Abstract
The structure of glucose dehydrogenase from the extreme halophile Haloferax mediterranei has been solved at 1.6-Å resolution under crystallization conditions which closely mimic the “in vivo” intracellular environment. The decoration of the enzyme’s surface with acidic residues is only partially neutralized by bound potassium counterions, which also appear to play a role in substrate binding. The surface shows the expected reduction in hydrophobic character, surprisingly not from changes associated with the loss of exposed hydrophobic residues but rather arising from a loss of lysines consistent with the genome wide-reduction of this residue in extreme halophiles. The structure reveals a highly ordered, multilayered solvation shell that can be seen to be organized into one dominant network covering much of the exposed surface accessible area to an extent not seen in almost any other protein structure solved. This finding is consistent with the requirement of the enzyme to form a protective shell in a dehydrating environment.
Keywords: Archaea, x-ray structure, water structure, hydrophobic surface, surface lysines
In highly saline environments, such as natural salt lakes, where salt concentrations can exceed 3 M, the main microorganisms present are extremely halophilic Archaea (1). These microbes are characterized by their ability to grow optimally in media containing 2.5–5.2 M NaCl, and the vast majority are unable to grow at salinities <2 M NaCl (2). To maintain a positive turgor pressure, these organisms either accumulate inorganic ions (K+ and Cl−) within the cell or synthesize compatible solutes to osmotically balance the external NaCl concentration. The proteins of the former group are thus specialized to function under high salt conditions (3). An understanding of the structural features that lead to the adaptation of proteins to such conditions could well be important in the rational modification of enzymes to function efficiently in other dehydrating conditions, such as those organic solvents commonly used in many industrial processes (4).
The growing sequence database of halophilic proteins is permitting more wide-ranging analyses of the structure–function relationships in this class of macromolecules and, although there are still relatively few structures of such molecules, it is now accepted that, for organisms that accumulate high intracellular concentrations of KCl, their molecular surfaces are decorated with an excess of acidic residues (5). However, to date, for those structures that have been determined, very few are at sufficient resolution to permit a detailed analysis of the protein–solvent interactions that are critical in understanding how the surface properties of the protein contribute to halophilicity. This paper reports the 1.6-Å structure determination of the glucose dehydrogenase (GlcDH) from Haloferax mediterranei (Hm), a member of the zinc-containing medium chain polyol dehydrogenase family, using crystals grown under conditions that closely mimic those experienced within the cell of the halophile (6–8). This work has allowed us to compare the structure with nonhalophilic homologues, locate bound counterions in the structure, and analyze the protein–solvent interactions to further our understanding of the molecular basis of halophilic adaptation.
Results
Secondary, Tertiary, and Quaternary Structure.
The structure of wild-type Hm GlcDH was solved by isomorphous replacement to 2.0-Å resolution and subsequently, to analyze the fine detail of the protein/solvent interactions, the structure of the D38C mutant was solved to 1.6 Å. Data collection and overall refinement statistics are summarized in Table 1.
Table 1.
Data collection, phasing, and refinement statistics
| Data collection and phasing | |||
| Data set | Native | Pb1 | D38C (KCl) |
| Space group | I222 | I222 | I222 |
| Unit cell parameters, Å | |||
| a | 61.8 | 61.8 | 60.5 |
| b | 110.2 | 110.9 | 109.3 |
| c | 152.3 | 151.7 | 151.9 |
| Temperature, K | 290 | 290 | 100 |
| Resolution, Å | 2.00 | 1.90 | 1.60 |
| Highest resolution shell, Å | 2.05–2.00 | 1.94–1.90 | 1.64–1.60 |
| Rmerge, %*† | 0.062 (0.309) | 0.079 (0.51) | 0.058 (0.592) |
| Completeness, %† | 92.0 (95.8) | 99.3 (99.9) | 99.1 (98.2) |
| No. unique reflections | 32,706 | 41,139 | 66,155 |
| I/σI† | 15.99 (3.05) | 17.06 (2.34) | 25.80 (2.70) |
| MFID, %‡ | 0.218 | ||
| Phasing power (acentric/ centric)§ | 1.56/1.23 | ||
| Rcullis, acentric/centric¶ | 0.75/0.66 | ||
| Number of sites | 1 | ||
| Refinement | |||
| No. of reflections used | 29,565 | 62,796 | |
| Rcryst, %/Rfree, % | 16.1/20.7 | 15.4/18.6 | |
| Ramachandran plot, % | 88.9/10.4/0.7 | 89.4/10.0/0.7 | |
| Rms deviation bond length, Å | 0.012 | 0.023 | |
| Rms deviation bond angle, ° | 1.58 | 1.55 | |
| Number in final model | |||
| Residues | 355 | 357 | |
| Protein atoms | 2749 | 2778 | |
| Catalytic Zn2+ | – | 1 | |
| Citrate ions | – | 1 | |
| NADP | 1 | 1 | |
| K+ | – | 5 | |
| Water molecules | 146 | 675 | |
| Mean B values, Å2 | |||
| Protein atoms | 34 | 25 | |
| Catalytic Zn2+ | – | 24 | |
| Citrate ion | – | 36 | |
| NADP | 32 | 24 | |
| K+ | – | 29 | |
| Waters | 44 | 41 |
*Rmerge = Σhkl|Ii − Im|/Σhkl1Im, where Im is the mean intensity of the reflection.
†Numbers in parentheses indicate values for the highest resolution shell.
‡Mean fractional isomorphous difference = Σ‖FPH| − |FP‖/Σ|FP|, where FPH and FP are the structure factor amplitudes for derivative and native crystals, respectively.
§Phasing power is defined as the rms value of heavy-atom structure factor amplitude divided by the rms value of lack-of-closure error.
¶Rcullis = 〈lack-of-closure〉/〈isomorphous difference〉.
Hm GlcDH exists as a dimer with each subunit consisting of a single polypeptide chain of 357 residues that folds into two domains separated by a deep cleft in which the active site is located. The secondary structure of the Hm GlcDH subunit (Fig. 3, which is published as supporting information on the PNAS web site) is very similar to that of the tetrameric Thermoplasma acidophilum (Ta) GlcDH with the dimer interface in the former representing that between subunits A and D of the latter (9).
Differences in Amino Acid Composition.
Hm GlcDH shows 39% and 41% sequence identity to the enzymes from the extreme thermoacidophilic archaebacterium, Sulfolobus solfataricus, and thermoacidophilic archaebacterium Ta, respectively. Previous comparison of the amino acid composition between halophilic proteins and their mesophilic counterparts has revealed an increase in acidic over basic residues (10). Comparison of the amino acid composition of the GlcDHs (Table 4, which is published as supporting information on the PNAS web site) shows similar trends, with 69% of the total number of charged residues being either aspartate or glutamate in Hm GlcDH compared with 54% and 50% of such residues in the Ta and Sulfolobus solfataricus enzymes, respectively, resulting in a net overall negative charge of −70 for the dimer of the halophilic protein. The increase in the number of acidic over basic residues in Hm GlcDH compared to the Ta enzyme is primarily due to an increase in glutamate combined with a corresponding decrease in lysine, consistent with the genome-wide reduction of lysine in the halophilic archaeon Halobacterium sp. NRC-1 (11) (www.ebi.ac.uk/proteome). The increase in glutamate is consistent with the suggestion that this residue has a superior water binding capacity over all other amino acids (12–14). However, it should be noted that such trends are not universal as, for example, in the glutamate dehydrogenase from Halobacterium salinarum, the increase in overall negative charge of this enzyme is related to a rise in aspartate together with a reduction in lysine (15). A further characteristic of the sequence of Hm GlcDH is an increase in alanine content (Table 4), which is also mirrored in the NRC-1 genome sequence.
Other reported sequence differences identified between halophilic proteins and their mesophilic counterparts are an increase in the “borderline” hydrophobic residues (serine and threonine) and a decrease in strongly hydrophobic residues (phenylalanine, isoleucine, valine and leucine) (10). However, in Hm GlcDH, there is no significant difference in the numbers of serine/threonine residues and, whereas the number of isoleucines is reduced by ≈50%, the numbers of phenylalanine, leucine, and valine all increase.
Analysis of the Solvent-Accessible Surface.
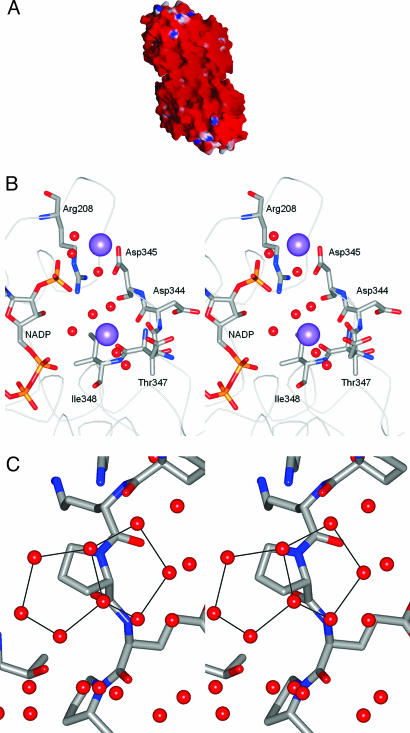
A comparison of the overall character of the solvent-accessible surface of the dimer of Hm GlcDH and tetramer of Ta GlcDH clearly shows an increase in acidic character and a decrease in nonpolar character for the halophilic enzyme (Table 2). Thus, the net charge density for the dimer of Hm GlcDH of −2.5 × 10−3 eÅ2, is far greater than the value of −0.6 × 10−3 eÅ2 for the tetrameric Ta GlcDH and comparable to the reported net charge densities of other halophilic proteins (15, 16) (Fig. 1A). This difference is not found in the surfaces that are buried at the dimer interface of the Hm enzyme (data not shown).
Table 2.
Characteristics of the overall solvent-accessible surface areas in the dimeric Hm GlcDH compared with the tetrameric Ta GlcDH
| Residue |
Hm |
Ta |
||
|---|---|---|---|---|
| Å2 | (%) | Å2 | % | |
| Non-polar | 12,050 | 44 | 26,500 | 51 |
| Polar | 6,800 | 25 | 12,700 | 24 |
| Negatively charged | 6,500 | 23 | 7,000 | 13 |
| Positively charged | 2,150 | 8 | 6,350 | 12 |
| Total area exposed | 27,500 | 52,550 | ||
Fig. 1.
The Hm GlcDH structure. (A) The molecular surface of the dimer of Hm GlcDH to show the electrostatic potential calculated at 0 M salt concentration, prepared by using the program grasp (17, 18). Red corresponds to a surface potential less than −10 kcal(mol·electron)−1; blue corresponds to a potential greater than +10 kcal(mol·electron)−1. (B) Stereo view of the location of two of the potassium ions (lilac spheres). Individual residues are shown in atom colors if they lie within 3.5 Å of each potassium ion. The remainder of the polypeptide chain is shown as an alpha carbon trace, whereas water molecules are depicted as red spheres. The bound cofactor, NADP, can be seen to lie close to a cation cluster involving two bound counterions. (C) A close up stereo view, using standard atom coloring for the protein, to show two fused pentagonal rings suspended above the hydrophobic chain of proline 21 and anchored by hydrogen-bonding interactions to the surrounding water molecules and polar protein atoms.
Analysis of the nature of the residues that contribute to the reduction in nonpolar surface (from 51% in Ta GlcDH to 44% in the Hm enzyme) clearly shows that this does not arise from any change in the distribution of strongly hydrophobic residues (Table 3). Rather, the fraction of the surface contributed by such residues in Hm GlcDH increases. Furthermore, despite the increase in alanine content in the halophilic enzyme, there is little difference in the average fractional exposure of this residue type to the solvent, and the overall contribution to the hydrophobic surface from alanine in both enzymes is small (Table 3). The origins of the decrease in nonpolar surface in Hm GlcDH, and one of the most striking differences between the surfaces of the halophilic and nonhalophilic enzymes, is a significant reduction in the percentage of hydrophobic surface accessible area due to lysine side chains from 9.4% in Ta GlcDH to 2.6% in the halophilic enzyme. Thus, overall, the 2-fold reduction in the proportion of lysines in the sequence leads to a 4-fold reduction in the exposed hydrophobic accessible surface area contributed by the associated alkyl component of the lysine side chain (Table 3). This change is the predominant factor in the overall difference in exposed hydrophobic surface and had previously been predicted from a comparative modeling study that compared the structure of another dehydrogenase, that of a mesophilic glutamate dehydrogenase, to a model of its homologous halophilic enzyme from Halobacterium salinarum (15). We have extended this analysis to compare the structure of a halophilic malate dehydrogenase (14) to that of a dogfish lactate dehydrogenase (19). Similarly, this finding reveals a significant overall reduction in the percentage of the surface accessible area due to lysine side chains (from 19.6% in lactate dehydrogenase to 3.6% in the halophilic malate dehydrogenase) and a corresponding reduction in the associated exposed alkyl groups for this residue type (12.8% vs. 2.0%). Interestingly, no significant trend can be detected for arginine.
Table 3.
Percentage of the solvent-accessible surface arising from different side chains and from the hydrophobic component of these side chains based on atom exposure
| Side chain | Total solvent-accessible area due to different side chains,* % |
Total solvent-accessible area due to the hydrophobic component of different side chains,* % |
||
|---|---|---|---|---|
| Hm | Ta | Hm | Ta | |
| Asp | 13.8 | 9.9 | 3.8 | 2.9 |
| Glu | 18.6 | 10.3 | 5.0 | 4.0 |
| Arg | 7.5 | 10.2 | 2.5 | 3.6 |
| Lys | 4.8 | 14.2 | 2.6 | 9.4 |
| His | 1.5 | 2.5 | 1.0 | 1.8 |
| Ser | 3.2 | 3.8 | 1.7 | 2.0 |
| Thr | 3.7 | 3.6 | 2.1 | 2.4 |
| Phe, Ile, Val, Leu | 9.7 | 9.3 | 9.7 | 9.3 |
| Ala | 2.0 | 1.0 | 2.0 | 1.0 |
*Percentages are expressed as a fraction of the total solvent-accessible surface of the dimer and tetramer of the Hm and Ta GlucDHs, respectively.
Identification of Bound Counterions.
Analysis of the electron density led to the identification of only five potassium ions, apparently leaving the enzyme surface with an overall negative charge (−30 per subunit). The existence of more mobile potassium ion binding sites cannot be ruled out, and such additional sites have been suggested in the structures of tRNA (20). Four of these bound cations have protein atoms amongst the ligands that form their coordination sphere (Fig. 1B). In the fifth, charge–charge interactions stabilize the cation-binding site, but all of the ligands to the potassium ions are water molecules in the hydration shell. Interestingly, despite the preponderance of carboxyl groups on the protein surface, of the 10 protein ligands to these five potassium ions, eight are carbonyl oxygens of the protein main chain, one is a threonine hydroxyl and only one involves a close interaction with a side chain carboxyl [D172 OD1/K+702 (2.74 Å)]. A citrate ion has also been identified on the surface of the protein where it forms interactions with two consecutive residues, K254 and H255, from helix α7 and the adenine ring of the NADP.
Analysis of the Solvent Structure and Its Interaction with the Protein Surface.
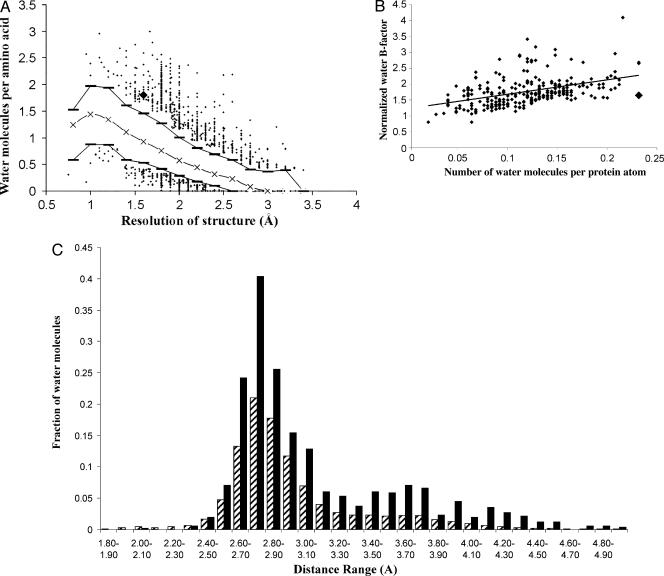
To compare the solvation shell of Hm GlcDH with that observed in other structures we have analyzed each of the 614 structures refined to resolutions between 1.65 and 1.55 Å in the July 2002 release of the PDB, considering all waters that appear in the coordinate sets with full occupancy. The number of waters per protein residue in Hm GlcDH is 1.9, much higher than the average (1.2) found in this subset of structures defined above (Fig. 2A). Only five structures show more waters per residue than Hm GlcDH (Protein Data Bank ID codes 3CAO, 1A1I, 1MBO, 2ILK, and 1A3J), all of which are significantly smaller than GlcDH and ranging in size from 21 to 160 residues. Therefore, restricting this analysis to the subset of 263 proteins refined at this resolution, and that are of equivalent or greater size, the halophilic GlcDH is the most heavily hydrated (Fig. 2A). In addition, a feature that emerges from this detailed analysis is that in the halophilic structure the temperature factors for the solvent molecules, normalized by the average B of the protein, is amongst the lowest for this subset of structures (Fig. 2B).
Fig. 2.
Comparison of the water structure around Hm GlcDH to that surrounding other proteins. (A) The dependence of the water to protein residue ratio (ordinate) against the resolution in Å (abscissa) for the structure determinations of all proteins solved between 3.5- and 0.5-Å resolution. Only the points in the lower 5% and above 95% are shown. The lower dashes, crosses, and upper dashes mark the 10, 50, and 90% boundaries for the data, respectively. The data point, corresponding to the Hm GlcDH, is shown by a large diamond. (B) A least squares line drawn through points that represent the B factors of the water structure normalized by the average B factor of the protein atoms (ordinate) plotted against the ratio of the number of water molecules to the number of protein atoms in a given structure (abscissa). The plot covers the 263 structures determined in the resolution range 1.55–1.65 Å for proteins that are of equivalent or greater size to Hm GlcDH. Each structure is represented on the plot by a diamond, except for the GlcDH structure, which is shown as a square. (C) A comparison of the distribution of the distance of the water molecules from the protein surface between the Hm GlcDH structure (black) and that of the average of the subset of 263 structures (hatched) as defined in B. The histogram shows the number of water molecules per residue that fall into specific distance bands from the protein surface. The abscissa is labeled with the midpoint of each range. Waters with partial occupancy were not included in the analysis.
Analysis of the distribution of the waters with respect to the protein surface shows that the number of waters per residue in the first solvation shell, and perhaps more importantly in the second shell, is higher than in any other protein in this class, revealing that the solvent structure in Hm GlcDH is also of higher complexity and hence the solvent shell is the most ordered (Fig. 2C).
An analysis of the spatial organization of the water molecules in the halophilic protein with respect to the protein side chains reveals a generally similar distribution to that observed in the collection of nonhalophilic proteins (data not shown). We have further analyzed the water structure of Hm GlcDH to identify any common geometrical arrangements of water clusters that might represent unusual patterns compared to those seen in proteins from nonhalophilic sources. Of the 675 water molecules in the Hm GlcDH structure, 9.6% are surrounded tetrahedrally by four nearest neighbor waters and 15.4% by water molecules or polar atoms of the protein. This fraction is similar to that for the other most heavily hydrated proteins in the subset. In previous work, pentagonal rings of water molecules have been identified on the surface of a number of proteins (21). Therefore, the solvent structure was analyzed for the presence of planar rings involving four, five, and six solvent molecules. This analysis has revealed that, in Hm GlcDH, the most common ring structure involves the formation of a pentagonal arrangement of water molecules, with 15 such rings being identified at 10 distinct sites on the protein surface. A number of these are constructed around a hydrophobic residue (Fig. 1C), as has been suggested elsewhere for the mode of interaction between water and hydrophobic groups (21). This fraction is higher than that observed elsewhere.
Of the 27,500 Å2 of the surface-accessible area in the dimer of Hm GlcDH, 10.4% is buried by crystal contacts to neighboring protein atoms. Of the remainder that is not covered by such contacts, 81.4% of the solvent accessible surface is covered by ordered solvent molecules. For the 675 water molecules that have been identified in the monomer, 544 form a network where the waters are either within 3.6 Å of each other or of a protein oxygen atom which is directly hydrogen bonded to other waters in the network. The remaining waters form much smaller clusters, the largest of which contains only 26 waters. Some of these clusters are necessarily separated by crystal contacts that, if removed, because they would be in an aqueous environment, might permit them to aggregate further. Thus, in the dimer as observed within the crystal, the two symmetry-related large networks are linked to each other to embrace a total of 1,126 water molecules. The presence of large water networks might have been expected given the high number of waters per residue for this protein structure (1.9). However, in the structure of pentaerythritol tetranitrate reductase (Protein Data Bank ID code 1H60), which has 1.83 waters per residue, the largest solvent network is much smaller accounting for 36% of the bound waters.
More recently, Nakasako (22) has also reported a large water network on the surface of a small killer toxin molecule from the halotolerant yeast Pichia farinosa, solved to 2.1 Å; this involves a total of 400 water molecules and 250 polar protein atoms and accounts for 90% of the total solvent. Rather than accumulate high intracellular concentrations of salt to counter the saline environment in which this species, and others like it, can be found these microorganisms rely on the accumulation of high concentrations of glycerol or other compatible solutes. Consequently, the proteins from such organisms do not appear to display the acidic sequence characteristics seen in haloarchaea. Nevertheless, the proteins from such species face the same challenges as those from the halophiles that accumulate salt in that they have to overcome the effects of the low activity coefficient of water. Therefore, it is interesting to observe similarities in the extent of the solvation shell.
The increased number of acidic residues in halophilic proteins gives rise to a higher frequency of surface carboxyl groups when compared to their mesophilic counterparts. Virtually all (>90%) of these acidic side chains are well ordered, with only five residues showing limited disorder. Thirty-three of the 63 acidic residues in the D38C mutant enzyme make a total of 40 interactions to the protein. These are made in the form of salt bridge partners (a total of 12), hydrogen bonds to backbone NH groups (a total of 16), interactions with threonine (four), serine (two), tyrosine (two), and histidine side chains (four), with further interactions to the NADP (one) and catalytic zinc ion (two). In addition, there is one carboxyl that forms a ligand to a bound potassium ion and a further two carboxyls forming long range interactions (≈5 Å) to other potassium counterions. Thirty of the carboxyl groups interact with the solvent alone.
In many protein structures, the side chains of surface lysines are frequently disordered. Unusually, however, of the 12 lysines per subunit in Hm GlcDH, 11 are ordered. Furthermore, as is clear from the comparison of the average solvent accessibility of lysines in the halophilic and thermophilic GlcDHs, the lysines in the former tend to be more buried and their corresponding hydrophobic alkyl tail, and therefore, necessarily, less exposed to solvent (Table 3). Given this finding, it might have been reasonable to speculate that these lysines represent a minimal, functionally conserved set. However, sequence comparisons show that only two are in common between the sequences of the Hm and Ta GlcDHs (Fig. 3). An analysis of the water structure around these lysine side chains shows, as might be expected, that the waters cluster around the exposed amino group of each side chain.
Halophilic Adaptations at the NADP-Binding Site.
Of the 30 residues that make contacts to the NADP, 10 are completely conserved between the halophilic and Ta enzymes including the glycine-rich motif, which characterizes the ADP-binding βαβ-fold of the cofactor binding site. In addition, packing interactions that do much to create the overall shape of the binding pocket are maintained. The 2′ phosphate of the adenine ribose is buried in a pocket formed by the side chains of conserved R207, R208, and by a nearby cation cluster, formed by two potassium ions. Both potassium ions lie in the second solvation shell surrounding the nucleotide cofactor, forming long-range interactions to the 2′ phosphate and the pyrophosphate moiety (K+701) and the 2′ phosphate alone (K+705) with the latter effectively providing one face of the 2′ phosphate-binding pocket. Important water-mediated interactions to the potassium ions of this cluster involve the carboxyl group of an aspartate (D345), which is the last of four consecutive such residues which form part of an acidic C-terminal tail to the protein. This latter tail is reminiscent of that seen in the halophilic 2Fe-2S ferredoxin, which has an additional hyperacidic domain close to the N terminus of the protein where 14 of the 33 residues are acidic and which has been suggested to represent a halophilic adaptation (16). In many dehydrogenases, the binding sites for the phosphates are mediated by positively charged side chains, such as lysine (23). We have already noted that this residue type is underrepresented in the genome of halophiles, and from our structural study appears to be responsible for a reduction of exposed hydrophobic surface. To our knowledge, this is the first reported use of a bound counterion to stabilize the phosphate groups of an NADP. The utilization of a bound counterion cluster in this way may well represent a novel adaptation to high salt that overcomes the penalty that halophilic enzymes appear to pay by the presence of such apparently disfavored residues. Whether this use of the counterions occurs in other halophilic proteins must await the emergence of more structural data on this class of enzymes.
Discussion
It has previously been suggested that the formation of a stabilizing hydration shell would be a feature of halophilic enzymes that have to survive in a highly saline environment (14). Similarly, a study of the dynamics of water molecules and ions near an aqueous micellar interface has suggested the existence of extended hydrogen bonding with the headgroups of the micellar assembly with the water molecules forming a bridge between the neighboring polar headgroups. The stability gained from the cooperative strengthening of the resultant hydrogen bonds has been proposed to lead to a slowdown of the rotation of these waters consistent with the formation of a stable hydration shell (24). Such bridges have also been observed in hydrated jet-cooled biomolecules (25). The extensive solvation shell seen in the structure of Hm GlcDH is consistent with this idea.
As observed in previous structures of halophilic proteins (16), the surface of Hm GlcDH is predominantly acidic in character. The question that therefore arises is to what extent is it a determinant of halophilicity? The overall geometric similarity in the arrangement of the water molecules around the halophilic GlcDH and other nonhalophilic proteins implies that there is no specific geometric effect on the first shell of water molecules created by the unusual surface character of the protein. Nevertheless, the size and order of the hydration shell in the halophilic enzyme is significantly greater. However, the analysis presented above shows that the differences in the characteristics of the molecular surface arise not only from an increase in negative surface charge but also from a number of distinct sequence changes that may be equally, if not more, important. The reduction in hydrophobic surface is very significant and results mainly from a loss of surface lysine. Unlike the nature of the acidic surface, this feature, although evident from the genome analysis of the halophile NRC-1, has been largely ignored to date. Indeed, this reduction more than outweighs an increase in hydrophobic surface arising from the subset of strongly hydrophobic side chains. Analysis of the structure clearly shows that the exposed hydrophobic groups act as contact surfaces for many water molecules. Unlike the side chain of lysine that is frequently disordered on the surface of proteins, the side chains of hydrophobic residues generally adopt fixed positions enabling networks of water molecules to be established. Consistent with this, our study also reveals that the lysines retained in the halophilic enzyme tend to be more buried than the average seen in nonhalophilic proteins, and consequently, are better ordered. That the guanidinyl side chain of arginine is more hydrophilic may explain the lack of any constraint on the use of this residue in the structure of halophilic enzymes, and therefore the absence of a genome-wide reduction of this residue.
A proper understanding of these preferences in sequence and structure is as yet unclear. However, it may well be that a dominant requirement for the formation of a stable hydration shell is an absence of very mobile side chains that might otherwise disturb the formation of a partially ordered array. Arising from this, the choice of an acidic rather than basic surface for a halophilic protein might be dictated by an absence of a small, positively charged side chain with these characteristics among the set of amino acids that make up proteins. Finally, despite the preponderance of negative charges on the surface of the protein, counterions from the solvent do not appear to play a dominant role in neutralization of such charge. However, the utilization of clusters of counterions in the recognition of negatively charged components of the NADP, as seen here, may represent a novel strategy for substrate recognition that can substitute for the lack of diversity in the subset of amino acids at nature’s disposal.
Methods
Protein Purification and Crystallization.
Wild-type Hm GlcDH was overexpressed in Escherichia coli, refolded, purified, and, in the presence of 1 mM NADP and absence of zinc, crystallized in two different forms by using either 2 M NaCl with 1.4–1.6 M sodium citrate or 2 M KCl and 1.4–1.6 M potassium citrate as the precipitant (8). A lead derivative of crystals of form II (space group I222; a = 61.8 Å, b = 110.2 Å, c = 152.3 Å, with a monomer in the asymmetric unit) grown by using sodium salts was prepared by soaking in a solution containing 10 mM lead chloride for ≈15 min.
Data Collection, Structure Solution, and Refinement.
Data were collected to 2-Å resolution at room temperature on a MAR345 detector with dual mirror focused CuKα X-rays, produced by a Rigaku RU200 generator, with a 0.3 mm × 3 mm filament, running at 50 kV and 100 mA. These data were processed and scaled by using the denzo/scalepack package (26) and subsequently handled by using ccp4 software (27). The position of a single lead site was determined automatically by using a genetic algorithm-based Patterson search program (unpublished data). The heavy atom position was refined and phases calculated by using mlphare (27, 28), and solvent flattening was applied by using dm (27, 29). An initial model was constructed by using the program warpntrace and extended by using the graphics package quanta98 (30). Iterative rounds of rebuilding and refinement were then conducted by using refmac5 (31).
Subsequently a D38C GlcDH mutant, constructed to analyze differences in the role of this residue in binding the catalytic zinc ion between Hm GlcDH (D38) and Ta GlcDH (C40) was used to produce crystals that were grown in the presence of zinc chloride and a buffer composed of potassium chloride and potassium citrate. Data on these crystals were collected to 1.5 Å and subsequently cut to 1.6 Å at 100 K without the need for additional cryoprotectant and using a Quantum4 charge-coupled device detector at the SRS Daresbury laboratory station 14.2. The structure of this mutant was solved by molecular replacement using the wild-type structure as the starting model. refmac5 (31) was used for the refinement with rebuilding carried out by using turbo-frodo (32). Water molecules were added with the program arp (33) and reorganized at the end of refinement by using watertidy (27). Those waters lying within 3.6 Å of the protein surface were defined as belonging to the first shell of solvent. Any waters making contact with this first shell and not with the protein were classed as being in the second solvation shell with further shells defined similarly. Counterions were identified by visual inspection on the basis that they were associated with strong peaks in the electron density map and that they were surrounded by ligands of appropriate charge and chemical characteristics in an octahedral arrangement.
The final model is composed of a single subunit made up of all 357 residues, an NADP molecule, one zinc ion, five potassium ions, one citrate ion, and 675 water molecules. Analysis of the stereochemical quality of the models was accomplished by using the program procheck (34). The solvent-accessible surface areas of Hm and Ta GlcDHs were calculated by using the algorithm of Lee and Richards (35). The definitions of Miller et al. (36) for nonpolar, polar, and charged constituents of proteins were used to calculate the chemical characteristics of the solvent accessible surface (sulfur atoms were classed as polar). Calculation of net charge densities followed the method of Frolow et al. (16), which assumes that histidine residues are uncharged. Ion pair interactions were identified by using the criteria of Barlow and Thornton (37). Side chains of different residue types were analyzed for their water coordination patterns against a subset of 614 high-resolution structures (refined to 1.55- to 1.65-Å resolution) deposited in the July 2002 release of the PDB.
Supplementary Material
Acknowledgments
We thank G. Taylor for supplying us with the coordinates for the Thermoplasma acidophilum GlcDH. We thank Dr. Roman Prokopenko for his valuable help in discussions over the programs that were developed to analyze water geometry. This work was supported by grants from The Wellcome Trust, Biotechnology and Biological Sciences Research Council (BBSRC), New Energy and Industrial Development Organisation, and Comision Interministerial de Ciencia y Tecnologia Grants PB98-0969 and BIO 2002-03179. Financial support to J.F. is gratefully acknowledged from Secretaria de Estado de Universidades e Investigación and Royal Society Grant RS1999-0061. M.F. was funded by the BBSRC. J.E. was supported by project BIO 2002-03179 Ministerio de Ciencia y Tecnología Formación de Personal Investigador fellowship from Generalitat Valenciana.
Abbreviations
- GlcDH
glucose dehydrogenase
- Hm
Haloferax mediterranei
- Ta
Thermoplasma acidophilum.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2B5V (wild-type GlcDH) and 2B5W (D38C mutant GlcDH)].
References
- 1.Galinski E. A., Trüper H. G. FEMS Microbiol. Rev. 1994;15:95–108. [Google Scholar]
- 2.Kamekura M. Extremophiles. 1998;2:289–295. doi: 10.1007/s007920050071. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg H., Mevarech M., Zaccai G. Adv. Protein Chem. 1992;43:1–62. doi: 10.1016/s0065-3233(08)60553-7. [DOI] [PubMed] [Google Scholar]
- 4.Flam F. Science. 1994;265:471–472. doi: 10.1126/science.8036489. [DOI] [PubMed] [Google Scholar]
- 5.Madern D., Ebel C., Zaccai G. Extremophiles. 2000;4:91–98. doi: 10.1007/s007920050142. [DOI] [PubMed] [Google Scholar]
- 6.Bonete M. J., Pire C., LLorca F. I., Camacho M. L. FEBS Lett. 1996;383:227–229. doi: 10.1016/0014-5793(96)00235-9. [DOI] [PubMed] [Google Scholar]
- 7.Pire C., Esclapez J., Ferrer J., Bonete M.-J. FEMS Microbiol. Lett. 2001;200:221–227. doi: 10.1111/j.1574-6968.2001.tb10719.x. [DOI] [PubMed] [Google Scholar]
- 8.Ferrer J., Fisher M., Burke J., Sedelnikova S. E., Baker P. J., Gilmour D. J., Bonete M.-J., Pire C., Esclapez J., Rice D. W. Acta Crystallogr. D. 2001;57:1887–1889. doi: 10.1107/s0907444901015189. [DOI] [PubMed] [Google Scholar]
- 9.John J., Crennell S. J., Hough D. W., Danson M. J., Taylor G. Structure (London) 1994;2:385–393. doi: 10.1016/s0969-2126(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 10.Lanyi J. K. Bacteriol. Rev. 1974;38:272–290. doi: 10.1128/br.38.3.272-290.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy S. P., Ng W. V., Salzberg S. L., Hood L., DasSarma S. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuntz I. D. J. Am. Chem. Soc. 1971;93:516–518. doi: 10.1021/ja00731a037. [DOI] [PubMed] [Google Scholar]
- 13.Saenger W. Annu. Rev. Biophys. Biophys. Chem. 1987;16:93–114. doi: 10.1146/annurev.bb.16.060187.000521. [DOI] [PubMed] [Google Scholar]
- 14.Dym O., Mevarech M., Sussman J. L. Science. 1995;267:1344–1346. doi: 10.1126/science.267.5202.1344. [DOI] [PubMed] [Google Scholar]
- 15.Britton K. L., Stillman T. J., Yip K. S. P., Forterre P., Engel P. C., Rice D. W. J. Biol. Chem. 1998;273:9023–9030. doi: 10.1074/jbc.273.15.9023. [DOI] [PubMed] [Google Scholar]
- 16.Frolow F., Harel M., Sussman J. L., Mevarech M., Shoham M. Nat. Struct. Biol. 1996;3:452–458. doi: 10.1038/nsb0596-452. [DOI] [PubMed] [Google Scholar]
- 17.Nicholls A., Sharp K. A., Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 18.Nicholls A., Honig B. J. Comput. Chem. 1991;12:435–445. [Google Scholar]
- 19.Abad-Zapatero C., Griffith J. P., Sussman J. L., Rossmann M. G. J. Mol. Biol. 1987;198:445–467. doi: 10.1016/0022-2836(87)90293-2. [DOI] [PubMed] [Google Scholar]
- 20.Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S.-H. Nucleic Acids Res. 1977;4:2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teeter M. M. Proc. Natl. Acad. Sci. USA. 1984;81:6014–6018. doi: 10.1073/pnas.81.19.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakasako M. Philos. Trans. R. Soc. London B. 2004;359:1191–1206. doi: 10.1098/rstb.2004.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker J. E., Saraste M., Runswick M. J., Gay N. J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balasubramanian S., Pal S., Bagchi B. Curr. Sci. 2002;82:845–854. [Google Scholar]
- 25.Zwier T. S. J. Phys. Chem. A. 2001;105:8827–8839. [Google Scholar]
- 26.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project No. 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 28.Otwinowski Z. In: Wolf W., Evans P. R., Leslie A. G. W., editors. Proceedings of the CCP4 Study Weekend; Warrington, U.K.: SERC Daresbury Laboratory; 1991. pp. 80–86. [Google Scholar]
- 29.Cowtan K. Joint CCP4 and ESF-EACBM Newslett. Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 30.Oldfield T. J. Acta Crystallogr. D. 2001;57:82–94. doi: 10.1107/s0907444900014098. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov G. N., Vagin A., A. Dodson E. J. Acta Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Roussel A., Cambillau C. turbo-frodo in Geometry Partners Directory. Vol. 88. Mountain View, CA: Silicon Graphics; 1991. [Google Scholar]
- 33.Lamzin V. S., Wilson K. S. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Lee B., Richards F. M. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 36.Miller S., Janin J., Lesk A. M., Chothia C. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 37.Barlow D. J., Thornton J. M. J. Mol. Biol. 1983;168:857–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.