Abstract
Autophagy plays a central role in regulating important cellular functions such as cell survival during starvation and control of infectious pathogens. Recently, it has been shown that autophagy can induce cells to die; however, the mechanism of the autophagic cell death program is unclear. We now show that caspase inhibition leading to cell death by means of autophagy involves reactive oxygen species (ROS) accumulation, membrane lipid oxidation, and loss of plasma membrane integrity. Inhibition of autophagy by chemical compounds or knocking down the expression of key autophagy proteins such as ATG7, ATG8, and receptor interacting protein (RIP) blocks ROS accumulation and cell death. The cause of abnormal ROS accumulation is the selective autophagic degradation of the major enzymatic ROS scavenger, catalase. Caspase inhibition directly induces catalase degradation and ROS accumulation, which can be blocked by autophagy inhibitors. These findings unveil a molecular mechanism for the role of autophagy in cell death and provide insight into the complex relationship between ROS and nonapoptotic programmed cell death.
Keywords: apoptosis, necrosis, nonapoptotic, reactive oxygen species
Programmed cell death plays an important role in embryonic development, immune regulation, and general cellular homeostasis (1–3). Apoptosis, the most thoroughly studied form of programmed cell death is defined by dependence on a family of proteases known as caspases. Activation of caspases leads to distinct morphological features in the cell, such as nuclear condensation, membrane blebbing, and cell shrinkage (4). Several studies have identified cell death programs that are clearly distinct from apoptosis (5, 6). These nonapoptotic cell death programs are genetically regulated and often have morphological features resembling necrosis, yet their underlying molecular mechanisms are unclear. Molecular insights into alternative cell death programs could lead to a better understanding of their role in normal cellular homeostasis as well as disease processes.
Autophagy is a cellular process that causes degradation of long-lived proteins and recycling of cellular components to ensure survival during starvation. During autophagy, cells exhibit extensive internal membrane remodeling, engulfing portions of the cytoplasm in large double-membrane vesicles. These autophagosomes dock and fuse with lysosomes, and the contents of the fusion vacuoles are eventually degraded (7). A group of genes known as ATG genes, which are conserved from yeast to humans, have been found to regulate autophagy (8). Genetic analyses in yeast, and recently in higher eukaryotes, have shown that autophagy plays a central role in the regulation of cell survival during nutritional deprivation (9, 10). In addition, autophagy has been shown to play a role in tumor suppression (11, 12), pathogen control (13), antigen presentation (14), and the regulation of organismal lifespan (15).
The involvement of autophagy in programmed cell death has been controversial. Autophagy has long been observed in dying cells during development, infectious processes, and neurodegenerative diseases. However, it has been uncertain whether autophagy functions to protect or kill cells under these circumstances. Recently, it has been shown that certain forms of cell death are prevented in either the presence of autophagic inhibitors or reduced ATG gene expression (16, 17), thus implying that autophagy initiates and participates directly in the death process. However, the mechanism by which autophagy programs cells to die is unknown.
Reactive oxygen species (ROS) are a group of highly reactive molecular forms of oxygen containing unpaired electrons (18). They include molecules such as singlet oxygen, hydroxyl radicals, superoxide, and hydrogen peroxides. ROS are continuously produced as a byproduct of the mitochondrial respiratory chain in normal, healthy cells. It is estimated that 2% of oxygen consumed by mitochondria is converted to ROS (19). Most ROS have extremely short half-lives (nanoseconds), but some ROS, such as hydrogen peroxide, have millisecond half-lives. Due to their high reactivity, ROS can oxidize cell constituents such as lipids, proteins, and DNA, thus damaging cell structures and compromising function (20). Because of these potentially lethal effects, cells maintain ROS at a tolerable level by means of antioxidants such as the redox system, superoxide dismutase (SOD), and catalase (21). Imbalance due to either increased ROS production or a decrease in ROS degradation can cause ROS accumulation and cell damage. ROS damage has been shown to cause various types of cell death (22, 23), but whether it plays a role in autophagic cell death has not been determined.
Previously, we observed that inhibition of caspase 8 by treatment with either benzyloxycarbonyl-valyl-alanyl-aspartic-acid (O-methyl)-fluoromethylketone (zVAD) or specific caspase 8 RNA interference (RNAi) triggers autophagy and cell death (16). zVAD-induced death was arrested by either chemical autophagy inhibitors or by knocking down the protein products of key autophagy genes. This finding provided definitive evidence of a signaling pathway leading to autophagic cell death. However, the molecular mechanism by which autophagy causes cell death remains a key unanswered question. We now show that zVAD treatment causes selective autophagic degradation of one of the main cellular antioxidants, catalase. The depletion of catalase causes a severe imbalance of ROS metabolism, leading to dramatic ROS accumulation, lipid peroxidation, and ultimately nonapoptotic death.
Results
ROS have been previously shown to cause cell damage and death (24, 25). To investigate whether ROS play a role in autophagic cell death, we first examined zVAD-treated cells for ROS accumulation in cells treated with zVAD to inhibit caspases. Cells were exposed to either 40 μM zVAD in DMSO or an equal volume of DMSO for 12 h, and then stained with Image-iT LIVE Green Reactive Oxygen Species Detection dye and counterstained with the chromatin binding dye DAPI (Fig. 1A). We found that zVAD treatment caused a dramatic increase in ROS. Next, we tested whether ROS were causally related to cell death by treating cells with a panel of ROS inhibitors, including propyl gallate (PG), octyl gallate (OG), nordihydroguaiaretic acid (NDGA), tert-butylhydroquinone (TBHQ), ethoxyquin, 2,6-di-tert-butyl-4-hydroxymethyl-phenol (Ionox 100), or butylated hydroxyanisole (BHA) (Fig. 1B). We found that, despite their chemical differences, all of these ROS inhibitors potently inhibited zVAD-induced cell death (Fig. 1B). This finding suggests that zVAD-induced cell death depends on ROS.
Fig. 1.
ROS inhibitors prevent autophagic cell death. (A) L929 cells were treated with 1 μl DMSO (Aa and Ab) or 40 μM zVAD (Ac and Ad) for 12 h, stained with DAPI (Aa and Ac) and Image-iT LIVE Green Reactive Oxygen Species Detection dye (Ab and Ad) and examined by fluorescent microscopy. (Magnification, ×400.) (B) L929 cells were pretreated with 100 μM PG, 100 μM OG, 50 μM NDGA, 100 μM TBHQ, 40 μM ethoxyquin, 20 μM Ionox 100, or 100 μM BHA for 1 h and then exposed to 20 μM zVAD or DMSO for 24 h, after which the reduction in cell number was quantitated by flow cytometry. Results are representative of three experiments (mean + SD).
ROS accumulation has been previously reported to cause cell membrane damage and permeability changes (26). Transmission electron microscopy (TEM) examination of zVAD-treated L929 cells revealed that cells first become vacuolated and autophagic (Fig. 2Ab and Ae), and then necrotic with massive plasma membrane damage (Fig. 2 Ac and Af). We therefore compared ROS accumulation, lipid peroxidation, and changes in membrane permeability and cell loss after administration of zVAD for 0, 6, 12, and 18 h (Fig. 2 B–E). These experiments included a control cell culture treated with 100 μM of the ROS inhibitor BHA during zVAD exposure for 18 h. ROS accumulation was detectable as early as 6 h after zVAD treatment and then increased dramatically at 12 and 18 h, and BHA treatment completely abrogated zVAD-induced ROS accumulation (Fig. 2B). We also detected lipid peroxidation, which was delayed in kinetics compared with ROS accumulation. A dramatic increase in lipid peroxidation was observed only after 12 h of zVAD treatment, and this peroxidation was inhibited by BHA (Fig. 2C). Both the loss of membrane integrity, as measured by the release of cellular lactate dehydrogenase (LDH) into the medium (Fig. 2D), and overall cell loss (Fig. 2E) displayed kinetics similar to those of lipid peroxidation. These data support the hypothesis that ROS accumulation is an early event that precedes lipid peroxidation, loss of membrane integrity, and cell death. The fact that the ROS scavenger BHA completely blocked lipid peroxidation, loss of membrane integrity, and cell loss indicates that ROS accumulation is the cause of lipid membrane damage and autophagic cell death.
Fig. 2.
ROS production and cell damage is associated with autophagic cell death. (A) Transmission electron microscopy (TEM) of L929 cells treated for 0, 12, and 18 h with zVAD. [Scale bar: 1 μm (Aa–Ac) and 0.1 μm (Ad–Af).] (B–E) L929 cells were treated with either zVAD for 6, 12, or 18 h (filled diamonds), or 100 μM BHA during zVAD treatment for 18 h (open triangles). (B) Cells were treated with 20 μM zVAD and stained with DCF and quantified by flow cytometry. The values are presented as fold increase of DCF mean fluorescence intensity (DCF F. I.). (C) L929 cells were treated with 40 μM zVAD and lipid peroxidation was measured as quantity of LOOHs, which are a specific indictor of ROS damage. (D) LDH release, a measure of the loss of membrane integrity, was quantified in the cellular supernatant of the experiment in B, and the percentage increase is shown. (E) zVAD-induced cell death of L929 cells. The percentage cell loss was quantified as described in Experimental Procedures.
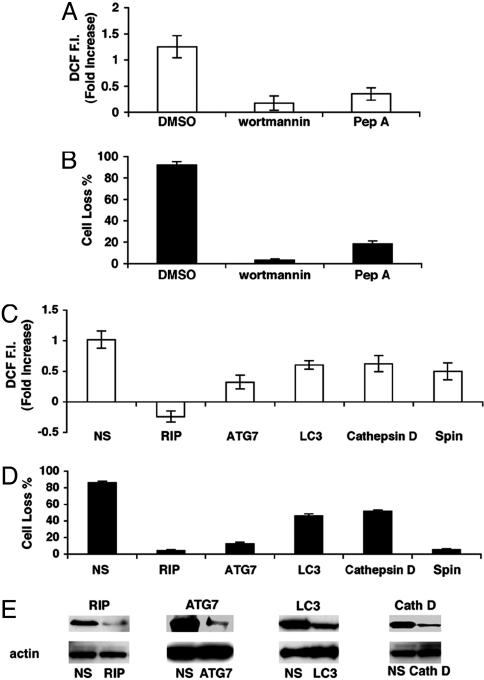
We next examined the molecular pathways that regulate zVAD-induced ROS accumulation. Previously, phosphoinositide 3-kinase (PI3K) inhibitors were shown to reduce zVAD-induced autophagy and cell death (16). Therefore, we pretreated cells with wortmannin, a PI3K inhibitor, and found that it completely blocked zVAD-induced ROS accumulation (Fig. 3A) and cell death (Fig. 3B). Furthermore, cells pretreated with pepstatin A (Pep A), an inhibitor of acidic lysosomal protease cathepsin D, which acts during the final degradation step of autophagy, were also protected from zVAD-induced ROS accumulation (Fig. 3A) and cell death (Fig. 3B). These findings suggest that zVAD-induced ROS production depends on autophagy. To further examine the role of autophagy in this process, we measured zVAD-induced ROS accumulation in cells transfected with RNAi for several key autophagy genes—ATG7, a key autophagy gene that encodes a ubiquitin E1-like protein; the microtubule-associated protein light chain 3 (LC3), which is mouse analog of yeast ATG8, and cathepsin D. Knocking down any of these autophagic genes inhibited ROS accumulation in proportion to the level of protein reduction (Fig. 3 C and E). Importantly, knocking down the receptor interacting protein (RIP), which is required for zVAD-induced autophagy and cell death, also blocked zVAD-induced ROS accumulation (Fig. 3C). A reduced level of cell death was correlated with the observed amount of ROS accumulation, further strengthening the association between autophagy, ROS, and cell death (Fig. 3D).
Fig. 3.
ROS accumulation depends on autophagy. L929 cells were pretreated with the phosphoinositide 3-kinase inhibitor wortmannin for 1 h or the cathepsin D inhibitor Pep A for 10 h and then with DMSO or 40 μM zVAD for 24 h. ROS accumulation (A) and percentage of cell loss (B) were quantified. L929 cells were treated for 24 h with zVAD or DMSO 72 h after transfection with RIP, ATG7 (mAPG7), LC3, cathepsin D (Cath D), Spinster (Spin) RNAi, or nonspecific (NS) oligoribonucleotides and accumulation of ROS (C) and reduction in cell number (D) were quantified. (E) Reductions in the corresponding proteins by RNAi compared with nonspecific (NS) oligoribonucleotides and actin as shown by Western blotting. Reduction of level of spin RNA was verified by Northern blotting.
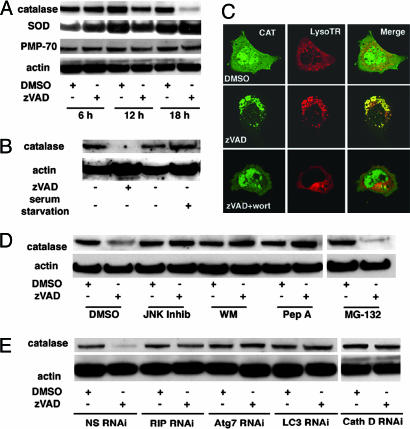
ROS accumulation may be caused by either an increase in ROS production or decreased ROS degradation. To determine whether autophagy impairs ROS degradation, we evaluated both catalase and SOD, the key enzymatic scavengers of ROS. The steady-state quantities of both catalase and SOD were measured after 6, 12, and 18 h of zVAD treatment (Fig. 4A). We found that the quantity of SOD protein remained unchanged or increased in zVAD-treated cells throughout all three time points (Fig. 4A). By contrast, a steady level of catalase decreased after 12 h of zVAD treatment and was greatly reduced after 18 h (Fig. 4A). The loss of catalase was specific because actin was not diminished after zVAD treatment. Because autophagy can engulf and degrade whole organelles, we also checked for possible degradation of peroxisomes, which are some of the main sites of catalase activity in the cell. The level of PMP-70, a peroxisomal membrane marker protein, remained unchanged, implying that peroxisomes remain intact throughout zVAD-induced autophagy. Also, direct staining of peroxisomes revealed no decrease in the organelles after zVAD exposure (data not shown). We thus concluded that cytoplasmic stores of catalase were depleted during autophagic cell death. To determine whether reduced steady-state quantities of catalase were due to catalase protein degradation or reduced gene expression, we measured catalase mRNA level by Northern blotting and found that zVAD treatment did not reduce the level of catalase mRNA (Fig. 7A, which is published as supporting information on the PNAS web site). We therefore measured catalase protein degradation by [35S]methionine pulse–chase analysis. After zVAD treatment, the level of catalase was reduced compared with the DMSO control (Fig 7B). Thus, the reduced steady-state quantity of catalase seems to be due to protein degradation, but not reduced mRNA level. Interestingly, serum starvation, which rapidly induces classic macroautophagy, did not induce catalase degradation (Fig. 4B), suggesting a different underlying mechanism between zVAD and nutrition starvation-induced autophagy.
Fig. 4.
zVAD induce catalase degradation. (A) L929 cells were treated with 40 μM zVAD or DMSO for 6, 12, 18 h and the protein levels of catalase, actin, SOD, and the 70-kDa peroxisomal membrane protein (PMP70) were measured by Western blotting. (B) L929 cells were treated with 40 μM zVAD or went through serum starvation for 18 h, and the protein levels of catalase, actin were measured by Western blotting. (C) L929 cells were transfected with GFP-catalase, and, 24 h after transfection, cells were treated with 40 μM zVAD or DMSO for 18 h with or without being pretreated with 1 μg/ml wortmannin for 1 h, stained to identify lysosomes, and imaged by confocal microscopy. Green fluorescence for catalase-GFP (CAT), red fluorescence for Lysotracker lysosomal stain (Lyso TR), and the merged images (merge) are shown. (D) L929 cells were pretreated with JNK inhibitor II (JNK Inhib), wortmannin (WM), or Pep A (Pep A) or MG-132 for 1 h and with DMSO or 40 μM zVAD for 18 h, after which the protein levels of catalase and actin were measured by Western blotting. (E) L929 cells were treated for 24 h with zVAD or DMSO after transfection with RIP, ATG7(mAPG7), LC3, cathepsin D (Cath D), Spin RNAi or nonspecific (NS) oligoribonucleotides and the protein levels of catalase and actin were measured by Western blotting. Reductions in the corresponding proteins were shown by Western blotting (see Fig. 3E, which is the same experiment).
To determine how catalase was degraded, we pretreated cells with a panel of inhibitors that prevent L929 autophagic cell death. Treatment with the inhibitors wortmannin, c-Jun N-terminal kinase (JNK) inhibitor II, and Pep A blocked zVAD-induced reduction in the steady-state level of catalase (Fig. 4C). By contrast, the proteasome inhibitor MG-132 did not prevent catalase degradation (Fig. 4C). We had previously shown that these inhibitors prevent accumulation of autophagosomes in L929 cells (16), suggesting that autophagy may be responsible for catalase degradation. Cells treated with zVAD after transfection with RIP, ATG7, LC3, and cathepsin D RNAi displayed no reduction in the level of catalase (Fig. 4D), further confirming a role for autophagy in catalase removal. We also verified this conclusion by transfecting cells with a catalase-GFP construct and monitoring its localization after treatment with zVAD. Before zVAD treatment, catalase was homogenously distributed in the cytoplasm. After zVAD treatment, the catalase-GFP formed punctate structures that colocalized with a lysosomal marker Lysotracker. By contrast, cells pretreated with the autophagic inhibitor wortmannin showed reduced colocalization of catalase with Lysotracker (Fig. 4E). Thus, catalase seems to be targeted to lysosomes by autophagy. Taken together, these data indicate that autophagy can contribute to ROS accumulation by selectively removing catalase, which is a lethal event for the cell. However, it is likely that other mechanisms may also contribute to ROS accumulation, because small amounts of ROS can be detected as early as 6 h, but catalase degradation becomes evident only after 12 h of zVAD treatment.
We previously reported that zVAD-induced autophagy and cell death are mediated specifically by inhibiting caspase 8 (16). To determine whether zVAD-induced catalase degradation and ROS accumulation can be caused by caspase 8 inhibition, we transfected L929 cells with either caspase 8 RNAi or nonspecific RNAi. Ninety-two hours after transfection, there was substantial accumulation of ROS (Fig. 5A) as well as almost total degradation of catalase in the cells in which caspase 8 had been suppressed (Fig. 5B). The actin control was unaffected under these conditions. We therefore concluded that caspase 8 inhibition is sufficient to induce catalase degradation and associated ROS accumulation.
Fig. 5.
Reduction of caspase 8 induces catalase degradation and ROS accumulation. (A) L929 cells were transfection with caspase 8 RNAi or nonspecific (NS) oligoribonucleotides, and, 96 h after transfection, cells were harvested and ROS accumulation was quantified by flow cytometry. (B) Protein levels of catalase, caspase 8, and actin of the corresponding samples were measured by Western blotting.
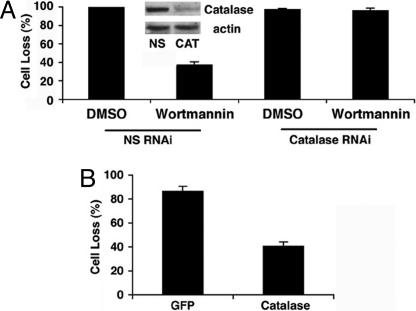
To further investigate the mechanism of autophagic cell death, we altered autophagy and catalase levels in L929 cells. After transfecting cells with either a nonspecific control RNAi or catalase RNAi, we pretreated the cells with either DMSO or wortmannin for 1 h and then zVAD for 24 h. We hypothesized that, if autophagy causes cell death by means of catalase degradation, then knocking down catalase should overcome the protection from autophagic cell death generated by wortmannin inhibition of autophagy. Indeed, we found that knocking down catalase completely surmounted the protective effect of wortmannin (Fig. 6A). We also noticed that wortmanin failed to prevent zVAD-induced ROS accumulation in catalase knock-down cells (data not shown). We then examined the effect of catalase overexpression on zVAD-induced cell death. We found that overexpression of catalase-GFP but not GFP alone could largely block zVAD-induced cell death (Fig. 6B). These experiments suggest that autophagy-mediated catalase degradation is necessary and sufficient for zVAD-induced autophagic cell death in L929 cells.
Fig. 6.
Autophagic cell death depends on catalase degradation. (A) L929 cells were treated for 1 h with DMSO or 1 μg/ml wortmannin and then 24 h with zVAD or DMSO 72 h after transfection with catalase RNAi or nonspecific (NS) oligoribonucleotides, and cell losses were quantified. (B) L929 cells were treated for 24 h with zVAD or DMSO after transfection with GFP or catalase-GFP. Percentage cell loss was quantified as described in Experimental Procedures.
Discussion
We had previously proposed a model for zVAD-induced autophagic cell death in which caspase 8 inhibition activates a pathway involving RIP and JNK that eventually leads to autophagy (16). However, the molecular mechanism behind autophagic cell death was not clear. We now show that autophagy can lead to the degradation of catalase, a key enzymatic ROS scavenger, which disrupts the intracellular ROS balance. The resulting accumulation of ROS in the cell leads to membrane peroxidation, loss of membrane integrity, and eventually cell death (Fig. 8, which is published as supporting information on the PNAS web site). Directly varying the cellular level of catalase recapitulates the mechanism of autophagic cell death. A reduction in cellular levels of catalase leads to cell death even if the cell has been treated with wortmannin, an autophagic inhibitor. By contrast, an increase in the cellular levels of catalase protects against autophagic cell death. Therefore, the regulatory effect of autophagy on catalase and thereby ROS and cell fate is direct and rate-limiting.
Autophagy was first observed in dying cells in the 1960s (27, 28); however, until recently, the relationship between autophagy and cell death has remained unclear. The recent identification of autophagic genes in yeast and technological innovations such as RNAi make it possible to examine autophagy and cell death as discrete processes and thereby study their relationship. Thus far, autophagy has been shown to either promote or inhibit cell death depending on the conditions. For example, in yeast undergoing starvation, autophagy works as an alternative energy source and is essential for the cell’s survival (29). On the other hand, various cytokines and chemicals that induce cell death (17, 30, 31) have been shown to be caused by autophagy. It has also been reported that autophagy is responsible for a certain type of developmental cell death, and it may play a role in some types of neurodegenerative diseases (32, 33). It is quite clear that autophagy is capable of either antagonizing or promoting cell death; however, the factors determining the cell fate by autophagy have not previously been elucidated. Our data show that autophagy can induce cell death by degrading catalase. Interestingly, starvation-induced autophagy does not lead to catalase degradation; therefore, catalase degradation, or lack thereof, may be a major factor deciding the outcome of autophagy. Our studies of this mechanism have been restricted to L929 cells, a widely studied model for TNF-α-induced cell death. Autophagy has been associated with cell death in several developmental contexts in different animals, and the mechanism of autophagic death may not be identical to what we have found in L929 cells. Nevertheless, our data provide a useful starting point to study the biochemical mechanisms of autophagy and its relationship to cell death in mammals.
The prevailing view is that autophagy is a nonselective degradation mechanism. The autophagic membrane is believed to envelop cellular contents nonselectively and deliver them to the lysosome for degradation. However, an increasing number of reports have shown that, under certain conditions, organelles such as the peroxisome (34) or proteins such as α-and β-synuclein (35) and Ald6p (36) can be specifically degraded by autophagy. For example, two forms of autophagy known as micropexophagy and macropexophagy can act to degrade the peroxisome. Although each form is morphologically distinct, both are regulated by important autophagy genes, such as ATG7 and ATG8. However, there are also certain proteins that are specific to each pathway. For example, micropexophagy requires proteins such as ATG26 and PFK1, even though macropexophagy does not (34). Thus far, it is not known how pexophagy achieves cargo specificity. One can speculate that peroxisomes must first be labeled and then recognized by the proteins involved in pexophagy. We know that some peroxisomal matrix proteins like PEX14 are required for pexophagy. Such proteins may serve as specific pexophagic markers (37); however, the gene involved in targeting these markers has not yet been identified. Similarly, little is known about which autophagy genes are involved in targeting specific proteins. Our results on the selective degradation of catalase indicate that cargo specificity may make the difference between life and death. It will be of great interest therefore to determine the molecular basis of selective action of autophagy.
Because ROS are an unavoidable byproduct of oxidative respiration, cells have developed a sophisticated antioxidant system that maintains the delicate balance between ROS production and degradation. One of the major enzymes in this antioxidant system, SOD, converts the extremely toxic superoxide anions into hydrogen peroxide and oxygen. Catalase then breaks down the less harmful hydrogen peroxide into harmless H2O and O2 (38). The importance of the SOD-catalase system is reflected in diseases that are caused by a disruption of SOD or catalase. For example, acatalasemia, an inherited catalase deficiency, is associated with an increased risk of diabetes (39, 40). Overexpression of SOD and catalase has been shown to have therapeutic benefit in models of neurodegeneration (41). Besides actively regulating the intracellular ROS level, catalase is important to the cell’s tolerance to extracellular ROS as well (42). Reduced levels of catalase have been directly linked to the development of necrosis in plants (43). Furthermore, overexpression of catalase can rescue cells from a variety of ROS-related deaths (44, 45). Our data further demonstrate the importance of this SOD-catalase antioxidant system as a means of mammalian programmed cell death.
The fact that a small increase in ROS levels can be detected before catalase degradation indicates that other mechanism(s) may contribute to caspase-triggered ROS accumulation. Intracellular ROS level is a balance between ROS production and degradation. Catalase degradation contributes to ROS accumulation by decreasing ROS degradation, but caspase inhibition may also induce ROS production. Little is known about how caspase inhibition can affect ROS production, and it is possible that ROS regulatory proteins may be directly regulated by caspases. The identify of such caspase substrate(s) that regulate ROS regulation needs to be investigated.
It has long been known that intracellular-ROS levels are in constant flux and are continuously balanced by the antioxidant system. Increasing the level of ROS can up-regulate key antioxidant enzymes such as SOD and catalase, which in turn keep ROS in check (46, 47). We and others have shown that oxidative damage can cause nonapoptotic cell death resembling necrosis. In the present case, our data reveal that death is due to oxidative membrane lipid damage. It is suppressed in most circumstances by the activity of caspases, the major enzymatic mediator of apoptosis. By directly linking autophagic cell death and catalase degradation, our data illustrate how autophagy can disrupt the delicate balance between ROS production and degradation, and how a mechanism that originally promotes survival can turn into an efficient mediator of cell death.
Experimental Procedures
Reagents and Antibodies.
Wortmannin, JNK inhibitor II (anthra[1,9-cd]pyrazol-6(2H)-one 1,9-pyrazoloanthrone), MG-132, and Pep A were purchased from Calbiochem. zVAD was purchased from Enzyme Systems Products (Livermore, CA). PG, OG, NDGA, TBHQ, Ionox 100, and BHA were purchased from Sigma. Image-iT LIVE Green Reactive Oxygen Species Detection kit, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), lysotracker, antibodies to mouse caspase-8, RIP, and Beclin-1 were purchased from Pharmingen. The LDH Detection Kit was purchased from Roche Applied Science (Indianapolis). Lipid hydroperoxide (LPO) Kit was purchased from Cayman Chemical (Ann Arbor, MI). Antibodies to RIP, cathepsin D, and caspase 8 were purchased from Becton Dickinson. The Atg7 antibody was a kind gift from W. Dunn (University of Florida College of Medicine, Gainesville).
Preparation of Small Interfering RNA (siRNA).
Nonspecific RNAi oligoribonucleotides and RNAi oligoribonucleotides corresponding to the following cDNA sequences were purchased from Dharmacon Research (Layfayette, CO) (mouse sequences): GTTTGTAGCCTCAAGTGTT for mouse ATG7; CCACTAGTCTGACTGATGA for RIP; GATCGAGGATTATGAAAGA for caspase-8. LC3 and cathepsin D smart pool RNAi were purchased from Dharmacon Research, and the sequences were not revealed by the manufacturer.
Transfection of siRNA.
The 0.5-nmol RNAi were transfected by means of Amaxa (Gaithersburg, MD) NucleofectionTM, by using solution V and program T-20. Cells were then cultured in growth medium for 96 h before further analysis.
Tissue Culture.
The mouse L929 cell line was obtained from the American Type Culture Collection (Rockville, MD). L929 cells were cultured in DMEM with 4.5 g/liter glucose. Media were supplemented with 2 mM l-glutamine, 1% penicillin/streptomycin solution, and 10% FBS.
Cell Death Analysis.
Cell viability was determined by staining with propidium iodide (2 μg/ml), and flow cytometric analysis was conducted by using facscan software. The percentage of cell death was quantified as described (16).
Electron Microscopy Analyses.
Cells were fixed in 3% glutaraldehyde in 0.1 M Mops buffer (pH 7.0) for 8 h at room temperature, 3% glutaraldehyde/1% paraformaldehyde in 0.1 M Mops buffer (pH 7.0) for 16 h at 4°C, postfixed in 1% osmium tetroxide for 1 h, embedded in Spurr’s resin, sectioned, double stained with uranyl acetate and lead citrate, and analyzed by using a Zeiss EM 10 transmission electron microscope.
LDH Assay.
The release of LDH was measured by using the Cytotoxicity Detection Kit (LDH) from Roche Applied Science. The results are expressed as the percentage of the cytosolic LDH activity of the total activity from 1% Trixon-100 permeabilized cells.
Lipid Hydroperoxide (LOOH) Assay.
The extent of lipid peroxidation was determined by measuring lipid hydroperoxides (LOOH) by using a Lipid Hydroperoxide (LPO) Kit from Cayman Chemical. Briefly, cells were treated as indicated, and cell membranes were extracted by using chloroform:methanol (2:1, vol/vol). Ferrous iron was added to the chloroform extract, and formation of ferric thiocyanate ion was measured with a 96-well plate reader. 13-Hydroperoxy octadecadienoic acid (13-HpODE) was used as the standard. The results are expressed as nmol LOOH/mg total protein.
ROS Measurements.
Dichlorodihydrofluorescein diacetate (DCF) was purchased from Molecular Probes. Intracellular ROS levels were measured as described elsewhere (48). Briefly, cells were treated with 25 μM DCF in PBS for 30 min, and flow cytometric analysis was conducted on a FACScan. For visualization of intracellular ROS, cells were stained with Image-iT LIVE Green Reactive Oxygen Species Detection kit according to protocol provided by the manufacturer.
Catalase Half-Life Determination.
L929 cells were pretreated with and without zVAD (20 μM) for 4 h and were then incubated in methionine medium containing 10% dialyzed FBS for 30 min. [35S]Methionine/[35S]cysteine (final concentration 100 μCi/ml, Translabel, ICN) (1 Ci = 37 GBq) was then added, and cells were incubated for 1 h. At this point (t = 0), the radioactive medium was removed, and cells were washed once in PBS solution and either lysed immediately in TNESV lysis buffer or recultured in complete medium for varying times in the presence and absence of zVAD before harvesting. Catalase was immune-precipitated from detergent-soluble cell extracts normalized for protein content and analyzed by SDS/PAGE. Gels were dried and exposed to Kodak X-Omat AR-5 x-ray film for 4 h at −70°C or for 24 h at room temperature.
Supplementary Material
Acknowledgments
We thank Tim Maugel for assistance in transmission electron microscopy and Richard Siegel, Lixin Zheng, and Nicolas Bidere for comments on the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases, and NIH Grant GM59136 (to E.H.B.). E.F. is an NIH–Oxford University Scholar in Biomedical Research.
Abbreviations
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- zVAD
benzyloxycarbonyl-valyl-alanyl-aspartic-acid (O-methyl)-fluoromethylketone
- RNAi
RNA interference
- PG
propyl gallate
- OG
octyl gallate
- NDGA
nordihydroguaiaretic acid
- TBHQ
tert-butylhydroquinone
- Ionox 100
2,6-di-tert-butyl-4-hydroxymethyl-phenol
- BHA
butylated hydroxyanisole
- Pep A
pepstatin A
- LC3
light chain 3
- RIP
receptor interacting protein
- JNK
c-Jun N-terminal kinase
- LDH
lactate dehydrogenase
- LOOH
lipid hydroperoxide
- DCF
dichlorodihydrofluorescein diacetate.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Degterev A., Boyce M., Yuan J. Oncogene. 2003;22:8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson M. D., Weil M., Raff M. C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson D. W., Thornberry N. A. Science. 2003;299:214–215. doi: 10.1126/science.1081274. [DOI] [PubMed] [Google Scholar]
- 4.Salvesen G. S., Dixit V. M. Proc. Natl. Acad. Sci. USA. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leist M., Jaattela M. Nat. Rev. Mol. Cell Biol. 2001;2:589–598. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
- 6.Fiers W., Beyaert R., Declercq W., Vandenabeele P. Oncogene. 1999;18:7719–7730. doi: 10.1038/sj.onc.1203249. [DOI] [PubMed] [Google Scholar]
- 7.Klionsky D. J., Emr S. D. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohsumi Y. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 9.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 10.Scott R. C., Schuldiner O., Neufeld T. P. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., et al. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue Z., Jin S., Yang C., Levine A. J., Heintz N. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 14.Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Munz C. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 15.Melendez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 16.Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E. H., Lenardo M. J. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C. B., Tsujimoto Y. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 18.Riley P. A. Int. J. Radiat. Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- 19.Balaban R. S., Nemoto S., Finkel T. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Holler N., Zaru R., Micheau O., Thome M., Attinger A., Valitutti S., Bodmer J. L., Schneider P., Seed B., Tschopp J. Nat. Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 21.Mates J. M., Sanchez-Jimenez F. Front. Biosci. 1999;4:D339–45. doi: 10.2741/mates. [DOI] [PubMed] [Google Scholar]
- 22.Laurent A., Nicco C., Chereau C., Goulvestre C., Alexandre J., Alves A., Levy E., Goldwasser F., Panis Y., Soubrane O., et al. Cancer Res. 2005;65:948–956. [PubMed] [Google Scholar]
- 23.Valencia A., Moran J. Free Radical Biol. Med. 2004;36:1112–1125. doi: 10.1016/j.freeradbiomed.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Martinvalet D., Zhu P., Lieberman J. Immunity. 2005;22:355–370. doi: 10.1016/j.immuni.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Pan J., She M., Xu Z. X., Sun L., Yeung S. C. Cancer Res. 2005;65:3671–3681. doi: 10.1158/0008-5472.CAN-04-2744. [DOI] [PubMed] [Google Scholar]
- 26.Madesh M., Hajnoczky G. J. Cell Biol. 2001;155:1003–1015. doi: 10.1083/jcb.200105057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scharrer B. Z. Zellforsch. Mikrosk. Anat. 1966;69:1–21. doi: 10.1007/BF00406264. [DOI] [PubMed] [Google Scholar]
- 28.Schin K. S., Clever U. Science. 1965;150:1053–1055. doi: 10.1126/science.150.3699.1053. [DOI] [PubMed] [Google Scholar]
- 29.Mizushima N., Sugita H., Yoshimori T., Ohsumi Y. J. Biol. Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 30.Kanzawa T., Zhang L., Xiao L., Germano I. M., Kondo Y., Kondo S. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 31.Shao Y., Gao Z., Marks P. A., Jiang X. Proc. Natl. Acad. Sci. USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baehrecke E. H. Cell Death Differ. 2003;10:940–945. doi: 10.1038/sj.cdd.4401280. [DOI] [PubMed] [Google Scholar]
- 33.Anglade P., Vyas S., Javoy-Agid F., Herrero M. T., Michel P. P., Marquez J., Mouatt-Prigent A., Ruberg M., et al. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 34.Farre J. C., Subramani S. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 36.Onodera J., Ohsumi Y. J. Biol. Chem. 2004;279:16071–16076. doi: 10.1074/jbc.M312706200. [DOI] [PubMed] [Google Scholar]
- 37.Bellu A. R., Komori M., van der Klei I. J., Kiel J. A., Veenhuis M. J. Biol. Chem. 2001;276:44570–44574. doi: 10.1074/jbc.M107599200. [DOI] [PubMed] [Google Scholar]
- 38.Sohal R. S., Orr W. C. Ann. N.Y. Acad. Sci. 1992;663:74–84. doi: 10.1111/j.1749-6632.1992.tb38651.x. [DOI] [PubMed] [Google Scholar]
- 39.Heales S. J. Lancet. 2001;357:314. doi: 10.1016/S0140-6736(05)71763-3. [DOI] [PubMed] [Google Scholar]
- 40.Goth L., Eaton J. W. Lancet. 2000;356:1820–1821. doi: 10.1016/S0140-6736(00)03238-4. [DOI] [PubMed] [Google Scholar]
- 41.Pong K. Expert Opin. Biol. Ther. 2003;3:127–139. doi: 10.1517/14712598.3.1.127. [DOI] [PubMed] [Google Scholar]
- 42.Willekens H., Chamnongpol S., Davey M., Schraudner M., Langebartels C., Van Montagu M., Inze D., Van Camp W. EMBO J. 1997;16:4806–4816. doi: 10.1093/emboj/16.16.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi H., Chen Z., Du H., Liu Y., Klessig D. F. Plant J. 1997;11:993–1005. doi: 10.1046/j.1365-313x.1997.11050993.x. [DOI] [PubMed] [Google Scholar]
- 44.Evens A. M., Lecane P., Magda D., Prachand S., Singhal S., Nelson J., Miller R. A., Gartenhaus R. B., Gordon L. I. Blood. 2005;105:1265–1273. doi: 10.1182/blood-2004-03-0964. [DOI] [PubMed] [Google Scholar]
- 45.Ito K., Nakazato T., Yamato K., Miyakawa Y., Yamada T., Hozumi N., Segawa K., Ikeda Y., Kizaki M. Cancer Res. 2004;64:1071–1078. doi: 10.1158/0008-5472.can-03-1670. [DOI] [PubMed] [Google Scholar]
- 46.Zelck U. E., Von Janowsky B. Parasitology. 2004;128:493–501. doi: 10.1017/s0031182004004895. [DOI] [PubMed] [Google Scholar]
- 47.Mari M., Cederbaum A. I. Hepatology. 2001;33:652–661. doi: 10.1053/jhep.2001.22521. [DOI] [PubMed] [Google Scholar]
- 48.Ding W. X., Ni H. M., DiFrancesca D., Stolz D. B., Yin X. M. Hepatology. 2004;40:403–413. doi: 10.1002/hep.20310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.