Abstract
Arrestins regulate signaling and trafficking of G protein-coupled receptors by virtue of their preferential binding to the phosphorylated active form of the receptor. To identify sites in arrestin involved in receptor interaction, a nitroxide-containing side chain was introduced at each of 28 different positions in visual arrestin, and the dynamics of the side chain was used to monitor arrestin interaction with phosphorylated forms of its cognate receptor, rhodopsin. At physiological concentrations, visual arrestin associates with both inactive dark phosphorylated rhodopsin (P-Rh) and light-activated phosphorylated rhodopsin (P-Rh*). Residues distributed over the concave surfaces of the two arrestin domains are involved in weak interactions with both states of phosphorhodopsin, and the flexible C-terminal sequence (C-tail) of arrestin becomes dynamically disordered in both complexes. A large-scale movement of the C-tail is demonstrated by direct distance measurements using a doubly labeled arrestin with one nitroxide in the C-tail and the other in the N-domain. Despite some overlap, the molecular “footprint” of arrestin bound to P-Rh and P-Rh* is different, showing the structure of the complexes to be unique. Strong immobilizing interactions with residues in a highly flexible loop between β-strands V and VI are only observed in complex with the activated state. This result identifies this loop as a key recognition site in the arrestin-P-Rh* complex and supports the view that flexible sequences are key elements in protein–protein interactions.
Keywords: site-directed spin labeling, protein-coupled receptors, electron paramagnetic resonance
G protein-coupled receptors (GPCRs) are the largest known family of signaling proteins. Arrestins play a key role in the regulation of the signaling and trafficking of the majority of GPCRs (1, 2). The visual amplification cascade has long served as a prototypical GPCR-driven signaling system. Visual arrestin demonstrates an exquisite selectivity for light-activated phosphorylated rhodopsin (P-Rh*), its cognate receptor (3). However, arrestin binding is not an “all-or-nothing” event: it binds with low affinity to light-activated unphosphorylated rhodopsin and inactive dark phosphorylated rhodopsin (P-Rh) (3, 4). These observations and the evidence that arrestin undergoes a conformational rearrangement in the process of receptor binding (5) led to the model of sequential multisite interaction that ensures arrestin selectivity for P-Rh* (6). The model posits that arrestin has two primary binding sites that serve as phosphorylation and activation “sensors” that interact with receptor-attached phosphates and the active receptor conformation, respectively. Only P-Rh* can simultaneously engage both sensors, triggering arrestin transition into its high-affinity receptor-binding state. Thus, arrestin functions as a molecular “coincidence detector,” swinging into action only when the receptor binds both sensors.
This model is based on ample indirect evidence (4, 5, 7–17), but direct structural data on the nature of the interaction with the receptor is absent. In this study, site-directed spin labeling (SDSL) was used to investigate the interaction of arrestin with P-Rh and P-Rh*. For this purpose, 28 single cysteine (Cys) substitution mutants of arrestin were prepared on a Cys-less background and spin-labeled with a methanethiosulfonate nitroxide reagent to produce the side chain designated as R1 (Fig. 1C Inset) (18). The electron paramagnetic resonance (EPR) spectra of the spin-labeled proteins encode information on the dynamics of R1 and thus provide a means of mapping protein–protein interaction surfaces through changes in R1 motion (19, 20). The SDSL data presented here support the multisite-binding model, identify the parts of the arrestin molecule engaged by different states of rhodopsin, and provide direct evidence for binding-induced conformational rearrangements in arrestin itself.
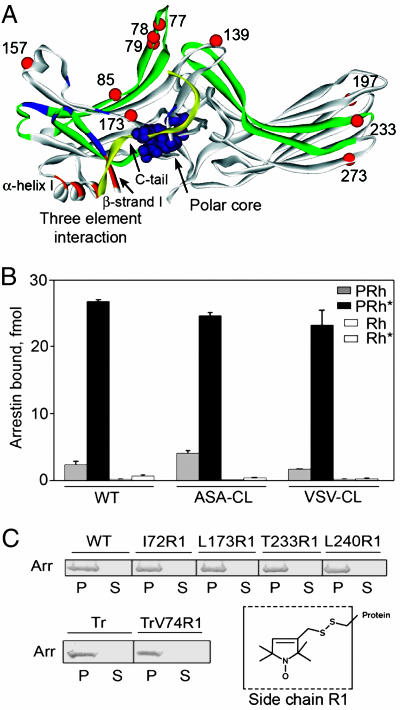
Fig. 1.
Functional characterization of Cys-less base mutants and spin-labeled arrestins. (A) Visual arrestin crystal structure showing the location of Cys substitutions that significantly perturb arrestin binding to P-Rh* as red spheres at the Cα atoms. For reference, sequences containing phosphate-binding residues [Lys-14, Lys-15, Arg-18, Lys-20, Lys-55, Arg-56, Lys 166, Arg-171, and Lys-300 (9, 10, 12, 13, 23, 29, 41)] are shown in blue; sequences implicated in receptor binding (9, 30–32) are shown in green. Residues of the polar core are shown space-filled in purple. Sequences of the three-element interaction (13) are shown in orange and yellow (C-tail). (B) Direct binding of WT arrestin and two Cys-less base mutants ASA-CL and VSV-CL to the four forms of rhodopsin was performed as described in ref. 43. (C) The functionality of all spin-labeled arrestins used in this study was tested in a pull-down assay. (Upper and Left Lower) Purified spin-labeled (R1) arrestin (25 μM) was incubated with 50 μM P-Rh* for 5 min at 37°C. Arrestin bound to rhodopsin-containing membranes was pelleted by centrifugation for 10 min through a 30-μl 0.2 M sucrose cushion. One-third of each pellet (P) and supernatant (S) was subjected to 10% SDS/PAGE. The gels were stained with Coomassie blue and scanned. Data for five representative mutants are shown [Tr, truncated arrestin (1–378)]. All spin-labeled mutants demonstrated similar (>98%) binding to P-Rh*. (Right Lower) Structure of the R1 side chain.
Results
Functional Characterization of Cys-Less Base Mutants and Spin-Labeled Derivatives of Arrestin.
The regions of arrestin implicated in receptor binding identified so far map to the concave sides of both arrestin domains (reviewed in ref. 6). These data identify a fairly extensive surface containing the side chains of >70 residues (Fig. 1A). Therefore, attention was focused on this surface and the regulatory elements that hold arrestin in its basal conformation: the polar core [a network of interacting solvent-excluded charges that serves as a phosphate sensor (12)] and the three-element interaction between β-strand I, α-helix I, and the C-tail (13) (Fig. 1A). Application of SDSL requires the elimination of reactive native Cys residues from the protein, the introduction of a Cys residue into the position of interest, and chemical modification of the unique Cys with a spin-labeling reagent (reviewed in refs. 21 and 22). Visual arrestin has three native Cys residues. In the crystal structure, Cys-63 and -143 are in a fairly hydrophobic environment, whereas Cys-128 is not (23). Therefore, a serine (Ser) was placed in position 128, whereas the other two Cys residues were substituted with either alanines (Ala) or valines (Val), producing two Cys-less base mutants, ASA-CL and VSV-CL, respectively. Both Cys-less arrestins were fully functional, demonstrating essentially wild-type (WT) binding levels and selectivity profiles (Fig. 1B).
A systematic “scan” of the putative receptor-binding surface was carried out by introducing individual Cys substitutions into one of the Cys-less backgrounds at 32 different sites. The functionality of the mutants expressed in cell-free translation was tested in a direct binding assay. Ten mutations (positions identified with red spheres in Fig. 1A) significantly affected arrestin binding to P-Rh*: F85C and F197C enhance binding by 13–15%; S78C, V139C, T157C, L173C, T233C, and S273C reduce binding by 14–23%; and L77C and F79C reduce binding by 58% and 42%, respectively, compared with WT (see Fig. 4, which is published as supporting information on the PNAS web site). These data implicate previously unappreciated elements of arrestin in receptor binding and identify the loop between β-strands V and VI containing Leu-77 and Phe-79 (Fig. 1A) as a major player.
Twenty-eight different single Cys mutants and one double Cys mutant were expressed in Escherichia coli, purified, and spin-labeled, and their ability to bind P-Rh* was tested at the concentrations used for EPR studies. All spin-labeled mutants were fully functional as evidenced by virtually quantitative binding to P-Rh*, similar to WT arrestin (Fig. 1C). Therefore, these arrestin mutants are suitable for directly studying arrestin–rhodopsin interactions.
Mobility of the R1 Side Chain in Free Arrestin.
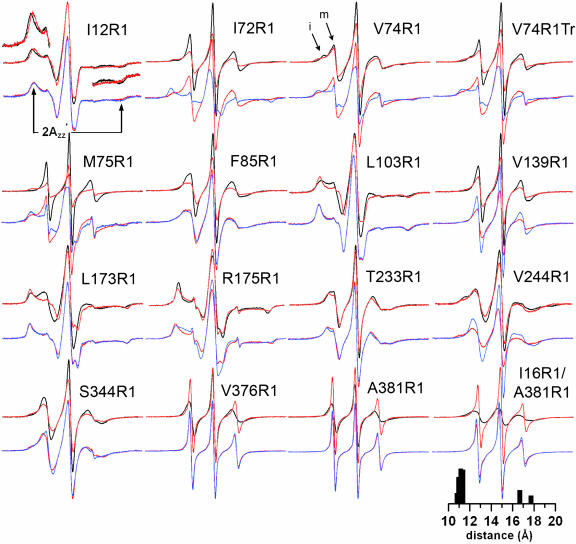
Fig. 2 shows the EPR spectra for R1 at sites in arrestin where significant changes in R1 mobility were observed upon interaction with phosphorylated forms of rhodopsin; spectra for other sites where little (16, 111, 157, 160, 162, 197) or no changes (60, 89, 240, 267, 272, 272, 348) were observed are provided in Fig. 5, which is published as supporting information on the PNAS web site. In each case, the spectra of the spin-labeled arrestins alone (black traces) are consistent with R1 mobility/structure correlations previously established for α-helical (18) and β-sheet proteins (24) and provide information on protein dynamics in solution (25). For example, the side chains of 12R1, 103R1, and 175R1 are buried in the protein interior (Fig. 3A), and the broad EPR spectra reflect the immobilization of R1. Of particular interest are the unusually sharp features of the single-component spectra for solvent-exposed sites 72R1, 75R1, 139R1, 376R1, and 381R1, indicating high R1 mobility and a flexible protein structure in these regions. Residues 72R1 and 75R1 are in the loop connecting β-strands V and VI (the “finger” loop); residue 139R1 is in the adjacent loop connecting strands VIII and IX; and 376R1 and 381R1 are in the C-tail (Fig. 3A). All of these residues are in sequences that have a high B factor in the crystal structure (11). The spectrum of 74R1, also in the finger loop, has two components (arrows, Fig. 2) corresponding to states of high and low mobility, the latter of which may reflect tertiary interactions of the side chain with the adjacent loop toward which it points. For Tr74R1 [74R1 in truncated arrestin (1–378)], there is an increase in the mobility of R1 relative to full-length arrestin, evidenced by a decrease in the more immobilized population.
Fig. 2.
EPR spectra of spin-labeled arrestin in solution and bound to P-Rh and P-Rh*. For each spin-labeled arrestin, normalized spectra in the absence (black) or presence (red) of P-Rh are compared in the top row, and spectra in the presence of P-Rh (red) and P-Rh* (blue) are compared in the bottom row. Portions of the overlaid spectra for I12R1are magnified to better illustrate the spectral changes and the location of the hyperfine splitting (2A′zz; arrows). Arrows for V74R1 indicate immobile (i) and mobile (m) components. The interspin distance distribution derived from the P-Rh spectrum of I16R1/A381R1 is shown (see text).
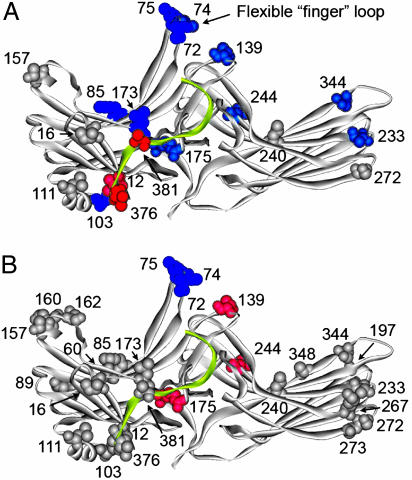
Fig. 3.
Summary of the changes in spin-label mobility induced by arrestin interactions with phosphorhodopsin. The magnitude of the detected changes in spin-label mobility (Figs. 2 and 5) is color-coded on the visual arrestin crystal structure as follows: gray, no change; pink/red, small and large increases in mobility, respectively; light blue/dark blue, small and large decreases in mobility, respectively. (A) Changes upon interaction with P-Rh. (B) Additional changes induced by light activation of phosphorylated rhodopsin relative to those induced by binding to dark P-Rh. This image includes additional sites found not to change upon P-Rh* interaction (Fig. 5B). The C-tail is shown in yellow.
Interaction of Spin-Labeled Arrestins with Phosphorhodopsin.
The interaction of arrestin with P-Rh or P-Rh* was investigated by using near-physiological concentrations of both proteins: 25 μM arrestin and 50 μM rhodopsin. Arrestin binding to phosphorylated forms of rhodopsin produced distinct changes in the mobility of R1 that are discussed below. Both arrestin and its complexes with rhodopsin are of sufficiently high molecular weight that overall rotational diffusion does not contribute significantly to R1 motion (except for the most immobilized states), and observed changes do not result from a decrease in overall rotational diffusion because of complex formation. The interaction is not a nonspecific absorption to the membrane, because no changes in the EPR spectra were observed with liposomes (50% phosphatidylcholine, 40% phosphatidylethanolamine, and 10% phosphatidylserine) or nonphosphorylated rhodopsin in disk membranes. Thus, changes in the mobility of R1 reflect either contact with phosphorylated rhodopsin or conformational changes in the arrestin molecule itself.
For the present purpose, simple spectral measures served to reveal mobility changes in R1. For example, changes in spectral amplitude for the normalized spectra reflect changes in linewidth and hence in R1 mobility on the nanosecond time scale; decreases in intensity reflect decreases in mobility and vice versa. In cases of low R1 mobility, where outer hyperfine extrema are resolved in the spectra (12R1, 103R1, 175R1), an increase or decrease in the splitting 2A′zz (Fig. 2) reflects a decrease or increase in mobility, respectively. Based on these criteria, a qualitative assessment of mobility changes can be made.
Binding to P-Rh.
A distinctive pattern of spectral changes is observed upon arrestin binding to P-Rh (Fig. 2, red traces). Fig. 3A shows the location of these sites in the structure, color-coded according to the magnitude and direction of the mobility changes. In the N-domain, residues 72R1, 74R1, Tr74R1, and 75R1 in the finger loop, 85R1 and 173R1 on the solvent-exposed surface of a β-strand, and residue 139R1 in the loop adjacent to the finger loop all experience reductions in mobility. On the concave surface of the C-domain, 344R1 shows a reduction in mobility, with smaller but significant reductions for 233R1 and 244R1. For 233R1, the change reflects an increase in order of R1 motion (25).
Important changes also occur in elements that hold arrestin in the basal state. For example, 175R1, buried in the polar core, becomes more immobilized upon interaction with P-Rh (Δ2A′zz > 0). Although a neutral R1 replaces a charged Arg-175 at this site, the protein behaves normally with respect to phosphorhodopsin binding, and the spectral change shows that the structure of the polar core is modulated by the interaction. Changes also are observed in the three-element interaction between β-strand I, α-helix I, and the C-tail (13, 23). On β-strand I, the mobility of 12R1 increases (2A′zz < 0), whereas that of 103R1 in α-helix I decreases (2A′zz > 0). The spectra of the spin labels at positions 376 and 381 in the C-tail show relatively high mobility in free arrestin and become essentially dynamically disordered upon P-Rh binding. These data strongly suggest that structural constraints on the arrestin C-tail are removed upon binding to P-Rh.
To test this hypothesis directly, arrestin was doubly labeled at position 16 in β-strand I and 381 in the C-tail. The pronounced broadening of the 16R1/381R1 spectrum in solution indicates strong spin–spin interaction between the nitroxides (black trace, Fig. 2), where the two spins are expected to be ≈12 Å apart based on the crystal structure (23). The interspin distance distribution reveals two populations centered around 11 and 17 Å (Fig. 2). The two populations could arise from multiple rotamers of R1 or distinct conformational substates of the protein. The strong magnetic interaction between these two R1 residues in free arrestin disappears in the presence of P-Rh as indicated by the loss of spectral broadening (interspin distance > 20 Å). In the bound state the spectrum of the double mutant is approximated well by the sum of the two P-Rh-bound single-mutant spectra (data not shown). Thus, the residues move away from each other upon binding to P-Rh. Collectively, these data demonstrate that binding to receptor-attached phosphates in the dark releases the C-tail of arrestin from interactions with the body of the protein.
Additional R1 mobility changes induced by light activation.
Light activation of P-Rh to produce P-Rh* results in additional changes (Fig. 2, blue traces, and Fig. 3B). The most dramatic are the further decreases in mobility of residues in the finger loop (72, 74, Tr74, and 75), leading to essentially complete immobilization. On the same face of the molecule, only residues 139R1 and 244R1 show additional changes (Fig. 3B). Interestingly, the mobility of 139R1, which was partially immobilized by P-Rh binding, reverts to that resembling the original state in free arrestin. For 244R1, light activation of P-Rh removes constraints on the motion of R1, leading to a single-component spectrum reflecting fast anisotropic motion.
In the polar core region, 175R1 shows a small increase in mobility, suggesting further modulation in the structure by P-Rh*. In the vicinity of the three-element interaction, the mobility of the spin label at sites 12, 103, 376, and 381 does not change further. Likewise, the spectra of the 16/381 doubly labeled arrestin bound to P-Rh and P-Rh* are virtually identical, indicating that the C-tail released by P-Rh interaction stays detached in complex with P-Rh*.
Thus, R1 at 15 of the 27 sites investigated shows significant changes in mobility upon interaction with the phosphorylated forms of the receptor.
Discussion
The rhodopsin-binding surface of visual arrestin has been independently mapped by several laboratories using a variety of methods: truncation mutagenesis (3, 4), differential chemical modification and H/D exchange (26), site-directed mutagenesis (8, 12, 13, 27–29), construction of chimeric arrestins (9, 30), peptide competition (31, 32), and epitope insertion (33). The elements implicated by these studies invariably map to the concave surfaces of the two arrestin domains (12, 23), leaving little doubt as to which side of the arrestin molecule faces the receptor in the complex. The size of the putative receptor-binding surface indicates that either a small number of interacting residues is scattered throughout the whole span of the molecule or a large number of arrestin residues directly participate in receptor binding. Systematic probing of residues in all arrestin elements localized on this surface is necessary to discriminate between these two possibilities. Together, the systematic scan of 32 positions by Cys mutagenesis and of 20 positions by spin labeling on this surface indicate that >20 arrestin residues are affected by receptor binding (Figs. 1A and 3). These results imply that an equally extensive receptor surface involving multiple loops and the C terminus must be in proximity to arrestin in the complex, which agrees with the results of the studies of arrestin-binding elements of rhodopsin (34–36).
The fact that arrestin preferentially binds light-activated phosphorylated rhodopsin was established >20 years ago (37). The sequential multisite-binding model predicts that arrestin binds with lower affinity to nonpreferred forms of rhodopsin, including P-Rh (4); this hypothesis was confirmed by using a very sensitive binding assay employing radiolabeled arrestin (3). For technical reasons, only low nanomolar concentrations of arrestin can be used in this assay. In SDSL experiments, arrestin is present in high micromolar concentrations [similar to that in rod photoreceptors (38)], and even low-affinity interactions are expected to result in significant concentrations of complexes with nonpreferred forms of rhodopsin. Indeed, the EPR spectral changes demonstrate that arrestin binds P-Rh as well as P-Rh* under these conditions (Fig. 2).
The binding to both P-Rh and P-Rh* similarly modulates the mobility of R1 residues 85, 173, 233, and 344 located on the concave surfaces on β-strands in the two domains (Fig. 3). The complete lack of mobility changes in positions 60, 89, 267, 272, 273, and 348 in the presence of P-Rh* (Fig. 5) as well as position-dependent and rhodopsin state-dependent decreases or increases in mobility (Fig. 3) show that the changes are very specific. The fact that many sites distributed throughout the molecule show no change upon complex formation suggests that the global structure of the individual arrestin domains does not change significantly. These results suggest that the mobility changes on the concave surface are due to proximity of the receptor in the complex rather than structural changes in arrestin. Interestingly, none of the β-strand residues (except 173R1) are strongly immobilized in the complex, suggesting that they are not located at compact interaction sites involving multiple contacts between arrestin and the receptor. The reduction in R1 mobility at these sites could be due to dampening of backbone motions by nearby contacts with the receptor and/or very weak interactions with mobile receptor side chains. The fact that three of four of the corresponding Cys mutations (F85C, L173C, and T233C) perturb binding indicates that they contribute to receptor interaction, consistent with the latter possibility. In the case of 173R1, the strong immobilization of this solvent-accessible site could arise from direct close contact with the receptor, and the region around 173 may represent an important anchor point in both P-Rh and P-Rh* complexes. Notably, two adjacent residues in this β-strand, Arg-171 and -175, are implicated in direct binding of receptor-attached phosphates (8, 12).
Even though overlapping sets of residues participate in P-Rh and P-Rh* binding, light activation of phosphorhodopsin induces further changes at some positions (Figs. 2 and 3B). For example, 139R1, immobilized by P-Rh, reverts to its original high mobility upon light activation, and 244R1 shows a dramatic increase in mobility to a level exceeding that in free arrestin (Fig. 2). The difference in the “footprint” of active and inactive phosphorhodopsin suggests that the orientation of bound arrestin relative to phosphorhodopsin and/or the conformation of P-Rh- and P-Rh*-bound arrestin is different. Importantly, this finding is true even in the case of truncated arrestin (1–378) (Tr74R1; Fig. 2) that shows dramatically enhanced binding to dark P-Rh (10).
The data (Figs. 1A and 2) clearly identify the finger loop between β-strands V and VI that protrudes far above the central “crest” on the receptor-binding surface as a major player in the arrestin–receptor interaction. This loop adopts different conformations in different crystal forms of both visual arrestin (23) and arrestin2 (11), suggesting structural plasticity. The high mobility of R1 at positions 72, 74, and 75 within this loop demonstrates its dynamic flexibility in solution. Mobility reductions occur for 72R1, 74R1, and 75R1 in the P-Rh complex, but only upon activation to P-Rh* does each residue become strongly immobilized, suggesting that at least the tip of the finger loop is now buried at a rigid arrestin–rhodopsin interface. The importance of this loop in receptor binding is underscored by the fact that Cys substitutions of three uncharged residues in this region (Leu-77, Ser-78, and Phe-79) are the most detrimental for arrestin binding to P-Rh* (Figs. 1A and 4). Sommer et al. (39) used both a fluorescent and spin-label at site 72 in visual arrestin to explore binding to phosphorhodopsin and also concluded that this site is buried at an interface in the arrestin–P-Rh* complex. However, neither the fluorescent nor spin-label detected binding to P-Rh. In addition, the EPR spectra of 72R1 in arrestin and in the arrestin–P-Rh* complex are different in detail from those reported here. The differences may arise from the different arrestin backgrounds used in this study (untagged Cys-less arrestin) and that of Sommer et al. (39) (His-tagged WT).
The higher mobility of 74R1 in truncated arrestin (1–378) compared with the full-length protein (Fig. 2) supports the hypothesis that truncation, which enhances binding to the nonpreferred forms of the receptor (4, 8–10, 12), does so by “loosening up” the conformational constraints of the basal state of arrestin (6). Even though truncated arrestin binding to P-Rh is at least half that for P-Rh* (10) (compared with <10% for full-length; Fig. 1B), the spectra of Tr74R1 indicate that it is not fully engaged with the receptor until rhodopsin is both light-activated and phosphorylated. Accordingly, the spectra of 74R1 in both backgrounds in complex with P-Rh* are virtually identical. These data suggest that the rhodopsin elements that interact with this arrestin finger loop are exposed only in its light-activated form and that arrestin can be properly oriented to engage these partners only when it is also “anchored” via receptor-attached phosphates.
The release of the arrestin C-tail upon binding to P-Rh* was previously inferred from its increased sensitivity to limited proteolysis (14, 15). The striking increase in mobility of R1 at positions 376 and 381 indicates essentially complete removal of structural constraints on the C-tail upon binding to either P-Rh or P-Rh*. This conclusion is further supported by the increase in distance between 16R1 (in β-strand I) and 381R1 (in the C-tail) upon binding to both forms of rhodopsin (Fig. 2). These data directly demonstrate that the arrestin C-tail is released upon receptor binding and that interaction with receptor-attached phosphates is necessary and sufficient for this rearrangement. Residues 12R1 and 103R1 are located in β-strand I and α-helix I, respectively, and report changes in the conformation of the other two elements of the three-element interaction. Conceivably, the changes in these two positions arise from interactions of the phosphates in the flexible C terminus of rhodopsin (40) with Lys-14 and -15 in β-strand I. Thus, receptor-attached phosphates destabilize the three-element interaction that anchors the C-tail to the body of the arrestin molecule (13).
Several putative phosphate-binding residues were identified in visual arrestin by mutagenesis, including Lys-20, Lys-55, Arg-56, Lys-166, Lys-300 (29), Arg-171, the “phosphate sensor” Arg-175 (8), Lys 14, Lys-15 (13), and Arg-18 (41), all of which are localized in the N-domain in regions where the mobility of R1 changes in response to P-Rh and P-Rh* (Figs. 1A and 3). In the basal state, R175 is buried in the polar core, as reflected by the highly immobilized state of 175R1 in free arrestin. The mobility of 175R1 successively changes upon interaction with P-Rh and P-Rh*, showing that while the receptor-attached phosphates of dark P-Rh modulate the structure of the polar core, further changes occur upon light activation of rhodopsin.
Conclusions
The data demonstrate that arrestin binding to phosphorylated inactive rhodopsin removes intramolecular constraints on the flexible C-tail of arrestin. In this complex weak intermolecular contacts are made between the receptor and sites on the concave surfaces of arrestin and the highly flexible loop between β strands V and VI. The arrestin N-domain and this finger loop are apparently engaged in stronger interactions with the receptor than the C-domain. Upon receptor activation, the arrestin C-tail remains detached from the body of the molecule, and the strength of the interaction with the finger loop is dramatically increased, leading to its immobilization. This result leaves little doubt that this flexible loop is a primary site of recognition between arrestin and the phosphorylated activated receptor. Weak interactions at other sites also are modulated, indicating that the footprint on arrestin of the inactive and active phosphoreceptor is different.
Materials and Methods
In Vitro Transcription and Translation, Evaluation of Mutant Stability, and Receptor-Binding Assay.
pGEM2-based plasmids with arrestin coding sequence equipped with the “idealized” 5′-UTR (42) under the control of the SP6 promoter were transcribed and translated in vitro in the presence of [3H]leucine and [14C]leucine as described in ref. 43. The translation of every mutant used in this study produced a single labeled protein band with the expected mobility on SDS/PAGE. The relative stability of all mutants (assessed as described in ref. 10) was >80%. Direct binding of arrestin to rhodopsin and the separation of rhodopsin-bound and free arrestin on Sepharose 2B-CL columns were carried out as described in ref. 43.
Arrestin Expression and Purification.
Arrestin expression in E. coli and purification was performed as described in ref. 43. To achieve optimal expression, I12C, I16C, Y58C, S60C, I72C, V74C, M75C, D82C, F85C, Q87C, Q89C, V94C, L103C, L111C, F152C, E160C, D162C, F197C, T233C, L240C, V244C, E266C, K267C, S272C, S273C, L339C, S344C, and A348C were introduced into the ASA-CL (C63A, C128S, C143A) background, whereas L77C, S78C, F79C, V139C, T157C, L173C, R175C, V376C, A381C, V74C (1–378), and I16C/A381C were introduced into the VSV-CL (C63V, C128S, C143V) background.
EPR Sample Preparation.
Arrestin single Cys mutants were spin labeled with a 10-fold molar excess of the sulfhydryl-specific spin label reagent 2,2,5,5-tetramethylpyrroline-3-yl-methanethiosulfonate (MTSL; Toronto Research Chemicals, Downsview, ON, Canada) overnight at 4°C. Excess reagent was removed by dialysis, and arrestin was concentrated by using Microcon YM-30 concentrators (Amicon). Final protein concentrations were determined by the BCA protein assay (Pierce) using BSA as a standard. EPR samples contained 25 μM spin-labeled arrestin and a 50-μM concentration of the indicated form of rhodopsin in native disk membranes (4) in a final volume of 10 μl.
EPR Spectroscopy and Spectral Analysis.
X-band EPR spectra were recorded for samples in glass capillaries (≈10 μl) at room temperature over a 100-G range with an incident microwave power of 10 mW on an ELEXSYS E500 (Bruker, Billerica, MA) fitted with a super high Q cavity. Spectra were typically the average of 25–36 scans, baseline corrected, and normalized to the same area for the absorption spectra. Doubly spin-labeled protein samples also were recorded at room temperature over 100 G, and interspin distances were determined by using software developed by C. Altenbach (44) using the sum of the singly labeled protein spectra as the noninteracting spectrum during analysis.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants EY11500 and GM63097 (to V.V.G.), AI58024 and GM70642 (to C.S.K.), and EY05216 (to W.L.H.) and the Jules Stein Professorship Endowment (to W.L.H.). S.M.H. was supported by National Institutes of Health Training Grant GM07628.
Abbreviations
- GPCR
G protein-coupled receptor
- P-Rh*
light-activated phosphorylated rhodopsin
- P-Rh
dark phosphorylated rhodopsin
- SDSL
site-directed spin labeling.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lefkowitz R. J., Shenoy S. K. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 2.Gurevich V. V., Gurevich E. V. Structure (London) 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 3.Gurevich V. V., Benovic J. L. J. Biol. Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- 4.Gurevich V. V., Benovic J. L. J. Biol. Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 5.Schleicher A., Kuhn H., Hofmann K. P. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 6.Gurevich V. V., Gurevich E. V. Trends. Pharmacol. Sci. 2004;25:105–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Gurevich V. V., Chen C. Y., Kim C. M., Benovic J. L. J. Biol. Chem. 1994;269:8721–8727. [PubMed] [Google Scholar]
- 8.Gurevich V. V., Benovic J. L. J. Biol. Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich V. V., Dion S. B., Onorato J. J., Ptasienski J., Kim C. M., Sterne-Marr R., Hosey M. M., Benovic J. L. J. Biol. Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 10.Gurevich V. V. J. Biol. Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 11.Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. Structure (London) 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 12.Vishnivetskiy S. A., Paz C. L., Schubert C., Hirsch J. A., Sigler P. B., Gurevich V. V. J. Biol. Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 13.Vishnivetskiy S. A., Schubert C., Climaco G. C., Gurevich Y. V., Velez M. G., Gurevich V. V. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 14.Palczewski K., Pulvermuller A., Buczylko J., Hofmann K. P. J. Biol. Chem. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 15.Vishnivetskiy S. A., Hirsch J. A., Velez M. G., Gurevich Y. V., Gurevich V. V. J. Biol. Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 16.Xiao K., Shenoy S. K., Nobles K., Lefkowitz R. J. J. Biol. Chem. 2004;279:55744–55753. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 17.Pulvermuller A., Maretzki D., Rudnicka-Nawrot M., Smith W. C., Palczewski K., Hofmann K. P. Biochemistry. 1997;36:9253–9260. doi: 10.1021/bi970772g. [DOI] [PubMed] [Google Scholar]
- 18.Mchaourab H. S., Lietzow M., Hideg K., Hubbell W. L. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 19.Qin Z., Wertz S. L., Jacob J., Savino Y., Cafiso D. S. Biochemistry. 1996;35:13272–13276. doi: 10.1021/bi961747y. [DOI] [PubMed] [Google Scholar]
- 20.Crane J. M., Mao C., Lilly A. A., Smith V. F., Suo Y., Hubbell W. L., Randall L. L. J. Mol. Biol. 2005;353:295–307. doi: 10.1016/j.jmb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 21.Hubbell W. L., Cafiso D. S., Altenbach C. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 22.Klug C. S., Feix J. B. SDSL: A Survey of Biological Applications in Biological Magnetic Resonance. Hingham, MA: Kluwer Academic/Plenum; 2004. [Google Scholar]
- 23.Hirsch J. A., Schubert C., Gurevich V. V., Sigler P. B. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 24.Lietzow M. A., Hubbell W. L. Biochemistry. 2004;43:3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 25.Columbus L., Hubbell W. L. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 26.Ohguro H., Palczewski K., Walsh K. A., Johnson R. S. Protein Sci. 1994:2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich V. V., Benovic J. L. Mol. Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 28.Gray-Keller M. P., Detwiler P. B., Benovic J. L., Gurevich V. V. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 29.Hanson S. M., Gurevich V. V. J. Biol. Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vishnivetskiy S. A., Hosey M. M., Benovic J. L., Gurevich V. V. J. Biol. Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 31.Pulvermuller A., Schroder K., Fischer T., Hofmann K. P. J. Biol. Chem. 2000;275:37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- 32.Kieselbach T., Irrgang K. D., Ruppel H. Eur. J. Biochem. 1994;226:87–97. doi: 10.1111/j.1432-1033.1994.tb20029.x. [DOI] [PubMed] [Google Scholar]
- 33.Dinculescu A., McDowell J. H., Amici S. A., Dugger D. R., Richards N., Hargrave P. A., Smith W. C. J. Biol. Chem. 2002;277:11703–11708. doi: 10.1074/jbc.M111833200. [DOI] [PubMed] [Google Scholar]
- 34.Krupnick J. G., Gurevich V. V., Schepers T., Hamm H. E., Benovic J. L. J. Biol. Chem. 1994;269:3226–3232. [PubMed] [Google Scholar]
- 35.Raman D., Osawa S., Weiss E. R. Biochemistry. 1999;38:5117–5123. doi: 10.1021/bi9824588. [DOI] [PubMed] [Google Scholar]
- 36.Raman D., Osawa S., Gurevich V. V., Weiss E. R. J. Neurochem. 2003;84:1040–1050. doi: 10.1046/j.1471-4159.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn H., Hall S. W., Wilden U. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 38.Hamm H. E., Bownds M. D. Biochemistry. 1986;25:4512–4523. doi: 10.1021/bi00364a010. [DOI] [PubMed] [Google Scholar]
- 39.Sommer M. E., Smith W. C., Farrens D. L. J. Biol. Chem. 2005;280:6861–6871. doi: 10.1074/jbc.M411341200. [DOI] [PubMed] [Google Scholar]
- 40.Langen R., Cai K., Altenbach C., Khorana H. G., Hubbell W. L. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 41.Sutton R. B., Vishnivetskiy S. A., Robert J., Hanson S. M., Raman D., Knox B. E., Kono M., Navarro J., Gurevich V. V. J. Mol. Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 42.Gurevich V. V. Methods Enzymol. 1996;275:382–397. doi: 10.1016/s0076-6879(96)75023-1. [DOI] [PubMed] [Google Scholar]
- 43.Gurevich V. V., Benovic J. L. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
- 44.Altenbach C., Oh K. J., Trabanino R. J., Hideg K., Hubbell W. L. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.