Abstract
With current concerns of antibiotic-resistant bacteria and biodefense, it has become important to rapidly identify infectious bacteria. Traditional technologies involving isolation and amplification of the pathogenic bacteria are time-consuming. We report a rapid and simple method that combines in vivo biotinylation of engineered host-specific bacteriophage and conjugation of the phage to streptavidin-coated quantum dots. The method provides specific detection of as few as 10 bacterial cells per milliliter in experimental samples, with an ≈100-fold amplification of the signal over background in 1 h. We believe that the method can be applied to any bacteria susceptible to specific phages and would be particularly useful for detection of bacterial strains that are slow growing, e.g., Mycobacterium, or are highly infectious, e.g., Bacillus anthracis. The potential for simultaneous detection of different bacterial species in a single sample and applications in the study of phage biology are discussed.
Keywords: bacteriophage T7, BirA, Escherichia coli, water sample
The number and diversity of bacteriophages in the environment provide a promising natural pool of specific detection tools for pathogenic bacteria. Currently there are several phage-based methods for detection of pathogenic bacteria (1): a plaque assay for detection of Mycobacterium tuberculosis (2); fluorescence-labeled phage and immunomagnetic separation assay for detection of Escherichia coli O157:H7 (3, 4); phage-based electrochemical assays (5); a luciferase reporter mycobacteriophage and Listeria phage assays (6, 7); and detection of the phage-mediated bacterial lysis and release of host enzymes (e.g., adenylate kinase) (8).
Two limiting features when detecting pathogenic bacteria are sensitivity and rapidity. Common fluorophores (e.g., GFP and luciferase) used as reporters have two major disadvantages: low signal-to-noise ratio due to autofluorescence of clinical samples and of bacterial cells and low photostability, such as fast photobleaching. To overcome these disadvantages, we used new fluorescent semiconductor nanocrystals, quantum dots (QDs) (9). QDs are colloidal semiconductor (e.g., CdSe) crystals of a few nanometers in diameter. They exhibit broadband absorption spectra, and their emissions are of narrow bandwidth with size-dependent local maxima. The presence of an outer shell of a few atomic layers (e.g., ZnS) increases the quantum yield and further enhances the photostability, resulting in photostable fluorescent probes superior to conventional organic dyes. Recently, development in surface chemistry protocols allows conjugation of biomolecules onto these QDs to target specific biological molecules and probe nanoenvironments (10–12). The power to observe and trace single QDs or a group of bioconjugated QDs, enabling more precise quantitative biology, has been claimed to be one of the most exciting new capabilities offered to biologists today (13, 14).
Typically, the detection of small numbers of bacteria in environmental or clinical samples requires an amplification step involving the growth of bacteria in culture to increase cell number. This procedure considerably prolongs the detection time, especially for slowly growing bacteria. Here we report a sensitive, rapid, and simple method for detection of bacteria. This method combines in vivo biotinylation of engineered host-specific bacteriophage and conjugation of the phage to streptavidin-coated QDs. This phage-based assay reduces the “amplification” to a short time (20–45 min from infection to lysis) because each infected bacterium can result in a release of 10–1,000 phage that can be readily detected by the use of QDs.
Results and Discussion
Overall Strategy and Considerations for the Detection Method.
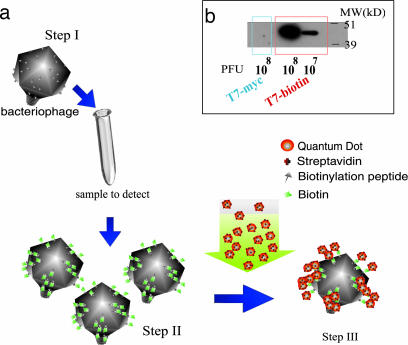
Our strategy of the detection method is shown in Fig. 1a. We engineered a phage to display a small peptide, which can be biotinylated (biotinylation peptide), fused to the major capsid protein. Our “reagent” phage (step I) contains the genetic information to display tagged head protein but is assembled either (i) in vivo in a nonbiotinylating mutant host to display nonbiotinylated biotinylation peptide or (ii) in vitro to contain the wild-type capsid protein (see below). If the bacteria sensitive to these phage are present in a sample, progeny virions produced from infection by the reagent phage will have been biotinylated by the host cell’s biotin–ligase protein (BLP; BirA in case of E. coli used in our experiments) (step II in Fig. 1a). Biotin (vitamin H), which is present in all living cells, is attached posttranslationally by BLP to a specific lysine residue in the tagged peptide (15). The biotinylation of the target protein(s) by BLP is extremely conserved throughout evolution (15, 16). Biotinylation is a highly conserved process in bacteria, so this approach is likely to be of general utility. For every bacterium in the sample, a high degree of amplification will occur depending on the burst size of the phage. In step III, the presence of biotinylated phage particles in the lysate, which reflects the presence of sensitive bacteria in the original sample, is detected by conjugation to streptavidin-functionalized QDs.
Fig. 1.
An overall strategy of bacterial detection using nanoengineered phage–QD complexes. (a) A schematic representation of the detection (see text for details). (b) Western blot analysis of T7-bio or control T7-myc phage particles using streptavidin-horseradish peroxidase.
Engineering the Reagent Phage.
As a model system, we engineered the coliphage T7 to express the major capsid protein gp10A, fused to the 15-aa biotinylation peptide (T7-bio): GLNDIFEAQKIEWHE (17). This cloning strategy (see Materials and Methods for details) results in the display of the given peptide on all 415 monomers of the major capsid protein. To differentiate the reagent phage from phage released from infected target cells, it is crucial that the reagent input phage is not biotinylated. We accomplished this in one of the two ways: (i) by packaging the engineered T7 phage DNA in vitro using wild-type virion proteins or (ii) by propagating our reporter phage on a biotin auxotroph in limiting biotin conditions. In the first method we used a commercially available packaging kit composed of wild-type phage proteins (T7Select, Novagen). When used with the engineered phage T7 DNA this system gave rise to 103 to 106 plaque-forming units per microgram of DNA. In the second method we prepared phage lysates by two rounds of growth on an E. coli biotin auxotroph that was starved for biotin, as was evident by an inhibition of bacterial growth without an evident decrease in plaque-forming units per milliliter. The absence of biotinylated phage resulting from either method of production was confirmed by Western blot analysis (using streptavidin-horseradish peroxidase) and fluorescence microscope using streptavidin QDs (Table 1, last row).
Table 1.
Detecting E. coli among several different bacterial cells
| No. of E. coli cells | No. of other cells | Expected no. of progeny phage per ml* | Percentage of QD-bound E. coli detected per ml† |
|---|---|---|---|
| 107 | 0 | 109 | 100 |
| 105 | 107 | 107 | 66 |
| 103 | 107 | 105 | 49 |
| 102 | 107 | 104 | 31 |
| 10 | 107 | 103 | 28 |
| 0 | 107 | 0 | 8 |
| 107‡ | 0 | 0 | 6 |
| 0§ | 0 | 0 | 1 |
*Based on burst size of 100 phage per cell.
†Only E. coli normalized to 100.
‡No phage added.
§Nonbiotinylated phage added at the detection step.
The resulting nonbiotinylated reagent phage, when making progeny particles after infection of wild-type E. coli bacterial cells, gets biotinylated at the displayed peptide by the host BLP, resulting in biotinylated phage referred to as T7-bio. As a negative control, we engineered T7 phage to express the major capsid protein fused to the 10-aa myc peptide (T7-myc), EQKLISEEDL, which results in a phage that displays the myc peptide but is not recognized by the host BLP.
The Phage-Displayed Peptide Is Biotinylated by the Phage-Specific Host in Vivo and Can Bind QDs.
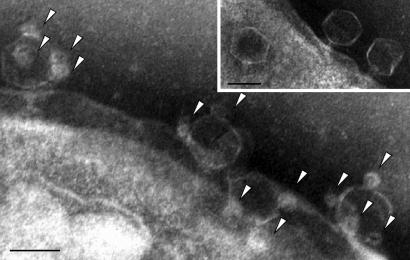
We tested the ability of the engineered phage to infect E. coli and become biotinylated by mixing the phage with a bacterial culture until cell lysis was visible. The presence of the biotin molecules on the phage’s displayed peptide was initially detected by Western blot analysis of the virion proteins from purified phage samples using streptavidin-horseradish peroxidase. The Western blot confirmed the in vivo biotinylation of the tagged capsid protein assembled on the phage head (Fig. 1b). We used transmission electron microscope (TEM) to estimate the number of biotinylated peptides on each phage. We adsorbed phage to carrier bacterial cells, conjugated streptavidin-coated QDs (in excess) to the phage, and removed free QDs by centrifugation and washing steps. Binding of QDs (arrowheads) to phage T7-bio was clearly demonstrable (Fig. 2), whereas control phage, T7-myc, did not show bound QDs (Fig. 2 Inset). We estimated the average number of QDs per phage to be 2.2 ± 1.3 (0.5% of the total peptides displayed), whereas ≈7% of the T7-bio phage had no QDs bound. Recombinant proteins carrying a biotinylation peptide expressed in low and high amounts are typically biotinylated at ≈30% and 6% efficiency, respectively, by endogenous levels of the BirA enzyme in E. coli (17). The low level of biotinylation obtained in our case, 0.5%, may be attributed to the very short (13-min) latent time of phage T7 and to the very high expression level of the capsid protein, which may exceed the capacity of BirA. In agreement with either explanation, when cells expressing BirA from a multicopy plasmid were used, a much higher level of bound biotin molecules per phage was obtained, as detected by Western blot analysis (data not shown). It might be useful to include the birA gene in the engineered phage to allow more of the displayed peptides to be biotinylated, thereby increasing the detection sensitivity. Biotin is a small molecule that does not seem to interfere with phage head assembly and stability (see below). Nevertheless, QD are ≈1/10th the size of the phage head, and modeling suggests that only ≈100 QDs could fit on the surface of the phage capsid. Importantly, the infective titer was not affected by binding of the QDs (data not shown), demonstrating that the presence of the QDs, at least at the level used here, causes neither inactivation nor aggregation of the virions.
Fig. 2.
T7-bio phage bound to streptavidin-functionalized QDs. TEM images of phage or phage–QD targeted bacteria are shown. The arrowheads point to QDs conjugated to the phage head. (Inset) Control T7-myc phage that are not biotinylated and therefore have no conjugated QDs. (Scale bars: 50 nm.)
Detection of QD–Phage Complexes by Flow Cytometry.
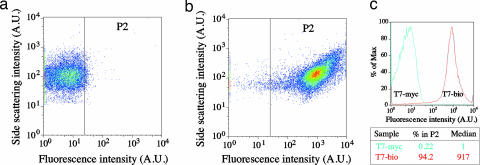
Biotinylated phage bound to QDs were initially detected and analyzed by flow cytometry. Flow cytometry allows scanning of a large number of particles; single cells flow in a fast stream through a focal volume of an excitation laser beam and light intensities due to side scattering (measured at 90° relative to the direction of the focused laser light), forward scattering (at 180°), and fluorescence light (at 90°) are monitored. Because phage–QD complexes are too small to be detected by the side scattering, we used carrier cells to which the phage T7 can bind. In Fig. 3 scatter plots of side-scattering vs. flourescence from each of the 30,000 cells infected either by T7-myc+QD (Fig. 3a) or by T7-bio+QDs (Fig. 3b), at a multiplicity of infection of 5, are compared. The results show that T7-bio-infected cells exhibit two orders of magnitude higher fluorescence than control as a result of the binding of streptavidin QDs to the biotins in the capsid of the T7-bio. Differentiation in fluorescence signal between the two populations is clear from the histograms of cells vs. fluorescence shown in Fig. 3c. Setting a threshold of m-2σ (22 arbitrary units in the fluorescence light channel) calculated from the histogram of the T7-myc-infected cells (designated P2), ≈94% of the T7-bio-bound cells showed fluorescence intensities above the threshold, whereas <1% of the T7-myc-bound cells did so. These results confirm our TEM observations in a larger population, validating no binding of QDs to the control phage, T7-myc, and preferential conjugation of streptavidin QDs to T7-bio. In addition, free QDs and/or phage–QD complexes that might be present in the sample are not detected because they do not trigger the detector channels. We estimate that the median in the flow cytometry measurement corresponds to four QDs: two QDs per phage (as estimated from TEM images) at a multiplicity of infection of 2. We believe that including the birA gene in the engineered phage, proposed above, would enhance the signal such that one phage per cell will be detected by using flow cytometry.
Fig. 3.
Phage–QD complexes detected by flow cytometry. Shown are scatter plots of bacteria targeted with T7-myc (a) or T7-bio (b) phage after addition of QDs. (c) Histograms of the number of cells vs. fluorescence with control phage (blue) and biotinylated phage (red), comparison of percentages within P2 range, and medians of the fluorescence intensities calculated from the histograms.
Single QD–Phage Complexes Are Readily Detected by Fluorescence Microscopy: Quantized Blinking State Allows for the Visualization of a Single Bound Phage.
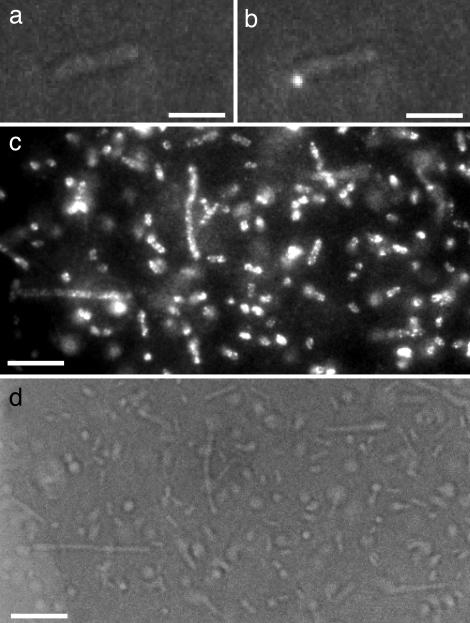
As a second method to detect QD labeled phage, we used fluorescence microscopy, which permits quantitative measurements with the sensitivity to detect a single QD conjugated to a single phage. As with the flow cytometry, we used carrier bacterial cells to allow removal of free QDs by washing. Fig. 4 shows optical micrographs of phage–QD complexes bound to cells. Fig. 4 a and b are typical images of a single cell decorated with a single phage–QD complex, obtained as a result of using a low number of biotinylated phage in the sample. Imaging at two different quantized blinking states in which the single QD is in either “on” or “off” verifies that a single QD was present. Fig. 4 c and d demonstrate that the number of phage–QD complexes on every cell was higher when a high number of biotinylated phages was added. Quantitative measurements of multiplicity of infection and the number of QDs on each phage may be possible, providing that the dispersion of phage–QD complexes is larger than the diffraction limit of the optical microscope, and as the optical characteristics of multiple QDs on a single phage–QD complex are the result of collective optical properties of single QDs. For instance, the number of quantized blinking steps in the fluorescence emission of a phage–QD complex will be directly correlated with the number of QDs in a single phage–QD complex (11). It is also noteworthy that no background fluorescence was observed from the cells or the medium (LB) and that the fluorescence emissions from QDs continued for hours without substantial photobleaching. No fluorescence signal was observed when phages were omitted or when T7-myc was used under the same conditions (data not shown).
Fig. 4.
Fluorescence microscope images of phage–QD complexes bound to cells. (a and b) A typical picture of E. coli cells exposed at low multiplicity to QD-tagged biotinylated phage. The field was simultaneously illuminated with a low-intensity white light source and a fluorescence excitation (447 ± 15 nm). The images are of two different quantized blinking states of a single QD: off (a) and on (b). (c) Fluorescence micrograph of cells with 100-fold excess of biotinylated phage. (d) Bright-field transmission micrograph of the same sample area obtained immediately after capturing the image in c. Note that some cells are immobilized on a substrate, but some are mobile in solution, resulting in out-of-focus fluorescence images when the focus is maintained on the cells on a substrate surface. [Scale bars: 1 μm (a and b) and 2 μm (c and d).]
Detection of E. coli in a Mix with Other Bacteria and in Environmental Samples.
To detect a small number of a given type of bacteria among several different bacterial cells, we used a culture with mixed bacterial strains on which phage T7 cannot propagate: Pseudomonas aeruginosa, Vibrio cholerae, Salmonella, Yersinia pseudotuberculosis, and Bacillus subtilis. We analyzed mixtures of 2 × 106 cells of each of the above strains mixed with different numbers of cells of E. coli (from 10 to 107 cells per ml). We followed the method as illustrated in Fig. 1 and scored the phage–QD conjugates by fluorescence microscopy. The results of such experiments, shown in Table 1, demonstrate that the non-E. coli strains provide a signal that is not significantly different from the background. The number of phage detected from a sample of only E. coli cells was normalized to 100% (all carrier cells contained QD). The number of phage detected depended on the number of E. coli cells in the sample. When 1,000 or 100 cells were detected, 52 of 107 (49%) or 48 of 155 (31%) carrier cells had conjugated T7-bio–QD, respectively. The signal from as few as 10 E. coli cells was significantly higher than the signal in controls with no E. coli or no phage added to the mix [124 of 450 (28%), 1 of 140 (<1%), and 45 of 736 (6%), respectively].
Finally, we tested water samples from the Potomac River. We determined, in ≈1 h, that there are at least 20 E. coli cells in 1 ml of the sample. In comparison, using the Coliscan MF kit (Micrology Laboratories, approved by the U.S. Environmental Protection Agency), it took 24 h to detect and identify 200 general coliforms in 1 ml of the same samples. The lower number of E. coli cells detected by our method is due to higher specificity of the phage to detect a particular coliform. These results demonstrate the rapid and specific nature of our assay.
Conclusions
The primary significance of the current work is the development of a simple and highly sensitive procedure for phage-based bacterial detection that achieves (i) enhanced detection limit, (ii) rapidity, and (iii) broad applicability. Sensitivity is shown by our method’s ability to detect and quantify low abundance targets of at least as few as 10 cells per milliliter. Our method takes ≈1 h to get results. The procedure uses biotinylation, a highly conserved pathway in nature, which can be applied to target a variety of bacteria in biological samples. Although we used a single phage–host system, the method may be expanded for the detection of multiple bacterial strains by their specific phages, each conjugated to QDs of different emission colors, in the same sample. Furthermore, higher specificity can be achieved by using multiple phages for one host in the same sample conjugated to QDs of different emission colors. The tools for detection can include microscopy, spectroscopy, and flow cytometry. It should be possible to use QD phage-based bacterial detection with hand-held instruments in the future. Additionally, because phage could not be seen by light microscopy previously and QD-labeled phage are infective, we believe that our method opens up new avenues to address phage biology-related questions on topics such as initial binding, phage localization, distribution, and more.
Materials and Methods
Engineering the T7-bio and T7-myc Phages.
We used the T7Select System (Novagen) for engineering and packaging of DNA into T7 phage particles. For the T7-bio we used two phosphorylated primers: 3′L6bio and 5′L6bio, containing overhang sequences for ligation with HindIII- and EcoRI-digested phage arms DNA, respectively (uppercase), and a 6-aa linker coding sequence (underlined), followed by the biotinylation peptide coding DNA (lowercase) and a stop codon (bold): 3′L6bio, 5′-AGCTTttagtgccattcgattttctgagcttcgaagatgtcgttcaggcctgaaccacgcggccgcaacG-3′; 5′L6bio, 5′-AATTCgttgcggccgcgtggttcaggcctgaacgacatcttcgaagctcagaaaatcgaatggcactaaA-3′. The primers were annealed to each other by heating at 95°C for 5 min in ligation buffer and cooling at room temperature, and ligation to T7 arms was done as recommended by the manufacturer. For the engineering of the T7-myc phage we used the primers MYC1 (5′-AATTCtggtggcagcggatctgagcagaagctgatcagcgaggaagatcttaattaaA-3′) and MYC2 (5′-AGCTttaattaagatcttcctcgctgatcagcttctgctcagatccgctgccaccaG-3′) containing overhang sequences for ligation with EcoRI- and HindIII-digested phage arms DNA (uppercase) and a 5-aa linker coding sequence (underlined), followed by the myc domain (lowercase) and a stop codon (bold).
Negative Stain for TEM.
Staining of phage was done as described elsewhere (18). Briefly, phage were incubated with E. coli for 4 min at 37°C in PBS buffer. A streptavidin-coated QD (QD 605) suspension (1 μM) (Quantum Dot, Hayward, CA) was diluted 100-fold in PBS. A total of 1 μl of the diluted solution was added, and incubation continued for 5 min at room temperature. After centrifugation at 200 × g for 5 min, 1 μl of the sample was placed onto a carbon-coated Formvar-filmed copper grid (Tousimis, Rockville, MD) and allowed to attach. The sample was negatively stained with 1% pH 7.0 phosphotungstic acid solution (Fisher Scientific, Fair Lawn, NJ). The grid was examined by an electron microscope operated at 75 kV (Hitachi H7000, Tokyo). Digital images were taken by a charge-coupled device camera (Gatan, Pleasanton, CA).
Flow Cytometry.
Phage were incubated with E. coli cells for 4 min at 37°C. A total of 1 μl of streptavidin-coated QDs (1 μM) was added, and incubation continued for 5 min at room temperature. After centrifugation at 200 × g for 5 min, 1 μl of the sample was resuspended to 6 × 104 cells per milliliter. Samples were analyzed by flow cytometry using BD FACSDiVa LSR II (Becton Dickinson) monitoring the ratio of 407/600-nm excitation/emission fluorescence from phage–QD-bound cells. Events shown in histograms in Fig. 3 were gated on fluorescence. All were detected in log scale, and events were triggered on side scattering. A total of 30,000 events were collected for each analysis.
Fluorescence Microscopy.
Samples were prepared as described for flow cytometry, except that an additional centrifugation was preformed and 2 μl of the sample was placed on a microscope slide, covered with a coverslip, and visualized on an Olympus Vanox-T microscope by using an Oriel 500-W Hg arc lamp running at 200 W, a fluorescence filter set [a bandpass exciter (447 ± 15 nm), a dichroic mirror (505-nm cutoff), and a longpass emission filter (560-nm cutoff)], and a 1.25-numerical-aperture oil-immersion objective (DPlan 100×, Olympus). Images were captured by an intensified cooled charge-coupled device camera (I-PentaMAX, Roper Scientific).
Detection of E. coli in a Mix of Bacteria.
A total of 2 × 106 cells of P. aeruginosa, V. cholerae, Salmonella enterica serovar typhimurium, Y. pseudotuberculosis, and B. subtilis were mixed with different numbers of E. coli BL-21 cells (10 to 107) as estimated by OD600 and confirmed by viable count (data not shown). After ≈10–15 min at 37°C, lysates were cleaned by centrifugation and assayed by using the fluorescence microscope.
Acknowledgments
We thank Lori Goldner, Peter Yim, Idan Mandelbaum, and Jeffrey Krogmeier for helpful discussions and Barbara J. Taylor for instruction in flow cytometry. We are also grateful to Michael Yarmolinsky for critical reading of the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, the Center for Cancer Research, the intramural Advance Technology Program of the National Institute of Standards and Technology (NIST), and the National Institute of Allergy and Infectious Diseases Biodefense Intramural Research Fund.
Abbreviations
- QD
quantum dots
- BLP
biotin–ligase protein
- TEM
transmission electron microscope/microscopy.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.McKinstry M., Edgar R. In: Phages: Their Role in Bacterial Pathogenesis and Biotechnology. Matthew K. W., Friedman D. I., Adhya S. L., editors. Washington, DC: Am. Soc. Microbiol.; 2005. pp. 430–440. [Google Scholar]
- 2.McNerney R., Kambashi B. S., Kinkese J., Tembwe R., Godfrey-Faussett P. J. Clin. Microbiol. 2004;42:2115–2120. doi: 10.1128/JCM.42.5.2115-2120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodridge L., Chen J., Griffiths M. Int. J. Food Microbiol. 1999;47:43–50. doi: 10.1016/s0168-1605(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 4.Goodridge L., Chen J., Griffiths M. Appl. Environ. Microbiol. 1999;65:1397–1404. doi: 10.1128/aem.65.4.1397-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neufeld T., Schwartz-Mittelmann A., Biran D., Ron E.-Z., Rishpon J. Anal. Chem. 2003;75:580–585. doi: 10.1021/ac026083e. [DOI] [PubMed] [Google Scholar]
- 6.Banaiee N., Bobadilla-del-Valle M., Bardarov S., Jr, Riska P. F., Small P. M., Ponce-de-Leon A., Jacobs W.-R., Jr, Hatfull G. F., Sifuentes-Osornio J. J. Clin. Microbiol. 2001;39:3883–3888. doi: 10.1128/JCM.39.11.3883-3888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loessner M. J., Rees C. E., Steward G. S., Scherer S. Appl. Environ. Microbiol. 1996;62:1133–1140. doi: 10.1128/aem.62.4.1133-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasco R., Murphy M. J., Sanders M. F., Squirrell D. J. J. Appl. Microbiol. 1998;84:661–666. doi: 10.1046/j.1365-2672.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- 9.Sukhanova A., Devy J., Venteo L., Kaplan H., Artemyev M., Oleinikov V., Klinov D., Pluot M., Cohen J. H., Nabiev I. Anal. Biochem. 2004;324:60–67. doi: 10.1016/j.ab.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 10.Dubertret B., Skourides P., Norris J. D., Noireaux V., Brivanlou H. A., Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 11.Yao J., Larson D. R., Vishwasrao H. D., Zipfel W. R., Webb W. W. Proc. Natl. Acad. Sci. USA. 2005;102:14284–14289. doi: 10.1073/pnas.0506523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn M. A., Tabb J. S., Krauss T. D. Anal. Chem. 2005;77:4861–4869. doi: 10.1021/ac050641i. [DOI] [PubMed] [Google Scholar]
- 13.Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., Sundaresan G., Wu A. M., Gambhir S. S., Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokumasu F., Fairhurst R. M., Ostera G. R., Brittain N. J., Hwang J., Wellems T. E., Dvorak J. A. J. Cell Sci. 2005;118:1091–1098. doi: 10.1242/jcs.01662. [DOI] [PubMed] [Google Scholar]
- 15.Kwon K., Beckett D. Protein Sci. 2000;9:1530–1539. doi: 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman-Smith A., Cronan J. E., Jr Biomol. Eng. 1999;16:119–125. doi: 10.1016/s1050-3862(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 17.Cull M. G., Schatz P. J. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- 18.Palmer E. L., Martin M. L. Electron Microscopy in Viral Diagnosis. Boca Raton, FL: CRC; 1988. [Google Scholar]