given relative to cells transfected with control GFP siRNA. The corresponding βA(G:U) siRNA effectively silenced βA mRNA but also efficiently silenced βS mRNA expression. (B) Similarly, the βS(G:U) siRNA silenced βS globin expression, as determined by a lack of βS-GFP fusion protein in transfected HeLa cells, but had no effect on the expression of βA-globin (βA-CFP). The βA(G:U) siRNA effectively silenced βA-globin, as well as βS-globin, expression. dsRed expression was used as a transfection control. (C) βS(G:U) siRNA silences βS in MEL cells stably expressing βS from the human globin promoter and locus control region, compared with control siRNA-treated cells.
To test silencing of β-globin protein, plasmids were designed to express βS or βA fluorescently tagged with yellow fluorescent protein (YFP) or cyan fluorescent protein (CFP), respectively (Fig. 4B). The fluorescently tagged β-globin expression plasmids were cotransfected with the βS, βA, or control siRNA and a dsRed expression plasmid to identify transduced cells. The βS-YFP fusion protein, but not the βA-CFP fusion protein, was silenced in the βS siRNA-transfected cells. A control siRNA did not silence βS-YFP (Fig. 4B). βA siRNA effectively silenced βA-CFP but also reduced βS-YFP expression. Although βS-YFP fluorescence was reduced in each cell transfected with the βA siRNA, no cells had completely silenced βS. The luciferase reporter assay, the RT-PCR analysis of mRNA, and fluorescent-tagged protein expression visualized by microscopy all gave similar results supporting the specificity of the βS siRNA for targeting the SNP of the sickle allele and the lack of specificity of the βA siRNA. To verify that the βS siRNA works in the context of endogenous mRNA expression, the βS(G:U) siRNA was tested in mouse erythroleukemia cells that stably express βS from the endogenous β-globin promoter and regulatory region, the locus control region (MEL-βS). βS mRNA was significantly decreased (>5-fold) in treated cells compared with control siRNA-treated cells (Fig. 4C).
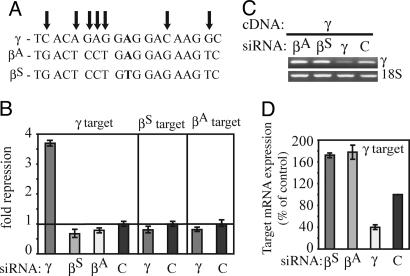
Because the severity of SCA is reduced by fetal (γ) globin gene expression, a therapeutic siRNA designed to reduce βS expression should not reduce fetal γ-globin gene expression. The γ-globin gene differs from βAor βS in the 19-nt targeted sequence by 7 or 8 nucleotide mismatches, respectively. It is therefore unlikely that either the βA or βS siRNA would silence γ-globin expression (Fig. 5A). To verify this conjecture, a corresponding 50-bp region of the γ-globin gene was cloned into the Renilla luciferase expression vector and tested for silencing when cotransfected with βS(G:U), βA(G:U), or control siRNA. None of these siRNAs silenced γ-globin (Fig. 5B). However, an siRNA designed to target the corresponding γ sequence silenced γ but not βA or βS. These findings were corroborated by RT-PCR and real-time PCR analysis of HeLa cells transfected with a γ-globin expression plasmid (Fig. 5 C and D). γ-globin mRNA was efficiently targeted by the γ siRNA but not by the βA or βS siRNAs. γ-globin mRNA was even somewhat higher in the βA- and βS-treated samples. Therefore, βS siRNA does not interfere with fetal globin gene expression.
Fig. 5.
βS siRNA does not reduce fetal γ-globin expression. (A) Schematic of the γ sequence targeted by the siRNAs against βS and βA. (B) The βS and βA siRNA do not repress γ expression of a luciferase reporter construct containing a 50-nt region of the γ-globin gene corresponding to the homologous region encoded by the βS and βA reporter constructs. Likewise, an siRNA against γ-globin failed to target βS and βA. (C and D) Expression of full length γ from the CMV promoter is silenced by a γ siRNA, but not by the βS(G:U) and βA(G:U) siRNAs, as shown by RT-PCR (C) and quantified by real-time RT-PCR (D). All values are relative to GFP siRNA-treated cells.
Discussion
A key factor that makes RNAi a powerful gene silencing tool and potential therapeutic is its specificity (5). Early studies demonstrated the possibility of specifically silencing a mutant allele differing by only a SNP and leaving the normal allele unaffected (10–12). However, it is now known that siRNAs also silence some targets with incomplete homology (5, 15). The central region of the siRNA between position 5 and 11 is the most sensitive to mismatches in the mRNA sequence, in particular positions 9, 10, and 11, which surround the mRNA cleavage site (5, 10, 16). The goal of this study was to design an allele-specific siRNA targeting the SNP of sickle βS without affecting normal βA or fetal γ-globin gene expression. By aligning the siRNA so that the SNP paired with position 10 of the siRNA guide strand, we were able to design an siRNA that silenced βS but left βA and γ-globin expression intact. This siRNA could be improved by introducing mismatches at the 5′ end of the guide strand to favor association of the guide, rather than the sense, strand with the RISC.
Our control βA siRNAs unexpectedly directed cleavage of the βS reporter gene and βS mRNA, despite a mismatch at the critical position of endonuclease cleavage. By examining gene silencing by siRNAs designed to target βA, βS, and another β-globin allele (βE) with various siRNA:mRNA mispairings at position 10, we found that an otherwise well matched siRNA can cleave its target mRNA, provided the mismatch is not composed of two bulky purine residues. siRNAs that are fully matched except for mismatched purine:pyrimidine or pyrimidine:pyrimidine residues at position 10 can still efficiently guide mRNA cleavage. This result suggests that the Ago2 catalytic site can accommodate some siRNA:mRNA mismatches, in particular, combinations of mismatches that involve the smaller pyrimidine residues. There are no structural studies of Ago2, but two groups have crystallized the endonuclease domain of a homologous bacterial protein bound to a small siRNA-like duplex (17, 18). Although the 5′ end of the siRNA guide strand was well ordered in the crystal and embedded in the bacterial Piwi protein, the region corresponding to position 10 was disordered, suggesting that the active site may allow some flexibility for binding. This observation is consistent with our finding that certain mismatches at the active site still permit significant mRNA cleavage.
Analysis of allele-specific silencing in other disease-causing SNPs supports our conclusion that an siRNA can specifically silence a gene containing an SNP without affecting the normal allele only when there is a purine:purine mismatch of the mutant allele siRNA and the normal target mRNA (see Table 1, which is published as supporting information on the PNAS web site). RNAi-mediated targeting of K-RASV12, resulting from a G→T transition in codon 12, significantly silenced K-RASV12 but not normal K-RAS (12). The K-RASV12 siRNA:normal K-RAS mRNA would contain a bulky purine:purine (A:G) mismatch at position 9 relative to the 5′ terminus of the guide siRNA strand, supporting the hypothesis that the juxtaposition of these bulky residues would impair silencing of the normal allele. In another example, an siRNA targeting a dominant SNP in SOD1, implicated in amyotrophic lateral sclerosis, showed minimal targeting of the normal allele because of the presence of a G:G mismatch (10). The specificity of silencing was strongest when the mismatch occurred at position 10 of the guide strand. However, the siRNA targeting the normal allele strongly cross-reacted to silence the mutant allele when the mismatch contained two pyrimidine (C:C) residues. In another study, allele-specific targeting was possible for mutant ataxin-3, which contains a G→C substitution immediately 3′ to the polyglutamine encoding CAG repeats but not for a disease-causing SNP in Tau (TauV337M, resulting from a G→A transition) (11). The mutant ataxin-3 siRNA that juxtaposed two purine residues (G:G) at position 11 when bound to the normal mRNA did not silence the normal allele. On the other hand, the siRNA designed to target TauV373M significantly inhibited expression of the normal allele. In that case, the mismatch with the normal mRNA consisted of a purine:pyrimidine G:U wobble. The Tau mutant allele could be specifically silenced when an additional mismatch was incorporated into the siRNA. A similar strategy has made it possible to increase the specificity of βE silencing (data not shown). In yet another example, an siRNA directed against HIV-1 vif silenced a variant gene containing a single nucleotide mismatch corresponding to position 9 of the siRNA guide strand (19). Again, this cross-reactivity could be explained by the presence of the non-Watson–Crick U:G pair. Interestingly, adding a methyl group to the U residue, N3-methyl-uridine, at position 11 of the siRNA guide strand inhibited silencing. The addition of this relatively small chemical group may have inhibited RNA–RISC interactions (20).
Our findings suggest that not only the position, but also the specific nucleotide combination, at mismatched sites helps determine the specificity of silencing. Significant silencing of the mRNA may occur with any combination of nucleotides that contains a pyrmidine, whether on the siRNA or the mRNA. Therefore, when designing siRNAs for the targeting of disease-causing alleles without impairing the expression of the normal allele, one wants to avoid purine:pyrimidine and pyrimidine:pyrimidine mismatches of the siRNA with the normal target mRNA, if silencing of the normal allele is not desirable. Although this rule should be generalizable to silencing other genes with other disease-related SNPs, how broadly applicable it is remains to be tested. This study, however, suggests that efficient allele-specific silencing of a mutant gene that differs from the normal allele by only a SNP can best be achieved in cases where the mutant residue is a pyrimidine and the normal residue is a purine. In this case, the siRNA targeting the mutant allele would contain a purine residue that would prevent silencing of the normal allele, as exemplified with the siRNA against sickle globin (Figs. 1 and 2). This situation is expected to occur in only one-fourth of cases. In the remaining situations, it may be necessary to add mismatches between the siRNA and mRNA, as was done to silence SNPs in Tau (11) and βE-globin (data not shown). However, the introduction of additional mismatches may come at the cost of less effective silencing because of reduced homology to the target sequence.
The rules that determine off-target silencing of partially homologous genes by siRNAs or that determine the endogenous targets of microRNAs are still not well defined. Although we have only investigated mismatches at position 10, it may be that the same principle applies at other sites, namely that a guide strand:mRNA purine:purine mismatch will result in significantly less (or potentially no) silencing compared with a mismatch containing at least one pyrimidine. This rule appears to be the case for nucleotides in the vicinity of the mRNA cleavage site, but it might also apply to the region important for mRNA target recognition, the “seed sequence” from positions 2–7 from the 5′ end of the guide strand.
Designing allele-specific siRNAs to target βS is only a first step toward testing whether RNAi can be harnessed to treat SCA. A key challenge is delivering siRNAs or their precursors into erythroid progenitors in vivo. There are two potential strategies: a drug approach in which siRNAs are delivered by a carrier or a gene therapy approach in which viral vectors express short hairpin (sh)RNAs, processed intracellularly into siRNAs. The latter approach has the potential advantage of providing life-long therapy (assuming transduction is efficient and expression is robust and sustained) but has the associated potential risks of insertional oncogenesis. A recent paper has shown that the coexpression of an shRNA against βS and a γ-globin transgene had therapeutic benefit in human erythrocytes derived from lentivirally transduced hematopoietic stem cells (21). A drug-like siRNA would require repeated treatments and a strategy for efficient systemic delivery into erythroid precursors. Recently some potential approaches for practical systemic siRNA delivery have been described, including targeting siRNAs in a cell-specific manner (22, 23). We are currently investigating both of these approaches in a mouse model of SCA.
Another potential concern in developing RNAi-based therapy for SCA is that silencing βS expression will lead to unbalanced α chain expression, α chain precipitation, and hemolysis. This imbalance might be mitigated if silencing β-globin resulted in compensatory enhanced expression of fetal globin. Alternatively, siRNA-mediated silencing could be done in conjunction with the therapeutic expression of βA87Thr→Gln-globin (4) or γ-globin (21) to provide the dual advantages of silencing and replacing the mutant allele with an antisickling form of globin. Because polymerization of deoxyHbS depends on its concentration, a large clinical benefit might be possible with modest panerythroid silencing.
Materials and Methods
Expression and Reporter Plasmids.
A 50-nt region of the βA and βS genes surrounding codon 6 and of βA and βE surrounding codon 26 was cloned downstream of the Renilla luciferase reporter gene in pRL-TK (Promega), which had been modified to contain XbaI and ApaI cloning sites in the 3′ UTR of the luciferase gene as described (8). The sequences are given in the figures. The βA and γ-globin cDNAs were from Origene Technologies (Rockville, MD). The βS variant was obtained by site-directed mutagenesis by using the QuikChange II site-directed mutagenesis kit (Stratagene) with complementary forward and reverse primers (forward primer, 5′-GCACCTGACTCCTGTGGAGAAGTCTG-3′). These sequences were PCR-amplified by using forward (5′-GGACTCAACTCGAGCAGACACCATGGTGCACCTGACTCC-3′) and reverse (5′-GTGTGGCGACCCGGGAGTGATACTTGTGGGCCAGGGCATTAGC-3′) primers and cloned by using XbaI and ApaI into pECFP-N1 or pEYFP-N1 (Clontech) to express fluorescently tagged fusion proteins.
siRNAs.
To maximize the specificity of silencing, the globin siRNAs were designed to place the mismatched nucleotide at position 10 relative to the 5′ end of the antisense strand. The 5′ terminal end of the siRNA guide strand was altered to maximize efficacy of silencing. All siRNAs were synthesized by Dharmacon Research (Lafayette, CO) with sequences indicated in the figures. The control siRNA against GFP was described in ref. 24.
Cells.
HeLa and MEL-βS cells were cultured in DMEM supplemented with 10% FCS, 1 mM l-glutamine, 10 mM Hepes, 50 μM 2-mercaptoethanol, penicillin, and streptomycin.
Luciferase Reporter Assay.
HeLa cells, plated at 2 × 105 cells per well in 12-well plates 1 day earlier, were cotransfected with 1 μg Renilla luciferase reporter plasmid, 1 μg of firefly luciferase vector (pGL3, Promega) and 200 nM of the indicated siRNA by using Lipofectamine 2000 transfection reagent as per the manufacturer’s protocol (Invitrogen). Luciferase activity was assessed by using the Dual-luciferase reporter assay system (Promega) and read on an Autolumat LB953 (Berthold Technologies, Bad Wildbad, Germany).
RT-PCR.
β-globin expression plasmids (0.5 μg) were cotransfected by using Lipofectamine 2000 (Invitrogen) with the indicated siRNA (100 nM) into HeLa cells (4 × 105 cells per well in 6-well plates). Total RNA was extracted 2 days later by using the RNeasy RNA extraction kit (Qiagen, Valencia, CA). First strand cDNA synthesis was performed by using random hexamer primers and Superscript III reverse transcriptase (Invitrogen). Globin gene segments were PCR amplified by using primers: βA forward, 5′-ATGGTGCATCTGACTCCTGA-3′; βS forward, 5′-ATGGTGCATCTGACTCCTGT-3′; βA and βS reverse, 5′-TAAAGGCACCGAGCACTTTC-3′; γ forward, 5′GGCTACTATCACAAGCCTG-3′; γ reverse, 5′-CCTTCTTGCCATGTGCCTTG-3′). The QuantumRNA Universal 18S Internal Standard was used as a positive control (Ambion, Austin, TX). Real-time PCR was used to quantify mRNA by using a Bio-Rad iCycler. β-actin was used as an internal control.
Real-Time RT-PCR.
Real-time RT-PCR was performed with the same primer pairs used above, except that a common β-globin forward primer was used, 5′-ATGGTGCATCTGACTCCTCG-3′. Real-time PCR was performed by using Platinum Taq polymerase (Invitrogen) and a Bio-Rad iCycler. SYBR green (Molecular Probes) was used to detect PCR products. Reactions were performed in 25λ in triplicate by using standard reaction conditions. Standard curves were generated, and the relative amount of mRNA was normalized to 18S mRNA. Specificity was verified by melt curve analysis.
Microscopy.
Microscopy was performed by using an Axiovert 200M inverted microscope (Zeiss) with YFP, CFP, and Cy3 filters (for dsRed) and analyzed by using slidebook v.4.0.1.32 (Intelligent Imaging Innovations, Denver, CO).
Supplementary Material
Acknowledgments
We would like to thank Dr. Eric Bouhassira (Albert Einstein College of Medicine, Yeshiva University, New York) for the kind gift of MEL-βS cells and members of the Lieberman laboratory for helpful suggestions and comments. This work was supported by National Institutes of Health Grants AI056900 (to J.L.) and P01 HL 055435-11 (to I.M.L.), and by a fellowship (to L.D.S.) under a National Institutes of Health Training Grant 5 T32 HL007556-19 to Dr. Ronald Nagel (Albert Einstein College of Medicine, Yeshiva University, New York).
Abbreviations
- CFP
cyan fluorescent protein
- HbS
sickle hemoglobin
- RNAi
RNA interference
- siRNA
small interfering RNA
- RISC
RNA-induced silencing complex
- SCA
sickle cell anemia
- YFP
yellow fluorescent protein.
Footnotes
Conflict of interest statment: No conflicts declared.
References
- 1.Stuart M. J., Nagel R. L. Lancet. 2004;364:1343–1360. doi: 10.1016/S0140-6736(04)17192-4. [DOI] [PubMed] [Google Scholar]
- Ferrone F., Nagel R. I. In: Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Steinberg M. H., Forget B. G., Higgs D. R., Nagel R. L., editors. Cambridge, U.K.: Cambridge Univ. Press; 2001. pp. 577–610. [Google Scholar]
- 3.Charache S., Dover G. J., Moyer M. A., Moore J. W. Blood. 1987;69:109–116. [PubMed] [Google Scholar]
- 4.Pawliuk R., Westerman K. A., Fabry M. E., Payen E., Tighe R., Bouhassira E. E., Acharya S. A., Ellis J., London I. M., Eaves C. J., et al. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 5.Dykxhoorn D. M., Lieberman J. Annu. Rev. Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir S. M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doench J. G., Petersen C. P., Sharp P. A. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena S., Jonsson Z. O., Dutta A. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 10.Ding H., Schwarz D. S., Keene A., Affar E. B., Fenton L., Xia X., Shi Y., Zamore P. D., Xu Z. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 11.Miller V. M., Xia H., Marrs G. L., Gouvion C. M., Lee G., Davidson B. L., Paulson H. L. Proc. Natl. Acad. Sci. USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brummelkamp T. R., Bernards R., Agami R. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 13.Rees D. C., Styles L., Vichinsky E. P., Clegg J. B., Weatherall D. J. Ann. N.Y. Acad. Sci. 1998;850:334–343. doi: 10.1111/j.1749-6632.1998.tb10490.x. [DOI] [PubMed] [Google Scholar]
- 14.Holen T., Amarzguioui M., Wiiger M. T., Babaie E., Prydz H. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S. K., Dykxhoorn D. M., Kumar P., Ranjbar S., Song E., Maliszewski L. E., Francois-Bongarcon V., Goldfeld A., Swamy N. M., Lieberman J., Shankar P. Blood. 2005;106:818–826. doi: 10.1182/blood-2004-10-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Q., Thonberg H., Wang J., Wahlestedt C., Liang Z. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma J. B., Yuan Y. R., Meister G., Pei Y., Tuschl T., Patel D. J. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker J. S., Roe S. M., Barford D. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacque J. M., Triques K., Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu Y. L., Rana T. M. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samakoglu S., Lisowski L., Budak-Alpdogan T., Usachenko Y., Acuto S., Di Marzo R., Maggio A., Zhu P., Tisdale J. F., Riviere I., Sadelain M. Nat. Biotechnol. 2006;24:89–94. doi: 10.1038/nbt1176. [DOI] [PubMed] [Google Scholar]
- 22.Song E., Zhu P., Lee S. K., Chowdhury D., Kussman S., Dykxhoorn D. M., Feng Y., Palliser D., Weiner D. B., Shankar P., et al. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 23.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 24.Novina C. D., Murray M. F., Dykxhoorn D. M., Beresford P. J., Riess J., Lee S. K., Collman R. G., Lieberman J., Shankar P., Sharp P. A. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.