Abstract
Distinctive Ig and T cell receptor (TcR) chains define the two major lineages of vertebrate lymphocyte yet similarly recognize antigen with a single, membrane-distal variable (V) domain. Here we describe the first antigen receptor chain that employs two V domains, which are generated by separate VDJ gene rearrangement events. These molecules have specialized “supportive” TcRδV domains membrane-proximal to domains with most similarity to IgNAR V. The ancestral NAR V gene encoding this domain is hypothesized to have recombined with the TRD locus in a cartilaginous fish ancestor >200 million years ago and encodes the first V domain shown to be used in both Igs and TcRs. Furthermore, these data support the view that γ/δ TcRs have for long used structural conformations recognizing free antigen.
Keywords: cartilaginous fish, evolution, γ/δ T cells, T cell receptor, V(D)J rearrangement
The hallmark characteristics of the adaptive immune system are specificity and memory, both conferred by the V domains of Ig and T cell receptors (TcR). All genes encoding antigen receptor V domains are in the same superfamily and are generated by the same gene rearrangement mechanism, yet all jawed vertebrates studied have discrete Ig and TcR loci. Cartilaginous fish are the oldest animals having an adaptive immune system centered on rearranging antigen receptors. They have all four types of TcR (α, β, γ, δ) (1), three Ig isotypes [IgM (2, 3), IgW (4), and IgNAR (new antigen receptor) (5)], the recombination-activating gene recombinase (6), and polymorphic MHC genes (7, 8). Studies of modern sharks may shed light on the origins of adaptive immunity (9).
Although sharks have much of the basic molecular hardware required for adaptive immunity, some differences have been noted between their antigen receptors and those of other vertebrates. Shark Ig loci are found in many “clusters” as opposed to the single translocon organization common to mammals. Each of the hundreds of Ig loci in the shark genome contains V, D (diversity), J (joining), and C (constant) genes and may be prejoined in the germ line (10), begging questions of the regulation of rearrangement (11). It was originally suggested that horned shark TRB was multicluster (12), but this species seems to be an exception, because all four TcR genes are single translocon loci in skate (1). In addition to monomeric and pentameric IgM, cartilaginous fish have at least two other IgH isotypes. The poorly understood IgW isotype occurs in multiple forms (13) [as does IgM from elasmobranchs (14)]. IgNAR, which is apparently found only in cartilaginous fish, binds antigen by means of a single V domain (15), which is no more similar to IgV than to TcR V (16, 17). The IgNARV gene undergoes extensive hypermutation resulting in affinity maturation (18). Only the conventional α, β, γ, and δ TcR chains with single C and V domains have been described from shark or other vertebrates (19).
Here we describe a unique antigen receptor chain that blends characteristics of (previously incompatible) Ig and TcR into a TcR chain with two V domains, each encoded by separate rearranging VDJ segments, on a membrane-anchored TcRδ C domain. The membrane-distal domain, christened NAR-TcRV, is most related to the IgNARV domain and was recombined into the TRD locus >200 million years ago. Considering the reported interaction with antigen by IgNARV domains, this TcR chain provides evidence for direct antigen binding by γ/δ TcRs.
Results
RACE PCR Reveals a NAR Domain.
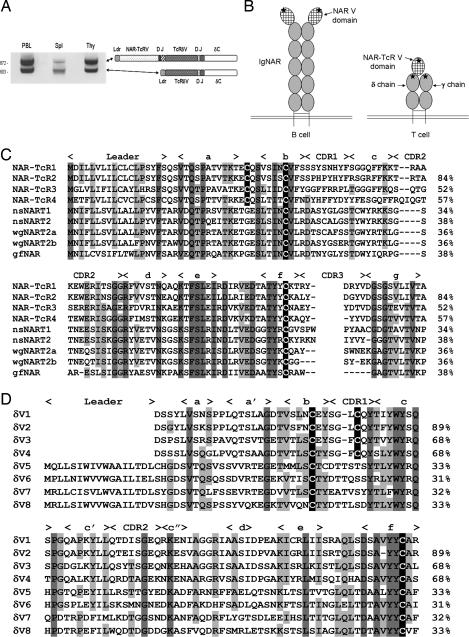
We discovered an Ig superfamily V domain while studying the genetics and expression of TcRδV families in the nurse shark Ginglymostoma cirratum. Using a reverse primer to the TcRδC domain, 5′ RACE PCR from RNA of several tissues amplified products of two sizes (Fig. 1A). Cloning and sequencing of the low-molecular-weight band yielded typical TcRδ leader-V-D-J-C-encoding transcripts, but the higher band unexpectedly contained transcripts encoding an additional V domain, N-terminal to TcRδV. This domain is most similar to V domains of IgNAR, the Ig class of cartilaginous fish that does not associate with Ig light (IgL) chains (Figs. 1B and 2). We named this V domain NAR-TcRV, which reflects its relatedness to IgNARV and its expression as a TcR, and we call the entire chain NAR-TcR (genes encoding the NAR-TcRV domain are denoted NT) (Fig. 3A). NT gene transcripts encode a typical leader peptide 5′ to the NTV segment and have a NTJ segment spliced directly to a TRDV segment. A parsimonious model of a γ/δ TcR complex including a NAR-TcR δ chain is depicted in Fig. 1B.
Fig. 1.
NAR-TcRV is a V domain supported on TcRδ. (A) Products of 5′ RACE PCR with TRDC primers using RNA from shark PBL, spleen, and thymus. (B) Cartoon depiction of IgNAR and the hypothetical γ/δ TcR with NAR-TcR antigen receptors. Stars mark CDR3s generated from somatic rearrangement. (C) Predicted amino acid sequences of four families of NAR-TcRV aligned with IgNARV from nurse shark (nsNART1, U18721; nsNART2, U18680), wobbeygong (wgNART2a, AF336092; wgNART2b, AF336091), and guitarfish (gfNAR, AY524298). Residues conserved are highlighted in gray, and Ig superfamily canonical intradomain and putative interdomain cysteines are highlighted in black. β-Strands and CDRs are noted above the alignment. Percent amino acid identity to the top sequence is shown on the right. (D) Predicted amino acid sequences of eight families of TcRδV, four of which support NAR-TcRV. Highlighting and notation are the same as in C.
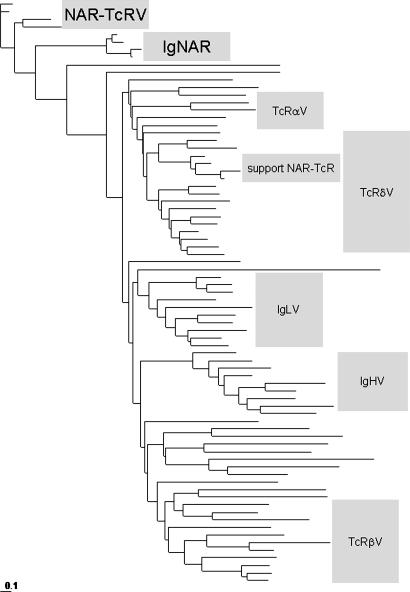
Fig. 2.
Phylogenetic analysis of Ig superfamily V domains. See Table 1 and Fig. 6, which are published as supporting information on the PNAS web site, for accession numbers of sequences used as well as the same data presented in a radial phylogram.
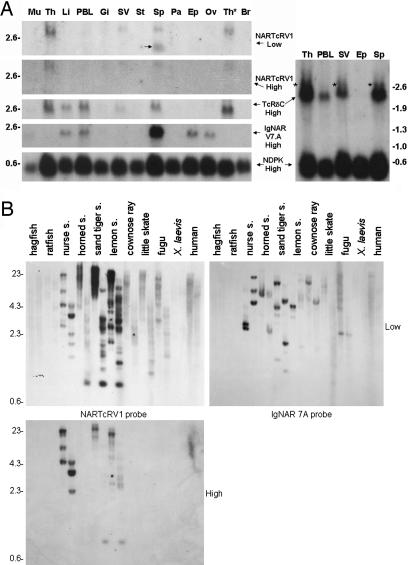
Fig. 3.
NAR-TcR is expressed in shark lymphoid tissues and is found in modern sharks. (A) Tissues used in the Northern blot are muscle, thymus, liver, PBLs, gill, spiral valve (intestine), stomach, spleen, pancreas, epigonal (a primary lymphoid organ of cartilaginous fishes), ovary, thymocytes, and brain. Marker (in kb) is shown flanking the blots. Probes were generated from V domain encoding genes of NTV and IgNAR, TRDC, and the loading control NDPK. An arrow marks the cross-hybridizing IgNAR band, and asterisks highlight the NAR-TcR encoding transcripts recognized by TRDC probing a blot allowed to migrate further. (B) Southern blot of HindIII- and PstI-digested genomic DNA probed with NTV1 and IgNAR 7A. Marker (in kb) is shown on left, and higher or lower wash stringency is indicated.
Several forms of NAR-TcRV were identified, all most related to IgNARV (≈30–40% identity; Fig. 1C) (20). β-Strands c and d deviate most from the Ig superfamily prototype V domain, and the canonical tryptophan of the WYRK motif is not present (21). NAR-TcRV lacks the cysteines in the c and d strands and complementarity-determining region (CDR)3 that make the additional disulfide bonds in some IgNARV domains (17). However, a conserved cysteine in the a–b loop is free of intradomain cysteine partners in all NAR-TcRV (see below).
Dedicated TcRVδ Domains Support NAR-TcRV Domains.
Genes encoding the same subfamilies of NAR-TcRV and TcRδV domains are always found in the same RNA transcripts (e.g., NTV1 is only found in transcripts 5′ of TRDV1). Alignment of four TcRδV subfamilies that support NAR-TcRV (families 1–4) with typical δV subfamilies (families 5–8) shows the supporting δV domains to lack leader peptides and share a cysteine residue in CDR1 (Fig. 1D). Threading these sequences onto crystal structures for IgNARV and human TcRδ predicts that the cysteine in the a–b loop of the NAR-TcRV domains and in CDR1 of supporting TcRδV domains are in close proximity and could form a disulfide bond (R. L. Stanfield, personal communication) (22, 23). Thus, the NAR-TcRV domain likely has two covalent anchors to the δV domain.
Genomic Organization of a Doubly Rearranging Receptor.
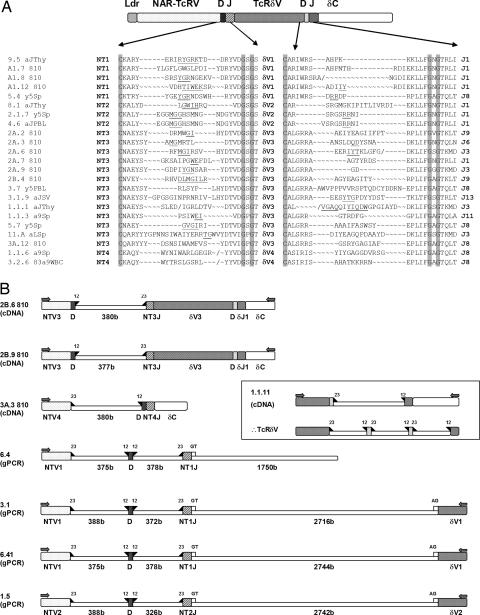
High variability in the sequence and length of CDR3 in NAR-TcRV sequences strongly suggested that these genes undergo V(D)J rearrangement events (Fig. 4A), prompting an investigation of NTV and supporting TRDV genomic organization. RT-PCR resulted in some incompletely rearranged transcripts that revealed the intergenic sequence between V and D, and D and J segments encoding the NAR-TcRV domain (Fig. 4B). In two transcripts, the associated TRDV gene is completely rearranged to the typical TcR DD and DJ gene segments. Genomic PCR using NTV forward primers and reverse primers to the corresponding TRDV revealed that a cluster comprising one NTV, one D, and one J segment is 5′ of each TRDV exon that supports NAR-TcRV. These TRDVs lost the exon encoding the leader peptide, and thus they can be expressed as protein only when attached to the N-terminal NAR-TcRV domain (Figs. 1A and 4B). The NTJ exon splices to a canonical splice site in the TRDV exon. The NTV, D, and J segments are bordered by recombination signal sequences (RSS) known to regulate recombination-activating gene-mediated rearrangement events (24, 25). Like in IgH loci heptamers and nonamers separated by 23-nucleotide spacers lie 3′ and 5′ of the NTV and J segments, respectively, and NTD segments are flanked by RSSs with 12-nucleotide spacers. Like in all other vertebrates examined, TRDD segments, which are flanked by RSS with 5′ 12-nucleotide and 3′ 23-nucleotide spacers, rearrange to a J segment RSS having a 12-nucleotide spacer. There is neither rearrangement between different NTV clusters (as is true of the IgH and IgL clusters of the cartilaginous fish) (10, 26) nor joining of NTV segments with DD or DJ segments. The incompletely rearranged transcripts provide preliminary evidence that the TRDV, D, and J segments rearrange before NTV, D, J joining. As mentioned, the CDR3 sequences at both junctions are diverse in length and sequence (Fig. 4A). Notably, the TcRδV that support NAR-TcRV are no less diverse in CDR3 than the typical TcRδVs (data not shown) despite the fact that the antigen-binding site of this domain might be occluded by NAR-TcRV. Both identified TRDD segments are used in the supporting TRDV rearrangements, and no preference for particular TRDJ segments has been observed.
Fig. 4.
NAR-TcR is doubly rearranging. (A) Focus on amino acid sequences of both NAR-TcRV and TcRδV CDR3s. Bold abbreviations indicate NAR-TcRV, TcRδV, and TcRδJ (unpublished data) families used in clones named at left. Conserved cysteine of the f strand and GXG motif of the g strand are highlighted flanking each CDR3. Known and predicted D-segment-encoded amino acids are underlined, “∼” are gaps introduced to align Vs and Js, and “/” marks frameshifts resulting from nonfunctional rearrangements. (B) Diagrams of clones showing NTV assembly mechanism. Recombination and splicing signals are marked by filled triangles for RSSs and open squares for GT/AG intronic splice sites. An incompletely rearranged 5′ RACE clone from PBL shows RSS 3′ of TRDD and 5′ of TRDJ. These data suggest that the mammalian RSS system for δ is also used in shark (Inset) (52, 53).
Tissue Expression.
Highest expression of NTV was detected in the thymus followed by spleen, spiral valve (shark intestine), and peripheral blood (Fig. 3A). The NTV1 probe cross-hybridized with the faster migrating IgNAR transcript under low-stringency conditions in the spleen. This band is not surprising considering the sequence homology between the two gene families and the high levels of IgNAR expression in spleen (27). Increasing the wash stringency resulted in loss of the cross-hybridizing IgNAR band. The NTV1 signal was also decreased because of the range of different NAR-TcRV family members (Fig. 1C). Hybridization with a typical TRDV probe that does not support NAR-TcRV revealed the same expression pattern (data not shown) as does a TRDC probe that recognizes all TRD transcripts regardless of V usage, with the exception of higher relative liver and peripheral blood leukocyte (PBL) expression for the TRDC probe and a higher relative expression of NTV1 in the spiral valve. Signal strength from the membrane-expressed, individual NTV family probes is, as expected, much less than that of secreted Ig or housekeeping probes and only a fraction of that of pan-V recognizing TRDC. Unlike what has been hypothesized for some unusual γδ TcR in mammals (28), the NAR-TcR tissue distribution is consistent with the great junctional diversity in CDR3 in suggesting that the chains have not been selected to recognize tissue-specific ligand. A second blot was prepared with immune tissue RNA, electrophoresed longer, and probed with only TRDC and NDPK to confirm that the higher 2.7-kb TRDC-positive transcript is indeed NAR-TcR and not old activity from previous NTV1 probings. This second blot shows significant enrichment of NAR-TcR in the spiral valve.
Phylogenetic Distribution of NAR-TcR and Number of Families.
The existence of NAR-TcR in other organisms was examined by genomic Southern blotting (Fig. 3B). All sharks tested (separated in phylogeny by up to 200 million years) were positive and had multiple copies of NTV, even under higher-stringency conditions. Weakly hybridizing bands were seen with the batoid species (ray and skate), fugu, and even human, which were washed away under high-stringency conditions; thus, it is not clear whether NAR-TcR existed in the ancestor of all cartilaginous fish and even perhaps all extant vertebrates. However, in silico investigations of fugu, Xenopus, and mammalian databases have failed in finding NTV homologues or telltale leaderless TRDV genes. An IgNARV probe hybridized to different bands on the same blot, dismissing cross-hybridization to the IgNAR loci as an explanation for NAR-TcR's existence in sharks (Fig. 3B).
We have identified four nurse shark NAR-TcRV families through cDNA and genomic cloning, but the Southern blot suggests that more members await discovery. Consistent with this proposal, screening of a 10× coverage nurse shark bacterial artificial chromosome (BAC) library with a NTV1 probe resulted in 208 clones, 168 of which also hybridized to TRDV probes. In contrast, no NAR-TcRV clone contained the single-copy TRDC gene, suggesting that the NTV genes are >150 kb away from TRDC. Five of the 11 TRDC-positive BAC clones were also positive for TRDV genes that do not support NAR-TcRV, which advocates a large, complex TRD locus with NTV-supportive TRDV miniclusters upstream of the traditional V, D, and J arrays (Fig. 5). No BAC clones positive for any TcR probes were also positive for IgNAR.
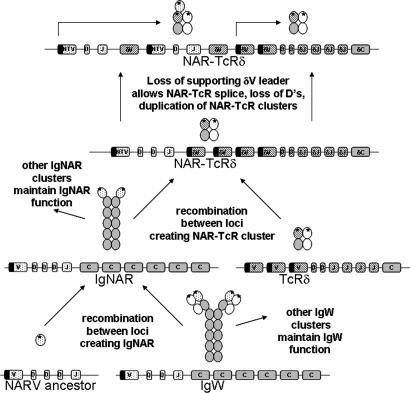
Fig. 5.
Hypothetical scheme for history of NAR variable domains. Depictions of genomic loci are simplified; filled bars indicate variable segments with leader exons upstream. Direction of cross-hatching distinguishes TRDV segments dedicated to supporting NAR-TcRV.
Initial characterization of nurse shark TcRβ and γ revealed no NAR-like mRNA transcripts by 5′ RACE or cDNA screenings (unpublished observations), nor did differential screening of a shark cDNA library with TRDC and NTV1 probes. Published studies of horned shark TcRβ also did not reveal NAR-TcR (12, 29).
Relatedness of NAR-TcRV to IgNARV and Other Antigen Receptors.
Amino acid alignment and phylogenetic analysis of many vertebrate V domains confirms the close relationship between NAR-TcRV and IgNARV and substantiates their distinction from TCR and Ig V domains (Fig. 2). It is peculiar that NAR-TcRV and IgNARV group not only outside of Ig and TcR V domains but also basal to diverse chordate V domains (30–35). We hesitate to infer from the tree an ancestral relationship between the NAR-TcR and IgNAR domains and those of conventional Ig and TcR, despite the parsimony of a single binding domain and the position of the IgNARV/NAR-TcRV cluster, because establishing alignments of such diverse V domains is challenging. Whatever the exact history of these gene families, V domains of NAR-TcR and IgNAR are clearly distinct from those of conventional Ig and TcR, as shown by genetic distance (Fig. 2), gross domain structure (Fig. 1 B and C), and mechanism of antigen binding (proven for IgNAR) (15).
It is also notable that the supporting TcRδV domains form a separate cluster within the major TcRδV group. The genetic distances between the supporting δV are similar to those of the NAR-TcRV, consistent with en bloc duplications of NTV/supporting TRDV genes over evolutionary time (Fig. 5).
Discussion
The maintenance of many functional NAR-TcR and their supporting TcRδV with conserved cysteines (presumably for interdomain stability), many NTV genes in shark genomes divorced hundreds of millions of years ago, and high expression in lymphoid tissues are all consistent with NAR-TcR playing an important role in the shark immune system. NAR-TcR transcripts comprise 21 of 99 of the TcRδ repertoire by sequenced full-length 5′ RACE clone count, and the higher band accounts for 51%, 42%, and 53% of the amplification products from PBL, spleen, and thymus by semiquantitative densitometry in Fig. 1A. Because the IgNARV domain recognizes antigen as a single chain (15, 22), and additional domains have not been found on shark TcRγ, it is likely that the NAR-TcRV domain will recognize antigen in a similar fashion. Crystallography of human γ/δ has shown a smaller angle between the C and V domains than is seen in α/β heterodimers (23); such an orientation in shark could accommodate steric constraints of an additional V domain in an immunological synapse. This angle could lend accessibility to ligand binding at either V domain, so we cannot rule out use of the typical γV/δV binding site. However, we propose that the NAR-TcRV uses the typical γ/δ TcR as a scaffold (Fig. 1B), and thus after antigen recognition by the NAR-TcR V domain the T cell signaling machinery is directed to induce cytokine secretion or cell killing. Quaternary structure modeling predicts that the TcRδV CDR3 might be occluded by the NAR-TcRV domain, which would replace the TcR γ/δ Vs in antigen recognition. Like other Igs, IgNAR has both transmembrane and secreted forms, and its gene undergoes extensive somatic mutation after antigenic stimulation. NAR-TcR neither has a secreted form (single band by Northern blotting and absence of alternative splicing from sequence data) nor is somatically mutated (extensive sequence analysis of particular families); thus, although the NAR-TcRV sequence is most related to IgNARV, its other basic characteristics are the same as those of TcRs.
Hallmark transmembrane charged residues of TcRδC, as well as its short cytoplasmic tail void of obvious signaling potential, predict that the NAR-TcR δ chain is part of a γ/δ/CD3 complex for cell surface expression and signal transduction. This notion is consistent with the short cytoplasmic tails found on IgNAR and other antigen receptor chains from cartilaginous fish to man. CD3 and CD79 genes have been found in diverse vertebrate groups, including the amphibian Xenopus and fish Takifugu (36, 37), and we predict that these signaling chains will be found in elasmobranchs. Thus, it is likely that the evolutionarily mobile NAR V domain signals via CD79 orthologues on B cells (as IgNAR) and CD3 orthologues on T cells (as NAR-TcRδ).
An IgNAR VDJ cluster may have recombined with TRDV genes sometime early in the evolution of modern sharks, 200 million years ago (Fig. 5). The original NTV-DV gene cluster then duplicated many times and is apparently used to a different extent in different shark taxa (as exemplified by fewer bands in the horn shark digests in Fig. 3B). We previously suggested that an IgNAR VDJ cluster recombined with an IgW cluster (an extant elasmobranch Ig isotype), which then gave rise to the IgNAR isotype gene, a possibility that is supported by these data (16). The origins of the NAR V domain is a mystery (38), because it does not show high similarity to any of the known antigen receptor V domains; the simplest interpretation is that the NAR V arose as a gene encoding a single domain, which was distributed to different antigen receptor families (Fig. 5).
Lateral gene transfer and endosymbiosis has forced major reconsideration of prokaryotic evolution (39). Horizontal gene transfer mechanisms have occurred in eukaryotes (40) but are often excluded from evolutionary hypotheses in favor of simpler Darwinian vertical-descent models. The concept of lateral gene flow is also a common theme for evolutionary immunologists, in which the demonstrable transposase activity of the recombination-activating gene products suggested an invading transposon in antigen receptor evolution (41, 42). The NAR domain now requires consideration as a nomadic gene shuttled intragenomically by means of conventional recombination. The exon-shuffling theory asserts that exon-encoded domains facilitated evolutionary swapping and experimentation of functional protein domains (43), and the best empirical evidence for this theory has been obtained from analysis of exon–intron structure of complex eukaryotic genes in relation to the proteins they encode (44). The use of the NAR domain in two very different antigen receptors on two very different lymphocytes is the most explicit example of the exon-shuffling theory of which we are aware.
Comparative antigen receptor studies throughout vertebrates have thus far shown contrasting natural histories for Ig and TcR. Although Ig have used various genomic arrangements, isotypes, and somatic diversification mechanisms in different vertebrates to achieve repertoire diversification (reviewed in ref. 45), TcR studies have yielded relatively few surprises (19). This dichotomy has been linked to the different stringencies required for function as secreted binders of free antigen and recognition of peptide presented by MHC molecules (46). However, because most γ/δ T cells are not MHC-restricted (47), they may have been afforded evolutionary freedom more similar to that of the direct binding B cell receptors (Igs). Therefore, perhaps it is not surprising that a mobile genetic element encoding an antigen-binding domain would thrive on TcRδ, giving it an additional tool for antigen recognition by means of a protruding single domain in addition to the planar recognition landscape possible with the TcRδ and TcRγ V domains. The NAR-TcR on a clonally expandable T cell could conceivably recognize cell-bound antigen of fungi, parasites, or virally infected cells and direct cellular cytotoxicity in a manner akin to antibody-dependent cellular cytotoxicity by natural killer cells through the TcR instead of Ab and FcR.
Materials and Methods
RACE PCR.
5′ RACE PCR products were amplified by using TcRδC primer (5′-GCTGGCCAGACAGACTGCAGCTTGGACAGC-3′) and nested TcRδC primer (5′-TTGGTGGATAAAAAGCCGAC-3′) from adult shark PBLs, spleen, and thymus total RNA and then separated in 1% agarose and visualized with ethidium bromide. The SMART RACE system (BD Biosciences) was used according to the manufacturer's protocol with 2 × 20 cycles annealing at 68°C. All products were cloned into pCR2.1 with TA Cloning kit (Invitrogen) and sequenced by the University of Maryland's Biopolymer Core facility.
RT-PCR.
Clones in Fig. 4A ending in 810 and incompletely rearranged cDNA clones in Fig. 4B were gathered with NAR-TcR to TcRδC RT-PCR by using the nonnested δC primer (Fig. 1A) and primers specific for NAR-TcRV1 (5′-ACGCGTGCAGCGAAGGAGTGG-3′), NAR-TcRV2 (5′-CATCCCTATCATTATTTTAGGAGT-3′), NAR-TcRV3 (5′-TTCAAACAAAGCCAGACAGGATCAGAG-3′), and NAR-TcRV4 (5′-CTCAGGCAAACCCAGACCAAGACAGAC-3′). Oligo-dT-primed cDNA was made from 5 μg of RNA as described and used as template for PCR amplification (14).
Genomic PCR.
PCR from genomic DNA isolated from shark erythrocytes using described NAR-TcRV primers (Fig. 2A) and δV1 (5′-TCTCTGCTCTCCTGAAATGTCTGT-3′) and δV2 (5′-TCTCTGCTCTCCTGAAATTTCTGT-3′) reverse primers shows the arrangement of two entire NAR-TcRV clusters. One microgram of genomic DNA was used as template in a PCR of five cycles annealing at 50°C followed by 25 cycles annealing at 54°C.
Northern and Southern Blotting.
Total RNA was prepared for Northern blotting as described (48), and 15 μg was loaded for each lane. All probes were labeled with 32P dCTP PCR as described (49), and free nucleotides were removed with Quick-Spin Sephedex G-50 columns (Roche). Probes were amplified by using primers to include most of the V segment (NAR-TcRV1, 5′-GACACAGAGCCCGGCAACGGTG-3′ and 5′-GTGCTTGATTCGTGGACAC-3′; IgNARV7A, 5′-GCTCGAGTGGACCAAACA-3′ and 5′-ACCGCAACGATACGTGCCAC-3′; TcRδC, 5′-TTAAGCCGAAATTGTCGGCT-3′ and 5′-AGAAATCCAGACTCGGGCAG-3′) or the loading control nucleotide diphosphate kinase (5′-AACAAGGAACGAACCTTC-3′ and 5′-TCACTCATAGATCCAGTC-3′). Probes routinely labeled to 3 × 107 cpm. Southern blot was performed on HindIII-digested and PstI-digested (Roche) genomic DNA as described (50). Organisms included are Pacific hagfish, spotted ratfish, nurse shark, horned shark, sand tiger shark, lemon shark, cownose ray, little skate, Japanese pufferfish, African clawed frog, and human. Final low-stringency wash conditions were 20-min agitation in 2× SSC/0.1% SDS at 55°C, and final high-stringency wash conditions were 20-min agitation in 0.2× SSC/0.1% SDS at 65°C.
Phylogenetic Analysis.
Multiple alignment of 78 chordate V domains, with emphasis on IgH, IgL, and the four TcR from diverse groups of vertebrates was performed in clustalx (51) and then trimmed to 114 informative columns (see Fig. 7, which is published as supporting information on the PNAS web site). The phylip suite was used for subsequent analysis. A distance matrix was created with protdist and used to draw a phylogram with neighbor. Bootstrap values after 1,000 iterations generated by seqboot are shown.
Supplementary Material
Acknowledgments
We thank Yuko Ota for RNA preparation; Helen Dooley and Robin Stanfield for helpful discussions of NAR-TcR structure; Becky Lohr for the identification of the nurse shark TcRδC domain gene; and J. Cerny, M. Diaz, L. Du Pasquier, and D. Nemazee for critique of the manuscript. The work was supported by grants from the National Institutes of Health (to M.F.F. and M.F.C.).
Abbreviations
- TcR
T cell receptor
- RSS
recombination signal sequence
- CDR
complementarity-determining region
- PBL
peripheral blood leukocyte.
Footnotes
References
- 1.Rast J. P., Anderson M. K., Strong S. J., Luer C., Litman R. T., Litman G. W. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 2.Marchalonis J., Edelman G. M. J. Exp. Med. 1965;122:601–618. doi: 10.1084/jem.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchalonis J., Edelman G. M. Science. 1966;154:1567–1568. doi: 10.1126/science.154.3756.1567. [DOI] [PubMed] [Google Scholar]
- 4.Harding F. A., Amemiya C. T., Litman R. T., Cohen N., Litman G. W. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenberg A. S., Avila D., Hughes M., Hughes A., McKinney E. C., Flajnik M. F. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 6.Greenhalgh P., Steiner L. A. Immunogenetics. 1995;41:54–55. doi: 10.1007/BF00188438. [DOI] [PubMed] [Google Scholar]
- 7.Kasahara M., McKinney E. C., Flajnik M. F., Ishibashi T. Eur. J. Immunol. 1993;23:2160–2165. doi: 10.1002/eji.1830230917. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto K., Nakanishi T., Kurosawa Y. Proc. Natl. Acad. Sci. USA. 1992;89:2209–2212. doi: 10.1073/pnas.89.6.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flajnik M. F., Rumfelt L. L. Curr. Top. Microbiol. Immunol. 2000;248:249–270. doi: 10.1007/978-3-642-59674-2_11. [DOI] [PubMed] [Google Scholar]
- 10.Hinds K. R., Litman G. W. Nature. 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 11.Eason D. D., Litman R. T., Luer C. A., Kerr W., Litman G. W. Eur. J. Immunol. 2004;34:2551–2558. doi: 10.1002/eji.200425224. [DOI] [PubMed] [Google Scholar]
- 12.Rast J. P., Litman G. W. Proc. Natl. Acad. Sci. USA. 1994;91:9248–9252. doi: 10.1073/pnas.91.20.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi K., Tomonaga S. Mol. Immunol. 1988;25:115–120. doi: 10.1016/0161-5890(88)90058-2. [DOI] [PubMed] [Google Scholar]
- 14.Rumfelt L. L., Avila D., Diaz M., Bartl S., McKinney E. C., Flajnik M. F. Proc. Natl. Acad. Sci. USA. 2001;98:1775–1780. doi: 10.1073/pnas.98.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanfield R. L., Dooley H., Flajnik M. F., Wilson I. A. Science. 2004;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg A. S., Hughes A. L., Guo J., Avila D., McKinney E. C., Flajnik M. F. Eur. J. Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 17.Roux K. H., Greenberg A. S., Greene L., Strelets L., Avila D., McKinney E. C., Flajnik M. F. Proc. Natl. Acad. Sci. USA. 1998;95:11804–11809. doi: 10.1073/pnas.95.20.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dooley H., Flajnik M. F. Eur. J. Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 19.Charlemagne J., Fellah J. S., De Guerra A., Kerfourn F., Partula S. Immunol. Rev. 1998;166:87–102. doi: 10.1111/j.1600-065x.1998.tb01255.x. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Lesk A. M., Chothia C. J. Mol. Biol. 1982;160:325–342. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- 22.Streltsov V. A., Varghese J. N., Carmichael J. A., Irving R. A., Hudson P. J., Nuttall S. D. Proc. Natl. Acad. Sci. USA. 2004;101:12444–12449. doi: 10.1073/pnas.0403509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison T. J., Winter C. C., Fournie J. J., Bonneville M., Garboczi D. N. Nature. 2001;411:820–824. doi: 10.1038/35081115. [DOI] [PubMed] [Google Scholar]
- 24.Max E. E., Seidman J. G., Leder P. Proc. Natl. Acad. Sci. USA. 1979;76:3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakano H., Huppi K., Heinrich G., Tonegawa S. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 26.Litman G. W., Anderson M. K., Rast J. P. Annu. Rev. Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 27.Rumfelt L. L., Diaz M., Lohr R. L., Mochon E., Flajnik M. F. J. Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]
- 28.Garboczi D. N. Science. 2005;308:209–210. doi: 10.1126/science.1111657. [DOI] [PubMed] [Google Scholar]
- 29.Hawke N. A., Rast J. P., Litman G. W. J. Immunol. 1996;156:2458–2464. [PubMed] [Google Scholar]
- 30.Pancer Z., Mayer W. E., Klein J., Cooper M. D. Proc. Natl. Acad. Sci. USA. 2004;101:13273–13278. doi: 10.1073/pnas.0405529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T., Shin I., Fujiyama A., Kohara Y., Kasahara M. J. Immunol. 2005;174:2885–2891. doi: 10.4049/jimmunol.174.5.2885. [DOI] [PubMed] [Google Scholar]
- 32.Cannon J. P., Haire R. N., Litman G. W. Nat. Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- 33.Dietrich J., Cella M., Seiffert M., Buhring H. J., Colonna M. J. Immunol. 2000;164:9–12. doi: 10.4049/jimmunol.164.1.9. [DOI] [PubMed] [Google Scholar]
- 34.Yoder J. A., Mueller M. G., Wei S., Corliss B. C., Prather D. M., Willis T., Litman R. T., Djeu J. Y., Litman G. W. Proc. Natl. Acad. Sci. USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chretien I., Robert J., Marcuz A., Garcia-Sanz J. A., Courtet M., Du P. L. Eur. J. Immunol. 1996;26:780–791. doi: 10.1002/eji.1830260409. [DOI] [PubMed] [Google Scholar]
- 36.Guselnikov S. V., Najakshin A. M., Taranin A. V. Immunogenetics. 2003;55:472–479. doi: 10.1007/s00251-003-0599-0. [DOI] [PubMed] [Google Scholar]
- 37.Guselnikov S. V., Bell A., Najakshin A. M., Robert J., Taranin A. V. Dev. Comp. Immunol. 2003;27:727–733. doi: 10.1016/s0145-305x(03)00055-7. [DOI] [PubMed] [Google Scholar]
- 38.Richards M. H., Nelson J. L. Mol. Biol. Evol. 2000;17:146–155. doi: 10.1093/oxfordjournals.molbev.a026227. [DOI] [PubMed] [Google Scholar]
- 39.Rivera M. C., Lake J. A. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 40.Benveniste R. E., Todaro G. J. Nature. 1974;252:456–459. doi: 10.1038/252456a0. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal A., Eastman Q. M., Schatz D. G. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 42.Hiom K., Melek M., Gellert M. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert W. Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 44.Liu M., Grigoriev A. Trends Genet. 2004;20:399–403. doi: 10.1016/j.tig.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Flajnik M. F. Nat. Rev. Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- 46.Eason D. D., Cannon J. P., Haire R. N., Rast J. P., Ostrov D. A., Litman G. W. Semin. Immunol. 2004;16:215–226. doi: 10.1016/j.smim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Jameson J., Witherden D., Havran W. L. Curr. Opin. Immunol. 2003;15:349–353. doi: 10.1016/s0952-7915(03)00045-1. [DOI] [PubMed] [Google Scholar]
- 48.Bartl S., Baish M. A., Flajnik M. F., Ohta Y. J. Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- 49.Mertz L. M., Rashtchian A. Anal. Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- 50.Greenberg A. S., Steiner L., Kasahara M., Flajnik M. F. Proc. Natl. Acad. Sci. USA. 1993;90:10603–10607. doi: 10.1073/pnas.90.22.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hata S., Satyanarayana K., Devlin P., Band H., McLean J., Strominger J. L., Brenner M. B., Krangel M. S. Science. 1988;240:1541–1544. doi: 10.1126/science.3259726. [DOI] [PubMed] [Google Scholar]
- 53.Elliott J. F., Rock E. P., Patten P. A., Davis M. M., Chien Y. H. Nature. 1988;331:627–631. doi: 10.1038/331627a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.