Abstract
Variola virus (VaV) is the causative agent of smallpox, one of the most devastating diseases encountered by man, that was eradicated in 1980. The deliberate release of VaV would have catastrophic consequences on global public health. However, the mechanisms that contribute to smallpox pathogenesis are poorly understood at the molecular level. The ability of viruses to evade the host defense mechanisms is an important determinant of viral pathogenesis. Here we show that the tumor necrosis factor receptor (TNFR) homologue CrmB encoded by VaV functions not only as a soluble decoy TNFR but also as a highly specific binding protein for several chemokines that mediate recruitment of immune cells to mucosal surfaces and the skin, sites of virus entry and viral replication at late stages of smallpox. CrmB binds chemokines through its C-terminal domain, which is unrelated to TNFRs, was named smallpox virus-encoded chemokine receptor (SECRET) domain and uncovers a family of poxvirus chemokine inhibitors. An active SECRET domain was found in another viral TNFR (CrmD) and three secreted proteins encoded by orthopoxviruses. These findings identify a previously undescribed chemokine-binding and inhibitory domain unrelated to host chemokine receptors and a mechanism of immune modulation in VaV that may influence smallpox pathogenesis.
Keywords: immune evasion, viral pathogenesis, cytokine receptor, poxvirus, inflammation
The poxvirus variola virus (VaV) is the causative agent of smallpox, which was declared to be eradicated in 1980 as a result of the World Health Organization Smallpox Global Eradication Campaign, becoming the first and only viral disease eradicated by mass vaccination (1, 2). Thus, research on VaV terminated and the remaining smallpox samples were stored in two high security laboratories. The deliberate release of VaV would have catastrophic consequences on global public health considering that the majority of the human population has not been vaccinated or received a vaccination boost in recent years. Therefore, smallpox is considered one of the most dangerous threats as a biological weapon in bioterrorism, and there is an urgent need to define the mechanisms of smallpox pathogenesis (3). The available data suggest that the toxaemia reported in individuals suffering from severe smallpox may be immune-related. Considering recent advances in molecular pathogenesis with related poxviruses, it is likely that immune evasion strategies play a critical role as determinants of immunopathology and pathogenesis of smallpox (4, 5). Moreover, immune evasion mechanisms may modulate an immunopathological reaction responsible for adverse effects after smallpox vaccination (1).
The primary function of the immune system is to protect the host from invading pathogens such as viruses. To survive in the immunocompetent host, viral mechanisms that target specific immune pathways have evolved. A unique immune evasion strategy used by large DNA viruses (poxviruses and herpesviruses) is the production of secreted versions of host receptors or binding proteins that sequester cytokines and neutralize these regulatory molecules (4, 5). Viral TNF receptors (vTNFRs) encoded by poxviruses block the activity of this proinflammatory cytokine and are examples of viral decoy receptors with sequence similarity to the extracellular cytokine-binding domain of their cellular counterparts. Some poxviruses and herpesviruses encode secreted chemokine-binding proteins belonging to a second class of viral decoy receptors with unique structures unrelated to host receptors (6, 7) that bind with high affinity a broad range of chemokines, which are mediators of cell migration (5, 8–14). The secreted 35-kDa protein that binds CC chemokines is the only viral chemokine-binding protein identified in VaV and the vaccinia virus (VV) smallpox vaccine strains Lister and DryVax (Wyeth) (8–10).
Four genes encoding vTNFRs have been described in poxviruses and named cytokine response modifier B (CrmB), CrmC, CrmD, and CrmE, and their expression varies among viral species (Fig. 1; refs. 4 and 5; www.poxvirus.org). Cowpox virus (CPV), a rodent virus that infects other species sporadically, encodes all four vTNFRs (15–18). Ectromelia virus (EV) is a highly virulent mouse pathogen that causes mousepox, a disease similar to smallpox, and encodes CrmD only (19). VV Western Reserve and the strains used as smallpox vaccines Copenhagen, DryVax (Wyeth), and Tian-Tan, do not encode vTNFRs, but CrmC and CrmE are encoded by the vaccine strains Lister, USSR, and Evans (20, 21). VaV and monkeypox virus, which causes a smallpox-like disease in humans, encode CrmB only (22–26). The reasons for the variety of vTNFRs are not understood. In addition to the cysteine-rich domains (CRDs) characteristic of the ligand-binding region of cellular TNFRs, CrmB and CrmD have a C-terminal domain (CTD) unrelated to host proteins (Fig. 1).
Fig. 1.
Schematic representation of vTNFRs and SCPs and their distribution among orthopoxviruses. The name of the genes in different viruses is indicated. The complete genome sequence of VV Lister is not available, and those genes that have been sequenced are indicated (+). nd, not determined.
We have characterized the CrmB protein encoded by VaV and found that it functions not only as a decoy TNFR but it also interacts with chemokines through its CTD, which is unrelated to host proteins. This previously undescribed chemokine-binding domain uncovers a family of poxvirus-encoded secreted chemokine inhibitors with potential immunomodulatory activity.
Results
VaV CrmB Functions as a Secreted Decoy TNF Receptor (TNFR).
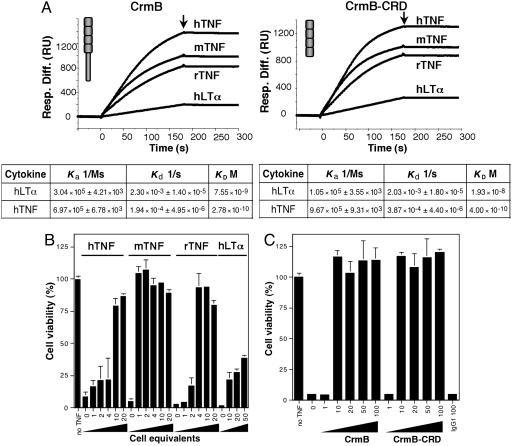
CrmB is the only vTNFR predicted to be active in the VaV strains sequenced to date (refs. 22–24; www.poxvirus.org). To explore the immunomodulatory activity of CrmB, we generated the VaV strain Bangladesh-1975 CrmB gene (22) by site-directed mutagenesis from the CrmB gene encoded by camelpox virus, a close relative of VaV (Fig. 8, which is published as supporting information on the PNAS web site; ref. 27). Recombinant CrmB was expressed by using the baculovirus system, and the purified protein was found by surface plasmon resonance (SPR) to bind with high-affinity TNF from human (Kd = 278 pM), rat (Kd < 20 pM), and mouse (Kd < 20 pM), as well as human lymphotoxin-α (LTα) with lower affinity (Kd = 7.55 nM) (Fig. 2A; see also Fig. 9, which is published as supporting information on the PNAS web site). Consistent with the binding data, VaV CrmB efficiently inhibited human TNF activity, was less potent at inhibiting LTα, and was a better inhibitor of mouse and rat TNF activity (Fig. 2B). As the sequence similarity of CrmB to human TNFRs is restricted to its N-terminal CRDs, we expressed a truncated version of CrmB containing the N-terminal CRDs (CrmB-CRD) in the baculovirus system. This protein retained the TNF-binding and -inhibitory activities of full-length CrmB (Figs. 2 A and C and 9), demonstrating that the N-terminal CRDs are sufficient for TNF binding and suggesting that the CTD may interact with other immune molecules. Our results were consistent with the finding that the N-terminal CRDs of the myxoma virus vTNFR M-T2 are sufficient for TNF binding (28).
Fig. 2.
TNF-binding and -inhibitory activity of VaV CrmB. (A) Sensorgrams showing binding of human (h), mouse (m), or rat (r) TNF or human LTα to purified recombinant CrmB or CrmB-CRD analyzed by SPR (Biacore X). Arrows indicate end of injection. Derived kinetic parameters and affinity constants are indicated below. (B) Inhibition of TNF- and LTα-induced necrosis in L929 cells by supernatants from insect cells infected with a recombinant baculovirus expressing CrmB. The numbers in the x axis indicate the amount of supernatant in cell equivalents (×1000). (C) Inhibition of mouse TNF-induced necrosis in L929 cells by increasing doses (ng) of purified recombinant CrmB or CrmB-CRD or control IgG1. In B and C, mean ± SD of triplicate samples of a representative experiment is shown.
The EV CrmD protein, a secreted vTNFR related to VaV CrmB (Fig. 1), was previously expressed from a bacterial vector and was shown to bind TNF (17). We expressed EV CrmD from VV Western Reserve, a strain that does not encode vTNFRs (20), and found that recombinant CrmD was secreted into the medium, bound human 125I-TNF, and inhibited human TNF biological activity (Fig. 10, which is published as supporting information on the PNAS web site). In addition, purified recombinant EV CrmD expressed in the baculovirus system bound human TNF by SPR and inhibited its activity (data not shown). To determine the region of CrmD required for TNF inhibitory activity, the N-terminal CRDs of CrmD (CrmD-CRD) were expressed in the VV system. Similar to full-length CrmD, CrmD-CRD-bound human TNF in soluble binding assays (data not shown) and inhibited the biological activity of human TNF (Fig. 10). Thus, similar to VaV CrmB, EV CrmD inhibited TNF through its N-terminal CRDs.
Previously Undescribed Chemokine-Binding Activity Encoded by VaV CrmB.
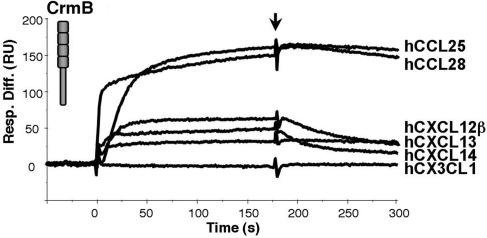
The data described above suggested an additional role of the CTD of the VaV CrmB protein. A screening with different cytokines by SPR identified chemokines as ligands of VaV CrmB. This result led us to screen by SPR the potential binding of purified CrmB to all 43 commercially available human chemokines. VaV CrmB interacted with some chemokines with binding affinities similar to that of TNF. The chemokines that best bound to VaV CrmB were as follows: CCL28, CCL25, CXCL12β, CXCL13, and CXCL14 (Fig. 3and Table 1). Similar studies by SPR with human and mouse chemokines showed that recombinant EV CrmD bound with high affinity the same set of chemokines (Table 2, which is published as supporting information on the PNAS web site). Recombinant CrmB from CPV expressed in the baculovirus system also bound these chemokines (data not shown).
Fig. 3.
VaV CrmB binds chemokines. Sensorgrams showing binding of the indicated human chemokines to purified recombinant VaV CrmB analyzed by SPR. Arrow indicates end of injection.
Table 1.
Kinetic parameters and derived affinity constants of the binding of VaV CrmB to the indicated human chemokines
| Chemokine | Ka, 1/Ms | Kd, 1/s | KD, M |
|---|---|---|---|
| hCCL28 | 1.11 × 106 ± 2.63 × 104 | 4.33 × 10−4 ± 1.44 × 10−5 | 0.30 × 10−9 |
| hCCL25 | 1.54 × 106 ± 2.58 × 104 | 7.74 × 10−4 ± 1.74 × 10−5 | 0.50 × 10−9 |
| hCXCL12β | 1.05 × 106 ± 1.91 × 104 | 4.48 × 10−3 ± 2.98 × 10−5 | 4.26 × 10−9 |
| hCXCL13 | 3.63 × 105 ± 1.47 × 104 | 2.16 × 10−3 ± 3.46 × 10−5 | 5.95 × 10−9 |
| hCXCL14 | 1.34 × 105 ± 3.41 × 103 | 8.40 × 10−4 ± 1.87 × 10−5 | 6.29 × 10−9 |
| hXCL1 | 9.06 × 104 ± 1.75 × 103 | 2.61 × 10−3 ± 2.14 × 10−5 | 28.8 × 10−9 |
| hCCL20 | 7.59 × 104 ± 1.02 × 103 | 2.22 × 10−3 ± 4.16 × 10−5 | 29.2 × 10−9 |
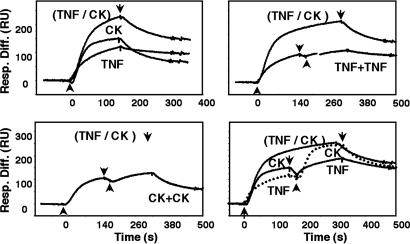
We next addressed whether VaV CrmB has two independent binding sites for TNF and chemokines (Fig. 4). In SPR experiments, TNF and CXCL12β were injected independently or simultaneously, and the signal was compared with that obtained when the cytokines were injected consecutively. The binding signal after the addition of TNF and CXCL12β together was equivalent to the sum of that obtained with TNF or CXCL12β alone. Moreover, consecutive addition of TNF and CXCL12β, or CXCL12β and TNF, gave maximum binding, whereas two successive injections of TNF or CXCL12β alone did not increase the binding detected with either of them. Thus, saturation of the TNF-binding sites did not affect binding of the chemokine to CrmB and vice versa, supporting the presence of two independent sites for TNF and chemokines in CrmB. Similarly, competitive inhibition studies indicated distinct binding sites for TNF and chemokines in EV CrmD (data not shown).
Fig. 4.
VaV CrmB binds chemokines and TNF through different sites. Sensorgrams showing the binding of chemokines and TNF to purified recombinant VaV CrmB by SPR. The binding of human TNF or CXCL12β (CK) alone, simultaneous injections of both cytokines (TNF/CK), or two consecutive injections of cytokines (TNF+TNF and CK+CK) are shown. In Lower Right, a discontinuous line indicates an injection of TNF followed by an injection of CK. Arrow indicate start (↑) and end (↓) of injection.
The CTD of VaV CrmB Represents a Previously Undescribed Protein Domain That Interacts with Chemokines.
The presence in CrmB of a CTD unrelated to host TNFRs and dispensable for TNF binding, together with the identification of independent binding sites for TNF and chemokines in CrmB, suggested that the previously undescribed chemokine-binding activity of CrmB may reside in the CTD. To test this hypothesis, we expressed the CTD of CrmB (CrmB-CTD) fused to the predicted N-terminal signal peptide from CrmB in the baculovirus system. Secreted CrmB-CTD purified from culture supernatants bound chemokines but not TNF, whereas CrmB-CRD bound TNF but not chemokines (Fig. 5A), demonstrating that CTD is a previously undescribed chemokine-binding domain that we named smallpox virus-encoded chemokine receptor (SECRET) domain. Similarly, EV CrmD-CRD did not bind chemokines (data not shown). Moreover, purified vTNFRs CrmC and CrmE encoded by CPV, which lack the CTD, did not bind chemokines (Fig. 5B and data not shown). The ability of the SECRET domain to confer chemokine-binding activity to vTNFRs was illustrated by expression in the baculovirus system of CPV CrmC and CrmE fused to CrmB-CTD. The fusion proteins retained TNF-binding activity but, in contrast to CrmC and CrmE, they also bound chemokines (Fig. 5B and data not shown).
Fig. 5.
VaV CrmB binds chemokines through their CTD. (A) Sensorgrams showing binding of human cytokines to VaV CrmB-CRD or CrmB-CTD by SPR. (B) Sensorgrams showing binding of human cytokines to CPV CrmC or CrmC fused to CrmB-CTD (CrmC-CrmB CTD) by SPR. The arrows indicate end of injections.
The SECRET Domain Identifies a Family of Poxvirus Secreted Chemokine Inhibitors.
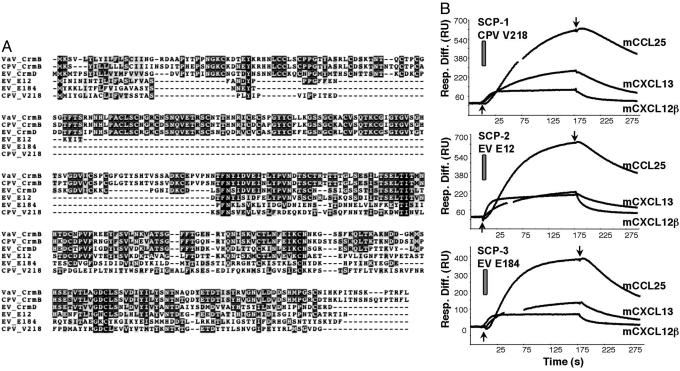
Analysis of poxviral genomes identified other gene products encoding SECRET domain-containing proteins (SCPs), which are predicted to be secreted (Figs. 1 and 6A). The multiple alignment of the SECRET domain from CrmB, CrmD, and the three SCPs showed 4% amino acid identity and 30% amino acid similarity, with two cysteine residues conserved in all of the proteins (Fig. 6A). The SECRET domain does not share any motifs with previously identified viral chemokine-binding proteins. CPV strain Brighton Red protein V218 (SCP-1) and EV strain Naval proteins E12 (SCP-2) and E184 (SCP-3) were expressed in the baculovirus system and found to be secreted. All three proteins bound the same set of mouse chemokines (CCL25, CCL27, CCL28, CXCL11, CXCL12β, CXCL13, and CXCL14) as determined by SPR with all 35 commercially available mouse chemokines (Fig. 6B and data not shown). These SCPs bound the same set of human chemokines as VaV CrmB and EV CrmD (data not shown), and CrmB and CrmD bound the same mouse chemokines as the SCPs (data not shown), indicating that the chemokine-binding specificity of the SECRET domain found in all members of the family is similar. Note that the mouse chemokines CCL27 and CXCL11, but not the human homologues, are recognized by the SECRET domain.
Fig. 6.
SCPs interact with chemokines. (A) Sequence alignment of VaV CrmB (gene G2R, strain Bangladesh 1975, UniProt/TrEMBL accession no. P34015), CPV CrmB (gene V005, strain Brighton Red, UniProt/TrEMBL accession no. Q85308), EV CrmD (gene E6, strain Naval), CPV SCP-3 (gene V218, strain Brighton Red, UniProt/TrEMBL accession no. Q8QMN0), EV SCP-2 (gene E12, strain Naval), and EV SCP-3 (gene E184, strain Naval). The sequences of EV strain Naval genes can be found at www.sanger.ac.uk/Projects/Ectromelia_virus. Black boxes indicate conserved residues; dark and light gray boxes indicate residues identical or similar in 50% or more of the represented sequences, respectively. (B) Sensorgrams showing the binding of mouse chemokines to the indicated proteins by SPR.
This family of secreted poxvirus proteins containing the SECRET domain interacted with chemokines, and, thus, they may function as soluble decoy receptors. The chemokine-inhibitory activity of the SECRET domain was shown by the inhibition of Molt4 cell migration in response to CCL25 in vitro by EV CrmD, CPV CrmB, VaV CrmB, and CPV SCP-1 (Fig. 7). As expected, no inhibition was observed with VaV CrmB-CRD or with CPV CrmC, a vTNFR lacking the CTD.
Fig. 7.
The SECRET domain inhibits chemokine activity. Inhibition of Molt4 cell migration is shown in response to mouse CCL25 in the absence or presence of the indicated molar excess of purified proteins (mean ± SD). Data are represented as the percentage of cell migration in the absence of inhibitor.
Discussion
The identification of immunomodulatory activities encoded by viruses is critical to understand molecular mechanisms of pathogenesis. Here we demonstrate anti-TNF and antichemokine activities in the TNFR homologue CrmB encoded by VaV, define a protein domain (SECRET domain) that binds chemokines and uncover a previously undescribed family of secreted viral immunomodulatory proteins containing the SECRET domain.
CrmB is the only predicted vTNFR active in VaV major and minor strains, which cause different fatality rates (1, 22–24), and in the four sequenced strains of monkeypox virus, which causes a smallpox-like disease in humans (22, 25, 26). Our demonstration that VaV CrmB is a potent inhibitor of TNF is consistent with previous evidence of down-regulation of TNF responses in an experimental macaque model of human smallpox (29). Although VaV is expected to have adapted to the human immune system, VaV CrmB was a better inhibitor of mouse TNF activity than of human TNF, as described for CPV CrmB (20) and EV CrmD (M.S. and A. Alcami, unpublished data). These binding properties may reflect the evolutionary origin of VaV (30) or structural constraints restricting changes in TNF specificity. A higher affinity for the mouse cytokine has also been observed with the VaV IL-18 binding protein (31).
The fact that the SCPs and the vTNFRs CrmB and CrmD bound the same chemokines, despite their relative low sequence similarity, reinforced the concept that the SECRET domain has a specific folding, allowing it to bind chemokines with high affinity, either independently or fused to TNFRs, and to inhibit chemokine activity. The modular nature of the SECRET domain was illustrated by its ability to confer chemokine-binding specificity to the CPV vTNFRs CrmC and CrmE when they were expressed fused to the SECRET domain. Thus, the SECRET domain described here, which has no amino acid sequence similarity to host chemokine receptors or previously described viral chemokine-binding proteins, is another example of unique protein domains evolved in viral genomes to efficiently neutralize host immune mediators (4, 5).
The chemokine-binding specificity of the SECRET domain is limited to a reduced set of chemokines and is in clear contrast to the broad spectrum binding specificity of previously identified viral chemokine-binding proteins in poxviruses and herpesviruses (4, 5). This binding activity points at a reduced set of chemokines, and cells expressing their specific receptors, that may play an important role in human smallpox and other poxvirus infections. The SECRET domain binds chemokines that are likely to be relevant in antiviral defense: (i) chemokines mediating T and B cell recruitment that are expressed by epithelial cells in mucosal surfaces (CCL25 and CCL28) or the skin (CCL27) (32–34), which constitute the sites of virus entry; (ii) CCL25 and CCL28 recruit IgA-producing B cells to mucosal sites (34, 35); (iii) CXCL14 is involved in dendritic cell migration to epidermal tissues (36); and (iv) CXCL13 attracts B cells to the spleen and lymph nodes (32).
The identification of the SECRET domain in five different poxvirus proteins is intriguing. This distribution may explain, in part, the variety of genes encoding vTNFRs in poxvirus genomes, some of which (CrmB and CrmD) encode this additional chemokine-inhibitory activity. It may also provide the virus the ability to differentially block chemokines involved in controlling distinct antiviral responses, inhibit chemokines at different stages of infection in the animal host, or simultaneously inhibit chemokines and TNF. It is likely that as poxviruses with narrow host species specificity adapted to particular hosts during evolution (i.e., VaV to humans or EV to mice), a particular set of genes were selected to facilitate viral replication and transmission in each host.
CrmB and CrmD inhibit the activity of both TNF, a potent proinflammatory cytokine, and chemokines involved in mucosal and skin inflammation. These viral proteins are likely to play an important role at the initial stages of infection with VaV, monkeypox virus, and EV, which are transmitted through the respiratory route and/or the skin. In addition, CrmB and CrmD also may promote virus replication during late stages of the disease characterized by generalized skin lesions in VaV, monkeypox virus, and EV. This possibility is supported by the finding that the chemokines recognized by CrmB and CrmD are implicated in mucosal and skin inflammatory responses, and their neutralization results in impaired leukocyte recruitment and suppression of inflammatory responses (33, 37, 38).
Soluble decoy TNFRs are used in the clinic to control immunopathological reactions that cause human diseases such as rheumatoid arthritis (39). Our finding that vTNFRs also target specific chemokines by incorporating the SECRET domain suggests that the addition of antichemokine activity to soluble TNFRs may increase their antiinflammatory properties and illustrates that the information found in viral genomes may be relevant for the development of new therapeutics.
In conclusion, we demonstrate TNF-binding activity in the CrmB protein encoded by VaV. We show that VaV CrmB and EV CrmD function not only as soluble decoy TNFRs, but they also bind and inhibit several chemokines. We also describe a previously undescribed chemokine-binding domain, expressed independently or fused to vTNFRs, that uncovers a family of poxvirus secreted chemokine inhibitors. These findings shed light on the molecular pathogenesis of smallpox.
Materials and Methods
Reagents.
Recombinant chemokines were purchased from PeproTech (London) or R & D Systems for migration assays. Recombinant TNF and LTα were from R & D Systems.
Cells and Viruses.
The growth of poxviruses in BSC-I cells and the source of VV, CPV, camelpox virus, and EV strains have been described (10, 19). Recombinant baculoviruses were grown in Hi5 insect cells.
Expression and Purification of Recombinant Proteins.
The cloning, generation of recombinant baculoviruses, and protein purification protocols are described in detail in Supporting Methods, which is published as supporting information on the PNAS web site. The VaV CrmB coding sequence was obtained by site-directed mutagenesis of the camelpox virus orthologue (27). Permission from the World Health Organization has been granted to hold VaV DNA, and its manipulation is performed in accordance to the established rules.
Characterization of Protein–Protein Interactions by SPR.
Cytokine-binding specificity and affinity constants were determined by SPR with a Biacore X biosensor. For ligand-screening experiments, purified recombinant proteins were amine- or thiol-coupled to CM5 chips to a level of ≈5,000 response units (RU) (5,000 pg/mm2) in each case. Recombinant cytokines were injected at 100 nM in HBS-EP buffer [10 mM Hepes/150 mM NaCl/3 mM EDTA/0.005% (vol/vol) surfactant P20, pH 7.4] at a flow rate of 10 μl/min, and association and dissociation were monitored. The surface was regenerated after each injection by using 10 mM glycine·HCl pH 2.0. For kinetic analyses, the recombinant proteins were immobilized at low densities (Rmax < 200 RU). Different concentrations of the corresponding cytokine were then injected at a flow rate of 30 μl/min over a 2-min period and allowed to dissociate for an additional 5 min. All Biacore sensorgrams were analyzed by using biaevaluation 3.2. Bulk refractive index changes were removed by subtracting the reference flow cell responses, and the average response of a blank injection was subtracted from all analyte sensorgrams to remove systematic artifacts. Kinetic data were globally fitted to a 1:1 Langmuir model.
TNF Activity Assay.
Cytoxicity assays of TNF and LTα were performed with L929 cells (18). TNF (20 ng/ml) was preincubated for 2 h at 37°C with purified recombinant proteins in 100 μl of complete DMEM supplemented with actinomycin D (4 μg/ml; Sigma). The mixture was then added to 2 × 104 cells seeded the day before in 96-well plates, and cell viability was assessed 16–18 h later by using the CellTiter 96 Aqueous One Solution cell proliferation assay (Promega) by following the manufacturer’s recommendations.
Chemokine Migration Assay.
The migration of Molt4 cells was assessed by using 24-well Transwell plates with 3-μm pore size filters (Costar) as described in ref. 40. Briefly, CCL25 (25 nM) alone or in the presence of increasing amounts of purified recombinant protein was placed in the lower compartment and incubated at 37°C for 30 min. After this period, 5 × 105 Molt4 cells were added in 100 μl of complete RPMI 1640 medium containing 0.1% FCS to the top well and the plate incubated at 37°C. Migration of Molt4 cells into the bottom compartment was determined after 4 h by flow cytometry.
Supplementary Material
Acknowledgments
We thank Rocío Martín for excellent technical support. This work was funded by the Wellcome Trust. M.B.R.-A. and A. Alejo also were supported by a Ramón y Cajal Fellowship (Spanish Ministry of Education and Science) and Comunidad de Madrid Fellowship, respectively. M.S. was supported by a Fundaçao para a Ciencia e Tecnologia-Praxis XXI Studentship.
Abbreviations
- CPV
cowpox virus
- CRD
cysteine-rich domain
- Crm
cytokine response modifier
- CTD
C-terminal domain
- EV
ectromelia virus
- LTα
lymphotoxin-α
- SCP
SECRET domain-containing protein
- SECRET
smallpox virus-encoded chemokine receptor
- SPR
surface plasmon resonance
- TNFR
TNF receptor
- VaV
variola virus
- vTNFR
viral TNF receptor
- VV
vaccinia virus.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fenner F., Anderson D. A., Arita I., Jezek Z., Ladnyi I. D. Smallpox and Its Eradication. Geneva: W.H.O.; 1988. [Google Scholar]
- 2.Smith G. L., McFadden G. Nat. Rev. Immunol. 2002;2:521–527. doi: 10.1038/nri845. [DOI] [PubMed] [Google Scholar]
- 3.Henderson D. A. Science. 1999;283:1279–1282. doi: 10.1126/science.283.5406.1279. [DOI] [PubMed] [Google Scholar]
- 4.Seet B. T., Johnston J. B., Brunetti C. R., Barrett J. W., Everett H., Cameron C., Sypula J., Nazarian S. H., Lucas A., McFadden G. Annu. Rev. Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 5.Alcami A. Nat. Rev. Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 6.Carfi A., Smith C. A., Smolak P. J., McGrew J., Wiley D. C. Proc. Natl. Acad. Sci. USA. 1999;96:12379–12383. doi: 10.1073/pnas.96.22.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander J. M., Nelson C. A., van Berkel V., Lau E. K., Studts J. M., Brett T. J., Speck S. H., Handel T. M., Virgin H. W., Fremont D. H. Cell. 2002;111:343–356. doi: 10.1016/s0092-8674(02)01007-3. [DOI] [PubMed] [Google Scholar]
- 8.Graham K. A., Lalani A. S., Macen J. L., Ness T. L., Barry M., Liu L. Y., Lucas A., Clark-Lewis I., Moyer R. W., McFadden G. Virology. 1997;229:12–24. doi: 10.1006/viro.1996.8423. [DOI] [PubMed] [Google Scholar]
- 9.Smith C. A., Smith T. D., Smolak P. J., Friend D., Hagen H., Gerhart M., Park L., Pickup D. J., Torrance D., Mohler K., et al. Virology. 1997;236:316–327. doi: 10.1006/viro.1997.8730. [DOI] [PubMed] [Google Scholar]
- 10.Alcami A., Symons J. A., Collins P. D., Williams T. J., Smith G. L. J. Immunol. 1998;160:624–633. [PubMed] [Google Scholar]
- 11.Parry C. M., Simas J. P., Smith V. P., Stewart C. A., Minson A. C., Efstathiou S., Alcami A. J. Exp. Med. 2000;191:573–578. doi: 10.1084/jem.191.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Berkel V., Barrett J., Tiffany H. L., Fremont D. H., Murphy P. M., McFadden G., Speck S. H., Virgin H. I. J. Virol. 2000;74:6741–6747. doi: 10.1128/jvi.74.15.6741-6747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant N. A., Davis-Poynter N., Vanderplasschen A., Alcami A. EMBO J. 2003;22:833–846. doi: 10.1093/emboj/cdg092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D., Bresnahan W., Shenk T. Proc. Natl. Acad. Sci. USA. 2004;101:16642–16647. doi: 10.1073/pnas.0407233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu F., Smith C. A., Pickup D. J. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 16.Smith C. A., Hu F. Q., Smith T. D., Richards C. L., Smolak P., Goodwin R. G., Pickup D. J. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 17.Loparev V. N., Parsons J. M., Knight J. C., Panus J. F., Ray C. A., Buller R. M. L., Pickup D. J., Esposito J. J. Proc. Natl. Acad. Sci. USA. 1998;95:3786–3791. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraiva M., Alcami A. J. Virol. 2001;75:226–233. doi: 10.1128/JVI.75.1.226-233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith V. P., Alcami A. J. Virol. 2000;74:8460–8471. doi: 10.1128/jvi.74.18.8460-8471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcami A., Khanna A., Paul N. L., Smith G. L. J. Gen. Virol. 1999;80:949–959. doi: 10.1099/0022-1317-80-4-949. [DOI] [PubMed] [Google Scholar]
- 21.Reading P. C., Khanna A., Smith G. L. Virology. 2002;292:285–298. doi: 10.1006/viro.2001.1236. [DOI] [PubMed] [Google Scholar]
- 22.Massung R. F., Esposito J. J., Liu L. I., Qi J., Utterback T. R., Knight J. C., Aubin L., Yuran T. E., Parsons J. M., Loparev V. N., et al. Nature. 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 23.Shchelkunov S. N., Massung R. F., Esposito J. J. Virus Res. 1995;36:107–118. doi: 10.1016/0168-1702(94)00113-q. [DOI] [PubMed] [Google Scholar]
- 24.Shchelkunov S. N., Totmenin A. V., Loparev V. N., Safronov P. F., Gutorov V. V., Chizhikov V. E., Knight J. C., Parsons J. M., Massung R. F., Esposito J. J. Virology. 2000;266:361–386. doi: 10.1006/viro.1999.0086. [DOI] [PubMed] [Google Scholar]
- 25.Shchelkunov S. N., Totmenin A. V., Safronov P. F., Mikheev M. V., Gutorov V. V., Ryazankina O. I., Petrov N. A., Babkin I. V., Uvarova E. A., Sandakhchiev L. S., et al. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen N., Li G., Liszewski M. K., Atkinson J. P., Jahrling P. B., Feng Z., Schriewer J., Buck C., Wang C., Lefkowitz E. J., et al. Virology. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gubser C., Smith G. L. J. Gen. Virol. 2002;83:855–872. doi: 10.1099/0022-1317-83-4-855. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber M., McFadden G. J. Immunol. 1996;157:4486–4495. [PubMed] [Google Scholar]
- 29.Jahrling P. B., Hensley L. E., Martinez M. J., Leduc J. W., Rubins K. H., Relman D. A., Huggins J. W. Proc. Natl. Acad. Sci. USA. 2004;101:15196–15200. doi: 10.1073/pnas.0405954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gubser C., Hue S., Kellam P., Smith G. L. J. Gen. Virol. 2004;85:105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- 31.Esteban D. J., Nuara A. A., Buller R. M. J. Gen. Virol. 2004;85:1291–1299. doi: 10.1099/vir.0.79902-0. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel E. J., Butcher E. C. Immunity. 2002;16:1–4. doi: 10.1016/s1074-7613(01)00261-8. [DOI] [PubMed] [Google Scholar]
- 33.Homey B., Alenius H., Muller A., Soto H., Bowman E. P., Yuan W., McEvoy L., Lauerma A. I., Assmann T., Bunemann E., et al. Nat. Med. 2002;8:157–165. doi: 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- 34.Lazarus N. H., Kunkel E. J., Johnston B., Wilson E., Youngman K. R., Butcher E. C. J. Immunol. 2003;170:3799–3805. doi: 10.4049/jimmunol.170.7.3799. [DOI] [PubMed] [Google Scholar]
- 35.Bowman E. P., Kuklin N. A., Youngman K. R., Lazarus N. H., Kunkel E. J., Pan J., Greenberg H. B., Butcher E. C. J. Exp. Med. 2002;195:269–275. doi: 10.1084/jem.20010670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaerli P., Willimann K., Ebert L. M., Walz A., Moser B. Immunity. 2005;23:331–342. doi: 10.1016/j.immuni.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 37.John A. E., Thomas M. S., Berlin A. A., Lukacs N. W. Am. J. Pathol. 2005;166:345–353. doi: 10.1016/S0002-9440(10)62258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohan K., Cordeiro E., Vaci M., McMaster C., Issekutz T. B. Eur. J. Immunol. 2005;35:1702–1711. doi: 10.1002/eji.200425885. [DOI] [PubMed] [Google Scholar]
- 39.Feldmann M., Steinman L. Nature. 2005;435:612–619. doi: 10.1038/nature03727. [DOI] [PubMed] [Google Scholar]
- 40.Zaballos A., Gutierrez J., Varona R., Ardavin C., Marquez G. J. Immunol. 1999;162:5671–5675. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.