Abstract
Viral signaling through retinoic acid-inducible gene-I (RIG-I) and its adaptor protein, IFN promoter-stimulator 1 (IPS-1), activates IFN regulatory factor-3 (IRF-3) and the host IFN-α/β response that limits virus infection. The hepatitis C virus (HCV) NS3/4A protease cleaves IPS-1 to block RIG-I signaling, but how this regulation controls the host response to HCV is not known. Moreover, endogenous IPS-1 cleavage has not been demonstrated in the context of HCV infection in vitro or in vivo. Here, we show that HCV infection transiently induces RIG-I- and IPS-1-dependent IRF-3 activation. This host response limits HCV production and constrains cellular permissiveness to infection. However, HCV disrupts this response early in infection by NS3/4A cleavage of IPS-1 at C508, releasing IPS-1 from the mitochondrial membrane. Cleavage results in subcellular redistribution of IPS-1 and loss of interaction with RIG-I, thereby preventing downstream activation of IRF-3 and IFN-β induction. Liver tissues from chronically infected patients similarly demonstrate subcellular redistribution of IPS-1 in infected hepatocytes and IPS-1 cleavage associated with a lack of ISG15 expression and conjugation of target proteins in vivo. Importantly, small-molecule inhibitors of NS3/4A prevent cleavage and restore RIG-I signaling of IFN-β induction. Our results suggest a dynamic model in which early activation of IRF-3 and induction of antiviral genes are reversed by IPS-1 proteolysis and abrogation of RIG-I signaling as NS3/4A accumulates in newly infected cells. HCV protease inhibitors effectively prevent IPS-1 proteolysis, suggesting they may be capable of restoring this innate host response in clinical practice.
Keywords: CARDIF, MAVS, VISA
Hepatitis C virus (HCV) is an enveloped virus of the Flaviviridae family that afflicts nearly 200 million people with chronic infection and is a leading cause of liver disease, including cancer (1, 2). The RNA genomes of HCV and related viruses contain regions of secondary and tertiary structure that are essential for function (3). Pathogen-associated molecular patterns embedded within these regions are recognized by the product of retinoic acid-inducible gene-I (RIG-I) or specific Toll-like receptors. RIG-I is a double-stranded RNA-binding DExD/H box RNA helicase (4, 5) and is essential for initiating the intracellular response to RNA virus infection (6). Once engaged by double-stranded RNA motifs within the viral genome, RIG-I signals the host response through its two amino-terminal caspase activation and recruitment domains (CARDs) by interacting with the amino-terminal CARD of IFN-β promoter-stimulator 1 (IPS-1) through a heterotypic CARD–CARD interaction (7). IPS-1, also known as MAVS, VISA, and CARDIF (8–10), is a mitochondrial outer membrane protein and an essential component of the RIG-I signaling pathway. It operates downstream of RIG-I to direct the activation of the IFN regulatory factor-3 (IRF-3) transcription factor to produce IFN-β and IFN-α and the expression of IFN-stimulated genes (ISGs), whose products limit virus infection by suppressing viral replication and modulating adaptive immunity (11).
RIG-I signaling imparts a host response that can control cellular permissiveness for HCV RNA replication (5), but HCV evades this response in part through its ability to antagonize signaling to IRF-3 (12). NS3/4A, the major serine protease expressed by HCV (2), disrupts the RIG-I pathway through proteolysis of essential signaling components of IRF-3 activation (13). NS3/4A also cleaves the TRIF adaptor protein to ablate Toll-like receptor-3 signaling of IRF-3 activation by extracellular double-stranded RNA (14). HCV therefore imparts major points of control over host defense that could be amenable to therapeutic modification by compounds that inhibit NS3/4A protease activity (12). The current study was undertaken to identify signaling partner(s) of RIG-I that are proteolytic substrates of NS3/4A and to determine their application for modulation by HCV protease inhibitors. We identified IPS-1 as a RIG-I signaling partner and found that both molecules are essential for triggering the host response to HCV infection. Our results show that IPS-1 is cleaved by NS3/4A both in vitro and in vivo within the liver during chronic infection and define the NS3/4A-IPS-1 interface as a previously unrecognized therapeutic target for restoration of the host response by HCV protease inhibitors.
Results
Regulation of IRF-3 During HCV Infection.
We examined the virologic and host response features of HCV infection using virus generated from the genotype 2a JFH-1 infectious clone of HCV (15). Human hepatoma (Huh7) cells and their derivatives are defective in Toll-like receptor-3 signaling (14) and are permissive for JFH-1 virus infection (16). Using immunofluorescence microscopy, we monitored these cells for IRF-3 activation after JFH-1 infection. IRF-3 was present in its inactive, cytoplasmic-bound state in mock-infected cells. After HCV infection, the active, nuclear isoform of IRF-3 was present only in Huh7 cells with low or undetectable levels of viral protein at early time points (24 and 36 h after infection; Fig. 1A). Nuclear IRF-3 was not present in Huh7 cells at later time points (48 h and after), corresponding to the period when HCV proteins and viral RNA accumulate. Nuclear IRF-3 was not observed in HCV-infected Huh7.5 cells (see Fig. 5A, which is published as supporting information on the PNAS website), which harbor a dominant-negative allele of RIG-I that ablates virus activation of IRF-3 (5). Compared with Huh7 cells, which have an intact RIG-I pathway, Huh-7.5 cells supported increased production of infectious virus (Fig. 1B) and more rapid virus spread (Fig. 5B). Thus, HCV infection can trigger RIG-I-dependent signaling events that transiently activate IRF-3 and limit viral replication and spread during initial rounds of replication. However, this host response is blunted once viral proteins accumulate in the cell.
Fig. 1.
NS3/4A cleaves IPS-1 to control the host response during HCV infection. (A) Huh7 cells were mock-infected or infected with JFH-1 HCV 2a at a multiplicity of infection of 3. At the indicated h after infection, cells were immunostained with HCV 2a patient serum (green) and anti-IRF-3 serum (red). Nuclei were visualized by staining the cells with DAPI (blue). Cells with nuclear IRF-3 (indicated by arrows) were counted and expressed as a percentage of the total cells in a field. The top left of A shows IRF-3 in Sendai virus-infected control cells. (B) Focus-forming units per ml of HCV within supernatant collected from cultures of Huh7 cells (white bars) or Huh7.5 cells (black bars) at 24 and 48 h after infection with JFH-1 virus. (C) Immunoblot of endogenous IPS-1, NS3, NS5A, and GAPDH in Huh7 cells that were mock-infected (lanes 1–4) or infected with the JFH-1 virus (lanes 5–8) for the time shown above each lane. Arrows mark the positions of full-length (FL) and cleaved (C) forms of IPS-1. (D) Immunoblot of endogenous IPS-1, NS3, and GAPDH in extracts from Huh7 (lane 1), Huh7-K2040 (lane 2), Huh7-HP (lane 3), and Huh7-JFHR (lane 4) cells and Huh7 cells that were mock-infected or infected with the JFH-1 virus (multiplicity of infection = 1) for 96 h (lanes 5 and 6, respectively). (E) Huh7 cells were cotransfected with constructs encoding the Myc-vector (−) or Myc-IPS-1 (+) and either the Flag-vector (lanes 1–2) or a Flag-tagged expression construct encoding WT NS3/4A protease from the HCV 1b Con1 clone (lanes 3–4), protease-deficient Con1 mutant (lanes 5–6), patient isolates (HCV 1a, lanes 7–8; HCV 1b, lanes 9–10; HCV 2b, lanes 11–12), or the JFH-1 clone (lanes 13–14). Expression of Myc-IPS-1, Flag-NS3, ISG56, and GAPDH is shown.
IPS-1 Signals the RIG-I-Dependent Host Response to HCV Infection.
To identify candidate signaling partners of RIG-I that may be involved in virus activation of IRF-3 and regulated by HCV, we searched the GenBank database (release no. 2.2.13) for CARD proteins, which could possibly interact with RIG-I CARDs (for details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). We identified the KIAA1271-expressed sequence tag that encodes a 540-aa protein that possesses a single amino-terminal CARD homologous to the CARDs of RIG-I and that has been implicated in the activation of nuclear factor κB (NF-κB), a transcription factor that, like IRF-3, is frequently activated in viral infections (17). The corresponding cDNA was cloned from total RNA recovered from Huh7 cells. We verified that ectopic expression of the encoded protein could induce IRF-3 activation and ISG expression independent of virus infection (data not shown). Similar results have now been reported by others (7–10); herein, we refer to this protein as IPS-1. Short-interfering RNA knock-down of IPS-1 mRNA prevented IRF-3 activation/nuclear accumulation by HCV and enhanced the spread and production of HCV from infected Huh7 cells (Fig. 6 A and B, which is published as supporting information on the PNAS web site). When overexpressed, IPS-1 directed a host response that reduced cellular permissiveness for HCV replication and suppressed viral protein expression (Fig. 6 C and D) and cellular abundance of distinct HCV RNA replicons (data not shown). These results indicate that HCV triggers IRF-3 activation through a RIG-I/IPS-1 signaling pathway to impart a host response that suppresses HCV infection. In the latter experiments, we reproducibly observed accumulation of a Flag-IPS-1 isoform with faster mobility on SDS/PAGE than the native Flag-IPS-1 species in replicon-bearing cells but not in control Huh7 cells, suggesting that the Flag-IPS-1 product was being proteolytically processed during HCV RNA replication. Thus, we further examined endogenous IPS-1 in Huh7 cells infected with JFH-1 virus. In mock-infected cells, IPS-1 was present as a single ≈87-kDa species. During HCV infection, a faster migrating form of IPS-1 began to accumulate from 24 h through 48 h after infection concomitant with accumulation of HCV proteins (Fig. 1C). The faster-mobility form of IPS-1 predominated in HCV-infected cells at 96 h after infection and in Huh7 cells harboring genotype 1b or 2a subgenomic replicons that encode the NS3-NS5B polyprotein segment (Fig. 1D). These results suggest that one or more nonstructural proteins, presumably the NS3/4A protease, cleave IPS-1 at an early point in infection.
The HCV NS3/4A Protease Cleaves IPS-1 at C508.
We evaluated the ability of NS3/4A to cleave IPS-1. Coexpression studies of Flag-NS3/4A variants with Myc-IPS-1 revealed that Myc-IPS-1 was processed into a faster-mobility form upon coexpression of NS3/4A from cDNAs cloned from the genotype 1b Con1 sequence (12), genotype 2a JFH-1 virus, or from RNA recovered from sera of patients infected with different subtypes of genotype 1 and 2 HCV (Fig. 1E). Myc-IPS-1 was not processed when coexpressed with Flag-vector, a proteolytically inactive S139A NS3/4A mutant (Fig. 1E), or with other HCV nonstructural proteins (data not shown). We also found that Flag-IPS-1 was processed to the faster-mobility form in osteosarcoma cells expressing NS3/4A (Fig. 2A) and was efficiently cleaved in a cell-free system by purified recombinant single-chain HCV 1b protease fused to maltose-binding protein (MB-scNS3) (see Fig. 7 A–C, which is published as supporting information on the PNAS web site) (14). Importantly, in these studies, IPS-1 cleavage was blocked by SCH6 or BILN 2061, which are peptidomimetic active site inhibitors of NS3/4A protease (18, 19), confirming that IPS-1 is a substrate of NS3/4A. To determine the site(s) at which IPS-1 is cleaved by NS3/4A, we used cell-based and in vitro assays to examine the ability of various IPS-1 mutants to be cleaved when coexpressed with NS3/4A. NS3/4A failed to cleave a C508Y mutant of IPS-1 (Figs. 2A and 5C), but proteolysis was retained when other potential cleavage sites (listed in Fig. 7D) were mutated to aa residues nonpermissive for NS3/4A cleavage. Therefore, NS3/4A cleaves IPS-1 between C508 and H509. Similar results were presented while this manuscript was in preparation (10).
Fig. 2.
NS3/4A cleavage of IPS-1 from the mitochondria is prevented by HCV protease inhibitors. (A) UNS3/4A cells, cultured to suppress (−) or induce (+) NS3/4A expression, were transfected with constructs encoding Flag-vector, WT Flag-IPS-1, or Flag-IPS-1 C508Y mutant. Expression of the Flag-IPS-1 constructs, NS3, and GAPDH is shown. (B Upper) aa sequence comparison of the NS3/4A cleavage sites (denoted by the arrow) of IPS-1 and the NS5A/5B junction in the HCV 1b polyprotein. Cleavage site positions and IPS-1 aa numbers are shown. Identical and conserved aa residues are boxed and underlined, respectively. (B Lower) Structural representation of IPS-1 denoting the aa positions of the CARD, Proline-rich region (Pro), transmembrane domain (TM), and the NS3/4A cleavage site at C508. (C) Huh7 cells that were mock-infected or infected with HCV for 48 h were immunostained for endogenous IPS-1 and HCV proteins. Nuclei were stained with DAPI. For the HCV-infected cells, the brightness of the IPS-1 panels was increased to 200% to allow visualization of the redistributed IPS-1. (Magnification: ×64.) (D) Immunoblot analysis of IPS-1 within mitochondria preparations. (Left) Mitochondria recovered from Huh7 cells (control, lane 1) or cells harboring the replicating full-length HCV-N genome (20) (lanes 2 and 4) and their IFN-cured counterparts (lanes 3 and 5). (Right) Mitochondria preparations from Huh7 cells were analyzed alone (lane 1) or after incubating with buffer (lanes 2–3), MB-scNS3 (lanes 4–5), or MB-scNS3 and 10 μM SCH6 (lanes 6–7). The mixtures in lanes 2–7 were further separated into soluble (S) and pellet (P) fractions before immunoblot analysis. Arrows denote the positions of IPS-1, which is recovered as multiple products from mitochondria preparations (8). The lower panels show the abundance of the mitochondrial membrane-associated 39-kDa subunit of cytochrome C oxidase I (CI 39) (21). (E) Huh7, Huh7-A7, or Huh7-HP cells were treated with DMSO (lanes 1–3), 10 μM SCH6 (lanes 4–6), or 1 μM ITMN-C (lanes 7–9) for 48 h before immunoblot analysis of endogenous IPS-1, NS3, and GAPDH.
NS3/4A Cleaves IPS-1 off the Mitochondrial Membrane but Is Prevented by NS3 Protease Inhibitors.
C508 lies adjacent to the transmembrane domain of IPS-1 (Fig. 2B), and cleavage at this position is expected to release it from its membrane anchor. Cleavage of endogenous IPS-1 in HCV-infected cells led to its redistribution from a primarily mitochondrial pattern to a more diffuse cytosolic distribution of the cleavage product (see Fig. 2C and Fig. 8A, which is published as supporting information on the PNAS web site). We also observed a reduction in the abundance of IPS-1 in HCV replicon cells (data not shown). Consistent with these observations, when expressed in Huh7 cells, a truncated IPS-1 mutant representing the 1-508 cleavage product localized throughout the cell independently of the mitochondria (Fig. 8B). Moreover, [35S]Met labeling of cells and pulse–chase/immunoprecipitation analyses demonstrated that the half-life of full-length IPS-1 (18 h in Huh7 cells) was reduced to 3.5 h in Huh7-K2040 cells harboring an HCV replicon, whereas the 1-508 cleavage product exhibited a half-life of 12 h (Fig. 8C).
To determine whether endogenous IPS-1 is cleaved and released from the mitochondria during HCV RNA replication, we examined the association of IPS-1 with mitochondria recovered from cell lines harboring replicating genome-length RNA replicons (20). Immunoblot analysis showed that IPS-1 was present within mitochondrial fractions from control Huh7 cells (Fig. 2D Left) but absent from similar fractions prepared from NNeo/C-5B clone 2-3 and 3 cells harboring replicating genotype 1b RNA (20). IPS-1 was restored to the mitochondria in their IFN-α-cured counterparts, 2-3c and 3c cells, respectively. To determine whether IPS-1 could be cleaved by NS3/4A after its insertion into the mitochondrial membrane or whether cleavage is possible only in the soluble form before membrane insertion, we added MB-scNS3 to crude mitochondrial preparations recovered from control Huh7 cells. IPS-1 was released from the insoluble mitochondrial fraction into the supernatant in a process that was blocked by SCH6 (Fig. 2D Right). Consistent with this finding, endogenous IPS-1 was cleaved in Huh7-A7 and Huh7-HP replicon cells in a manner that was blocked by chemically distinct inhibitors of the NS3/4A protease, SCH6 and ITMN-C (Fig. 2E). Thus, NS3/4A cleavage of IPS-1 releases it from the mitochondria even after its membrane insertion, and NS3/4A protease inhibitors protect IPS-1 from proteolysis and mitochondrial release by HCV.
IPS-1 Cleavage During HCV Infection Ablates RIG-I Interaction and Host Defense Signaling.
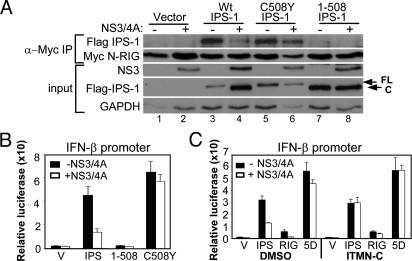
IPS-1 binds to the RIG-I CARDs in a reaction that signals the activation of IRF-3 and the induction of the IFN-β promoter (7). We thus examined the impact of IPS-1 cleavage upon its RIG-I binding and IRF-3 signaling properties. When expressed in Huh7 cells, the constitutively active N-RIG mutant (containing the RIG-I CARDs) (5) formed a stable complex with endogenous IPS-1 or Flag-IPS-1 that was disrupted by NS3/4A upon IPS-1 cleavage that conferred loss of signaling of IRF-3 activation (Fig. 3A and Fig. 9, which is published as supporting information on the PNAS web site). This finding suggests that the 1-508 IPS-1 cleavage product is unable to bind to RIG-I and mediate downstream signaling. Consistent with this notion, an IPS-1 1-508 mutant could neither bind to N-RIG nor signal to the IFN-β promoter, whereas these activities were preserved in an IPS-1 C508Y cleavage site mutant regardless of NS3/4A expression (Fig. 3 A and B). Promoter induction by Flag-IPS-1 was also fully restored by treating NS3/4A-expressing cells with ITMN-C (Fig. 3C). Thus, NS3/4A cleavage of IPS-1 disrupts its RIG-I binding and signaling actions, but these properties were restored by treatment with an HCV protease inhibitor.
Fig. 3.
Cleavage of IPS-1 blocks RIG-I interaction and disrupts signaling to IFN-β, but signaling is restored by protease inhibitor treatment. (A) UNS3/4A cells, cultured to suppress (−) or induce (+) NS3/4A expression, were cotransfected with constructs encoding Myc-N-RIG and Flag-vector (lanes 1–2), Flag-IPS-1 (lanes 3–4), Flag-IPS-1 C508Y (lanes 5–6), or Flag-IPS-1 1-508 (lanes 7–8). Extracts were subject to immunoprecipitation by using anti-Myc antibody and analyzed by immunoblotting for the presence of the WT or mutant Flag-IPS-1 protein and Myc-N-RIG (upper panels). The lower panels show the abundance of NS3, Flag-IPS-1, and GAPDH in the input extracts. (B and C) UNS3/4A cells, cultured to suppress (black bars) or induce (white bars) NS3/4A expression, were transfected with Renilla luciferase and IFN-β-luciferase plasmid constructs and a plasmid encoding the Flag-vector (V), Flag-IPS-1 (IPS), Flag-IPS-1 1-508 (1-508), Flag-IPS-1 C508Y (C508Y), Flag-N-RIG (RIG), or IRF-3–5D (5D). Cells were incubated for 24 h in medium alone (B) or medium with DMSO or 1 μM ITMN-C (C), and extracts were subjected to the dual luciferase assay. Bars show relative IFN-β-luciferase activity (±SD).
IPS-1 Is Cleaved and Redistributed in Vivo During Chronic HCV Infection.
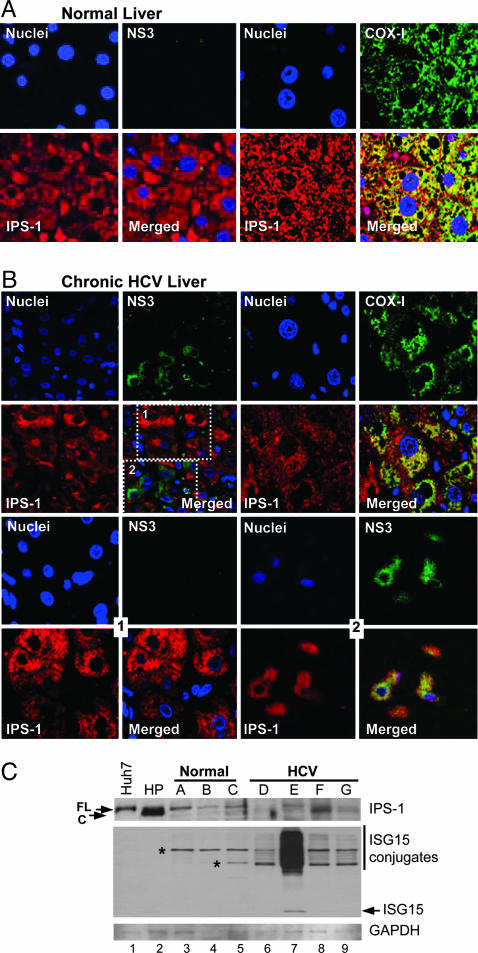
We examined the distribution and processing of IPS-1 in liver tissues recovered from uninfected donors and patients with chronic HCV infection. Confocal microscopy of immunostained normal liver sections showed that IPS-1 was codistributed with subunit I of the cytochrome C oxidase complex IV (COX-I), a mitochondrial membrane protein (21), confirming the mitochondrial distribution of IPS-1 in vivo (Fig. 4A). Similar findings were observed in tissues from a cohort of 12 patients with nonviral liver disease (D.T.-Y.L., P.M.F., and M.G., unpublished observations). Among a cohort of 18 patients with chronic HCV, IPS-1 was found distributed within focal clusters of cells independently of COX-I (Fig. 4B Upper Right). In these tissues, NS3 was variably detected in hepatocytes and appeared within discrete focal clusters in which the intensity of IPS-1 staining was reduced compared with neighboring hepatocytes staining negative for NS3 (Fig. 4B Upper Left). This staining pattern was similar to the distribution pattern of IPS-1 and NS3 observed in replicon- or JFH-1-infected Huh7 cells (see Fig. 2C). Further examination of HCV-infected liver sections by using confocal optical sectioning of regions containing NS3-positive cell clusters revealed a marked redistribution of IPS-1, atypical of mitochondria and overlapping with NS3 throughout the cytoplasm (Fig. 4B Lower Right). In contrast, a normal mitochondrial staining pattern for IPS-1 was present within neighboring cells lacking NS3 (Fig. 4B Lower Left).
Fig. 4.
In vivo redistribution and cleavage of IPS-1 during chronic HCV infection. Confocal micrographs of a 0.3-μm optical section of immunostained sections from normal donor liver (A) or liver biopsy from a patient with chronic HCV infection (B). (Magnification: ×40.) Shown are individual and merged images of sections stained with DAPI and antiserum specific to either IPS-1 (red) and NS3 (green) (Left) or IPS-1 (red) and COX-I (green) (Right). (B Lower) Images from the same tissue specimen shown in B Upper and were collected at a depth of 9 μm within the 32-μm-thick specimen. (B Lower Left) Images correspond to boxed area 1 (shown in the lower right corner of B Upper Left) of hepatocytes that stained negative for NS3. (B Lower Right) Images correspond to boxed area 2 (shown in the lower right corner of B Upper Left) of an adjacent focal area of hepatocytes that stained positive for NS3. (C) Immunoblot of protein extracts from Huh7 and Huh7-HP cells (lanes 1–2, respectively), specimens from three unused, normal donor livers (lanes 3–5), or specimens of liver from different patients chronically infected with HCV (lanes 6–9). Panels show the abundance of IPS-1, ISG15, or GAPDH. Arrows point to the positions of IPS-1 and native ISG15. The vertical line denotes ISG15 conjugation products; asterisks denote nonspecific bands that are not reproducibly observed.
Immunoblot analysis of protein extracts from a second set of tissue specimens demonstrated a single IPS-1 product in normal donor liver that comigrated with full-length IPS-1 present in extracts from control Huh7 cells (Fig. 4C). In contrast, full-length IPS-1 was only variably present in total protein extracts from liver needle biopsies from patients with chronic HCV infection, ranging from undetectable levels [patient D (patients A–G are indicated above the immunoblot in Fig. 4C)] to the presence of only the cleaved product of IPS-1 (patients F and G). Liver from one patient (patient E) appeared to have predominantly full-length, noncleaved IPS-1. We also determined the abundance of ISG15 in these liver extracts. ISG15 expression is induced by activated IRF-3 (22) and regulates cellular proteins by forming ISG15–protein conjugates, which are markers of the host response to virus infection (11). ISG15 was not detected in extracts from normal donor liver. Among the HCV-infected tissue panel, ISG15 and its protein conjugates were detected only in liver from patient E, which had predominantly full-length IPS-1. These results provide strong evidence that IPS-1 is targeted and proteolytically cleaved by NS3/4A in vivo in HCV-infected humans, resulting in its subcellular redistribution and the attenuation of the host response to infection.
Discussion
Viral persistence is a hallmark of HCV, wherein continual virus replication underlies the pathogenesis and liver disease associated with chronic infection. Our results provide evidence that links HCV persistence with viral control of the host response to infection. We found that HCV infection transiently signals the activation of IRF-3 and host cell responses in a RIG-I-dependent manner and that IPS-1 is an essential component of this signaling that is targeted and cleaved by NS3/4A during infection both in vivo and in vitro. These results confirm and extend the observations of others that have been reported during the preparation of this manuscript (10). Many viruses antagonize IRF-3 through varied mechanisms that support their replication (11), underscoring the central role of IRF-3 and IFN in antiviral immune defense (23). IPS-1 is rapidly cleaved at C508 by NS3/4A during HCV infection, resulting in release of IPS-1 from the mitochondrial membrane and destabilization and loss of its interaction with RIG-I. Thus, mitochondrial membrane association appears essential for IPS-1 function. IPS-1 likely requires interaction with other mitochondria-associated factors that participate in protein recruitment and/or signaling of IRF-3 activation. In support of this idea, recent studies suggest that IPS-1 may recruit a variety of effectors that confer signaling by RIG-I or the related helicase, MDA5, which may bind to distinct RNA pathogen-associated molecular patterns to initiate the host response independently of RIG-I (7). Importantly, NS3/4A blocks signaling by both RIG-I and MDA5 (24), consistent with a central role for IPS-1 in these respective pathways.
Our results support a model of dynamic control of the host response by HCV in which infection initially triggers IRF-3 activation through RIG-I- and IPS-1-dependent signaling during a window period of low intracellular viral protein abundance, slowing HCV replication and spread. However, as the infection progresses, the NS3/4A protease accumulates to a threshold level within the cell, ultimately resulting in cleavage of IPS-1 from the mitochondria. This process terminates RIG-I signaling of the host response, promoting viral persistence. Such a model predicts that IFN and ISG products would be continuously produced as a result of asynchronous viral spread within the liver during chronic infection, because the host response would be induced in newly infected cells until the point of IPS-1 cleavage. This prediction is consistent with previous studies showing expression of intrahepatic ISGs during chronic HCV infection (25) and is supported by our observations of varied IPS-1 abundance, cleavage, and distribution in patient tissues. Among the specimens from our HCV-infected patient cohort, partial cleavage of IPS-1 corresponded with an abundance of ISG15 products (see patient E in Fig. 4C), whereas complete cleavage or reduced levels of IPS-1 was associated with a lack of ISG15 expression. We conclude that IPS-1 is targeted and cleaved by NS3/4A during persistent HCV infection in vivo. The mechanism by which NS3/4A targets IPS-1 on the mitochondrial membrane is not yet known. Recent studies show that only a portion of total nonstructural proteins associate with the HCV replicase (26) and that NS3/4A can independently localize to the mitochondria (27), supporting roles for it both in viral replication and host defense regulation.
HCV protease inhibitors are currently being developed for clinical applications (28). We found that chemically diverse small-molecule inhibitors could protect IPS-1 from cleavage by NS3/4A, thereby restoring RIG-I and IPS-1 signaling to the IFN-β promoter. Our results suggest that these compounds may have dual therapeutic mechanisms, both suppressing HCV replication by preventing polyprotein processing, while concomitantly releasing the host response from the blockade imposed by NS3/4A cleavage of IPS-1. RIG-I signaling drives an IFN amplification loop that increases the level and diversity of ISG expression (4). In the context of IFN therapy for HCV infection, IPS-1 cleavage by NS3/4A would attenuate this loop and the expression of those ISGs normally amplified and/or induced through RIG-I- and IRF-3-dependent processes. Such a mechanism could limit the efficacy of IFN-based therapies. Targeting of the NS3/4A-IPS-1 interface by protease inhibitors may therefore enhance IFN actions. This therapeutic control of IPS-1 may explain the remarkable effectiveness and rapidity with which such compounds have been shown to suppress HCV replication and viremia in patients participating in early phase I clinical trials (19).
Materials and Methods
Cell Culture, Treatment, and Viruses.
Huh7 and Huh7.5 are human hepatoma cells (5). Huh7-HP, Huh7-A7, and Huh7-K2040 cells contain variants of an HCV 1b subgenomic replicon (12). 7-JFHR is a Huh7 cell line that harbors the JFH-1 HCV (genotype 2a) subgenomic replicon RNA (29) and was created by using in vitro-transcribed replicon RNA exactly as described in ref. 12. NNeo/C-5B clone 2-3 and 3 cells contain the genome-length HCV-N replicon (20); 2-3c and 3c cells represent their IFN-α cured progeny. UNS3/4A osteosarcoma cells express the HCV 1a NS3/4A protease under the regulation of a tetracycline-repressible promoter (12). Stocks of the NS3/4A-specific protease inhibitors, Schering 6 (SCH6) and ITMN-C [gifts from Schering-Plough and InterMune (Brisbane, CA), respectively] were prepared in DMSO. For treatment of cells, culture medium was replaced with medium containing 10–20 μM SCH6 or 1–2 μM ITMN-C for the indicated times. Immunofluorescence staining, immunoprecipitation, and immunoblot analyses were carried out as described in ref. 12. HCV was produced from the RNA from pJFH-1 encoding the JFH-1 HCV 2a infectious clone [a gift from T. Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo)] exactly as described in ref. 16. Sendai virus (Cantrell strain) was obtained from Charles River Laboratories. Virus infections and titrations were conducted and quantified as described in refs. 12 and 16.
Expression Constructs.
RIG-I, IRF-3, Renilla-luc, and IFN-β-luc constructs have been described (5, 15). IPS-1 cDNA was generated by PCR amplification of cDNA made from RNA recovered from Huh7 cells. PCR products were cloned into pEF-derived plasmids with amino-terminal Flag or Myc epitopes. IPS-1 mutants were created by using the QuikChange site-directed mutagenesis kit (Strategene). Primer sequences are available upon request.
Proteolytic Cleavage of IPS-1 from Isolated Mitochondria.
Huh-7, 2-3, 3, 2-3c, and 3c cells were resuspended in buffer (200 mM mannitol/70 mM sucrose/1 mM EGTA/10 mM Hepes, pH 7.5) and homogenized by 20 strokes in a tight-fitting Dounce homogenizer. Lysates were centrifuged at 500 × g and at 10,000 × g to collect crude mitochondria. Mitochondria were incubated at 37°C for 2 h in NS3/4A cleavage buffer (50 mM Tris·HCl/150 mM NaCl/20% glycerol/1 mM DTT, pH 7.5) without protease, with MB-scNS3 (3 μM) (14), or with MB-scNS3 plus 10 μM SCH6 and subsequently centrifuged at 10,000 × g to separate mitochondria from postproteolysis supernatant.
Human Liver Tissue.
Liver biopsy samples of chronic HCV patients and discarded tissue recovered from normal donor liver during transplantation surgery were obtained under Institutional Review Board-approved protocols and were immediately placed in liquid nitrogen storage or fixation buffer. Protein extracts from human liver tissue were prepared by grinding the minced liver sample in 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 0.5% Na-deoxycholate, 1% Triton X-100, 1 mM EDTA, and 0.1% SDS, supplemented with proteinase and phosphotase inhibitor cocktails (Sigma). Extracts were centrifuged at 16,000 × g at 4°C and the supernatant was analyzed by immunoblotting.
Immunostaining and Confocal Microscopy.
Paraffin-embedded liver tissues were deparrafinized in xylene and ethanol and subsequently immersed in 1× AR-10 solution (BioGenex Laboratories, San Ramon, CA) according to manufacturer's instructions. Slides were then incubated in 1% Triton X-100 and PBS, blocked with 10% normal goat serum, sequentially stained with the appropriate dilutions of primary and secondary antibodies, and washed, and Vectashield mounting medium (Vector Laboratories) was applied. Slides were sealed under coverslips and examined by multitrack confocal microscopy by using a Zeiss Laser Scanning Confocal Microscope in the University of Texas Southwestern Medical Center Pathogen Imaging Facility.
Antibodies and additional molecular biology reagents are described in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Wakita for the JFH-1 clone and Schering-Plough and InterMune for SCH6 and ITMN-C, respectively. This work was supported by National Institutes of Health Grants U01AI48235, R01AI060389 (to M.G.), U19AI40035 (to S.M.L.), R21DA018054 (to K.L.), R01AA012863 (to S.A.W.), and R01DK068598 (to D.T.-Y.L.) and grants from the Burroughs–Wellcome Fund (to M.G.).
Abbreviations
- IPS-1
IFN promoter stimulating factor-1
- IRF-3
IFN regulatory factor-3
- RIG-I
retinoic acid-inducible gene-I
- HCV
hepatitis C virus
- CARD
caspase activation and recruitment domain
- ISG
IFN-stimulated gene.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Shepard C. W., Finelli L., Alter M. J. Lancet Infect. Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach B. D., Rice C. M. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds P., Tuplin A., Evans D. J. RNA. 2004;10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 5.Sumpter R., Loo Y.-M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S. M., Gale M. J. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., et al. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 8.Seth R. B., Sun L., Ea C. K., Chen Z. J. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 11.Sen G. C. Annu. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 12.Foy E., Li K., Wang C., Sumpter R., Ikeda M., Lemon S. M., Gale M., Jr. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 13.Foy E., Li K., Sumpter R., Jr., Loo Y. M., Johnson C. L., Wang C., Fish P. M., Yoneyama M., Fujita T., Lemon S. M., et al. Proc. Natl. Acad. Sci. USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li K., Foy E., Ferreon J. C., Nakamura M., Ferreon A. C., Ikeda M., Ray S. C., Gale M., Jr., Lemon S. M. Proc. Natl. Acad. Sci. USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato T., Furusaka A., Miyamoto M., Date T., Yasui K., Hiramoto J., Nagayama K., Tanaka T., Wakita T. J. Med. Virol. 2001;64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 16.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. Proc. Natl. Acad. Sci. USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., et al. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 18.Arasappan A., Njoroge F. G., Chan T. Y., Bennett F., Bogen S. L., Chen K., Gu H., Hong L., Jao E., Liu Y. T., et al. Bioorg. Med. Chem. Lett. 2005;15:4180–4184. doi: 10.1016/j.bmcl.2005.06.091. [DOI] [PubMed] [Google Scholar]
- 19.Lamarre D., Anderson P. C., Bailey M., Beaulieu P., Bolger G., Bonneau P., Bos M., Cameron D. R., Cartier M., Cordingley M. G., et al. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda M., Yi M., Li K., Lemon S. M. J. Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalimonchuk O., Rodel G. Mitochondrion. 2005;5:363–388. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Grandvaux N., Servant M. J., tenOever B., Sen G. C., Balachandran S., Barber G. N., Lin R., Hiscott J. J. Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., et al. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 24.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., et al. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 25.Smith M. W., Yue Z. N., Korth M. J., Do H. A., Boix L., Fausto N., Bruix J., Carithers R. L., Jr., Katze M. G. Hepatology. 2003;38:1458–1467. doi: 10.1016/j.hep.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Quinkert D., Bartenschlager R., Lohmann V. J. Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasprzak A., Seidel J., Biczysko W., Wysocki J., Spachacz R., Zabel M. Liver Int. 2005;25:896–903. doi: 10.1111/j.1478-3231.2005.01109.x. [DOI] [PubMed] [Google Scholar]
- 28.Tan S. L., He Y., Huang Y., Gale M., Jr. Curr. Opin. Pharmacol. 2004;4:465–470. doi: 10.1016/j.coph.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Kato T., Date T., Miyamoto M., Furusaka A., Tokushige K., Mizokami M., Wakita T. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.