Abstract
Targeting tyrosine kinase receptors (RTKs) with specific Abs is a promising therapeutic approach for cancer treatment, although the molecular mechanism(s) responsible for the Abs’ biological activity are not completely known. We targeted the transmembrane RTK for hepatocyte growth factor (HGF) with a monoclonal Ab (DN30). In vitro, chronic treatment of carcinoma cell lines resulted in impairment of HGF-induced signal transduction, anchorage-independent growth, and invasiveness. In vivo, administration of DN30 inhibited growth and metastatic spread to the lung of neoplastic cells s.c. transplanted into immunodeficient nu/nu mice. This Ab efficiently down-regulates HGF receptor through a molecular mechanism involving a double proteolytic cleavage: (i) cleavage of the extracellular portion, resulting in “shedding” of the ectodomain, and (ii) cleavage of the intracellular domain, which is rapidly degraded by the proteasome. Interestingly, the “decoy effect” generated by the shed ectodomain, acting as a dominant negative molecule, enhanced the inhibitory effect of the Ab.
Keywords: Ab, metastasis, tyrosine kinase, receptor degradation, proteolytic cleavage
Scientific exploration of cancer immunotherapy began in the 1950s, and the first application relied on polyclonal antibodies. Today, after >5 decades, immunotherapy with mAbs continues to offer a promising alternative for cancer treatment (1, 2). Several antibodies targeting tyrosine kinase receptors (RTKs) are currently used in clinical practice (3), even if their mechanism of action is still poorly understood (4). Bevacizumab and Cetuximab target VEGF-VEGFR and EGF-EGFR respectively, and act by preventing ligand–receptor interaction (5, 6). The mechanism of action of Trastuzumab, a mAb specific for HER2 (a member of the EGFR family) is not completely clear, but it promotes HER2 degradation, thus decreasing receptor levels at the surface of tumor cells (7).
The MET oncogene, encoding the RTK for hepatocyte growth factor (HGFR), controls genetic programs leading to cell growth, invasion, and protection from apoptosis. Deregulated activation of HGFR is critical not only for the acquisition of tumorigenic properties but also for the achievement of the invasive phenotype (8). The role of MET in human tumors emerged from several experimental approaches and was unequivocally proved by the discovery of MET-activating mutations in inherited forms of carcinomas (9, 10). HGFR constitutive activation is frequent in sporadic cancers, and studies from this and other laboratories have shown that the MET oncogene is overexpressed in tumors of specific histotypes or is activated through autocrine mechanisms (for a list see www.vai.org/vari/metandcancer). Besides, the MET gene is amplified in hematogenous metastases of colorectal carcinomas (11). Interfering with MET activation is, thus, becoming a challenging approach to hamper the tumorigenic and metastatic processes. In the past years, several strategies have been proposed to block aberrant HGFR signaling, targeting either the HGFR itself or its ligand. These approaches include the use of HGF antagonists, HGF neutralizing antibodies, HGFR decoys, ATP-binding-site inhibitors of HGFR, or small molecules, such as geldanamycin, SH2-domain polypeptides, and ribozymes (reviewed in ref. 12). Although many of these approaches are attractive, their clinical application still remains elusive, mainly due to problems in efficient delivery.
In this work, we show that a monoclonal Ab directed against the extracellular domain of HGFR, is able to promote receptor down-regulation; the underlying molecular mechanism is different from that induced by ligand binding, and it involves proteolytic cleavage of the receptor, resulting in HGFR ectodomain release from the cell surface (“shedding”) and generation of the intracellular domain, which is rapidly degraded by the proteasome. As a consequence, Ab-induced receptor down-regulation impairs HGFR-activated signal transduction, abolishes the invasive growth response in vitro, and interferes with the tumorigenic and metastatic potential of cancer cells in vivo.
Results
The DN30 Ab Impairs HGFR Signal Transduction.
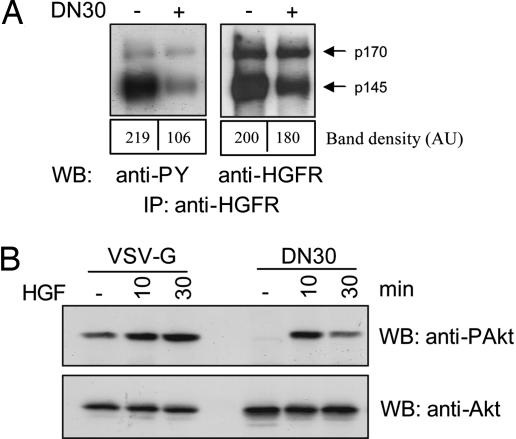
DN30 is a mAb directed against the extracellular domain of HGFR, where it recognizes an epitope distinct from that bound by the ligand (13). Previous work has shown that DN30 behaves as a partial agonist, because it induces phosphorylation of the receptor but is unable to trigger the whole set of downstream biological effects (13). To address the question of whether the DN30 mAb might represent a tool to interfere with constitutive HGFR activation, we first analyzed its biochemical and biological activity in tumor cells chronically exposed to the mAb. As a model, we used a human gastric carcinoma cell line (GTL16), where HGFR, as in many naturally occurring tumors, is overexpressed and constitutively activated (14). As shown in Fig. 1A, DN30 treatment induced a significant reduction in HGFR levels and tyrosine phosphorylation. We then analyzed the effect of this mAb on HGFR signal transduction. Because HGFR is known to promote a strong antiapoptotic program by stimulating Akt activation, we evaluated the level of Akt phosphorylation upon treatment with DN30 or an irrelevant isotype-matched Ab (VSV-G), as a control. As shown in Fig. 1B, Akt phosphorylation was inhibited in both basal condition and HGF-stimulated cells.
Fig. 1.
DN30 impairs HGFR activation and signal transduction. (A) Evaluation of HGFR activation. GTL16 cells were exposed to DN30 for 4 h. HGFR was immunoprecipitated from cell lysates, and Western blots were probed with the indicated Abs. The upper band corresponds to intracellular HGFR precursor (p170); the lower band (p145) is the mature form. DN30 treatment resulted in a decrease of receptor activation more pronounced than receptor down-regulation, as indicated by band density quantification. (B) Analysis of HGFR signaling. Cells were pretreated with either VSV-G or DN30 and stimulated with HGF for the indicated times. Akt phosphorylation was evaluated in total cell lysates. As shown, DN30 reduced both basal and HGF-induced Akt activation.
DN30 Inhibits the Transformed Phenotype of Cancer Cells in Vitro.
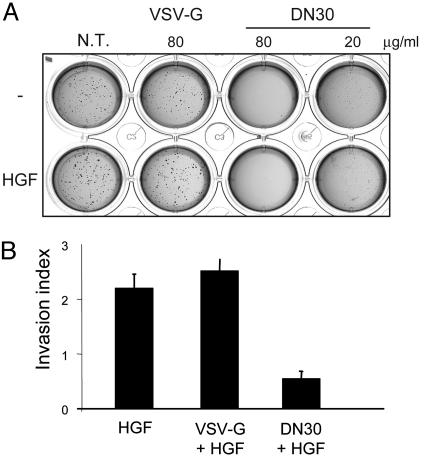
The effect of this mAb on the transformed phenotype was assessed by measuring the ability of cells to grow in the absence of anchorage and invade extracellular matrices. Anchorage-independent growth depends on the ability of cells to overcome apoptosis due to lack of anchorage, the so-called “anoikis” (15), and can be analyzed by evaluating the capability of cells to grow in soft agar. Because many reports have shown that HGFR activation is able to protect cells from anoikis (16–18), we seeded GTL16 cells in 0.5% agar and maintained the culture in the presence or absence of different amounts of DN30 or the control VSV-G mAb. Whereas VSV-G-treated and -untreated cells were able to form numerous colonies, DN30 drastically inhibited anchorage-independent growth of cancer cells in a dose-dependent manner (Fig. 2A). On the contrary, no difference in the ability of cells to grow in conditions of anchorage dependency was observed upon mAb treatment (see Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 2.
DN30 inhibits the transformed phenotype of cancer cells in vitro. (A) Anchorage-independent growth of GTL16 cells. Cells pretreated with either DN30 or VSV-G for 48 h were seeded in 0.5% agar and maintained in the presence of the indicated amounts of Abs with or without HGF (20 ng/ml). Anchorage-independent growth was drastically inhibited in the presence of DN30, even at low doses. (B) Invasion assay. MDA-MB-435 β4 cells were pretreated with the indicated antibodies for 24 h before seeding on a Matrigel-coated Transwell chamber. The lower chamber was filled with DMEM/2% FBS plus 100 ng/ml HGF. After 24 h, migrated cells were stained and counted. Invasive capacity in response to HGF is expressed as fold increase compared with nonstimulated cells. As shown, DN30 treatment significantly impaired cell invasion.
To evaluate the ability of the Ab to interfere with cell invasiveness, we studied MDA-MB-435β4, a mammary carcinoma cell line that represents a suitable model to study the invasive ability of cells in response to HGF (19). As shown in Fig. 2B, in vitro treatment of these cells with DN30 reduced the invasive properties in response to HGF.
DN30 Inhibits the Transformed Phenotype in Vivo.
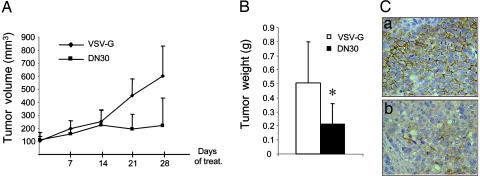
To assess the activity of DN30 on tumor growth, we inoculated s.c. GTL16 cells into the posterior flank of immunodeficient nu/nu female mice. The animals were treated twice a week with either DN30 or VSV-G, injected in situ, in the tumor (2 μg/g). The therapy started upon tumor appearance; animals bearing tumors of comparable size were treated for 4 weeks. Tumor volume was monitored during treatment, and a substantial decrease in growth was observed in DN30-treated mice (Fig. 3A). At the end of the experiment, mice were autopsied, and tumors were excised and weighed. In mice treated with DN30, tumors were significantly smaller than in controls (Fig. 3B). In these tumors, HGFR activation, shown by staining with specific antibodies against the phosphorylated form of the receptor, was reduced (Fig. 3C). Impairment of tumor growth was mainly due to increased apoptotic rate (see Fig. 11 A and B, which is published as supporting information on the PNAS web site).
Fig. 3.
DN30 inhibits tumor growth in vivo. (A and B) Tumorigenesis assay. Nude mice were injected s.c. with 1.5 × 106 GTL16 cells. After tumor appearance, mice displaying tumors of the same size were selected and then injected in situ in the tumor twice a week with 2 μg/g of either VSV-G or DN30. (A) Tumor volume was measured at different time points. Mice were killed after 4 weeks of treatment, and tumor weight was evaluated (B). In mice treated with DN30, tumors were significantly smaller than in control mice (P < 0.05). (C) Evaluation of HGFR activation. Tumor sections from mice treated with VSV-G (a), or DN30 (b) were stained with anti-human phospho-HGFR. HGFR activation was strongly decreased in mice treated with DN30. Magnification ×40.
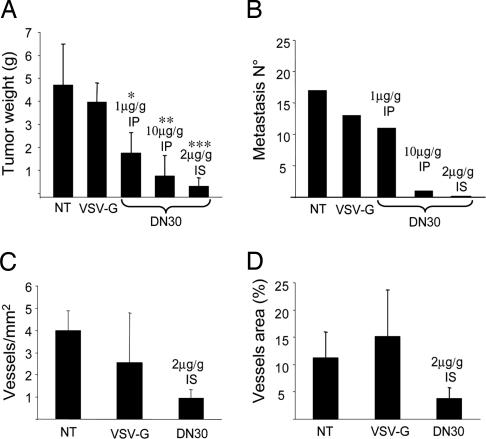
We performed the same kind of experiments on MDA-MB-435 β4 cells, a model system for in vivo spontaneous metastasis (19). Tumor-bearing animals were treated twice a week with different doses of DN30 or the control mAb, administered either systemically (1 μg/g or 10 μg/g i.p.) or into the tumor (2 μg/g in situ). The therapy started at the day of transplantation and was carried out for 8 weeks. After treatment, mice were autopsied, and analysis of primary tumors and lungs was performed. Spleen, bone marrow, liver, heart, bone, and kidney were also examined to rule out potential toxic effects. Macroscopic analysis showed that DN30 treatment resulted in growth inhibition of the primary tumor mass (Fig. 4A; and see Fig. 12 A–E, which is published as supporting information on the PNAS web site). Immunohistochemical staining with antibodies recognizing the tyrosine-phosphorylated form of HGFR showed, also in this case, a marked reduction of receptor activation (Fig. 12 F–J). Microscopic analysis of the lung sections revealed that both intratumor injection and systemic administration of DN30 prevented the appearance of distant metastases in the lung and in the other inspected organs (Fig. 4B).
Fig. 4.
DN30 treatment interferes with tumor progression in vivo. (A) Nude mice were inoculated s.c. with 2.5 × 106 MDA-MB-435 β4 cells and treated with the indicated doses of VSV-G or DN30, administered i.p. (IP), or in situ (IS). As shown, DN30 inhibited tumor growth. (B) Analysis of lung metastases. Metastases were counted by microscopic observation of the lung sections after hematoxylin/eosin staining. A dose-dependent reduction of metastases number was evident in DN30-treated mice. (C and D) Evaluation of tumor vascularization. Blood vessels staining on tumor histological sections was performed with an anti-mouse CD31 Ab. Number and area of vessels were evaluated by fluorescence microscopy. As shown, both the number and the area of vessels were reduced in response to DN30 treatment.
Because many works have shown that HGF is a potent angiogenic factor and that HGFR signaling contributes to tumor angiogenesis (20–23), we analyzed tumor vascularization upon DN30 treatment. In these tumors, we found a significant reduction of the number of vessels and of their area (Fig. 4 C and D). However, because DN30 mAb does not bind to mouse HGFR with high affinity (Fig. 12K), the observed result is due to an indirect effect of the mAb on tumor cells. It is known that HGFR promotes angiogenesis by inducing release of VEGF and of other angiogenic factors (20–23). Blocking HGFR activation in tumor cells can, thus, abrogate the release of these angiogenic factors.
DN30 Induces HGFR Down-Regulation.
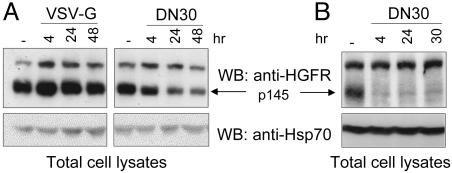
To study the mechanism through which DN30 interferes with HGFR activation, we treated GTL16 cells with either DN30 or VSV-G and analyzed receptor levels. As shown in Fig. 5A, the total amount of HGFR decreased in a time-dependent manner upon DN30 but not VSV-G treatment, suggesting that the anti-HGFR mAb specifically induced receptor down-regulation. It is interesting to emphasize that, in these cells, the ligand HGF was, instead, unable to induce receptor down-regulation (Fig. 6Lower).
Fig. 5.
DN30 induces HGFR down-regulation. GTL16 cells (A) and MDA-MB-435 β4 (B) were treated with DN30 for the indicated times. Equal amounts of total cell lysates were processed for Western blotting and probed with anti-HGFR or, as a loading control, with anti-Hsp70 antibodies. As shown, DN30 induced HGFR down-regulation in both overexpressing cells (GTL16) and in cells expressing normal levels of HGFR (MDA-MB-435 β4).
Fig. 6.
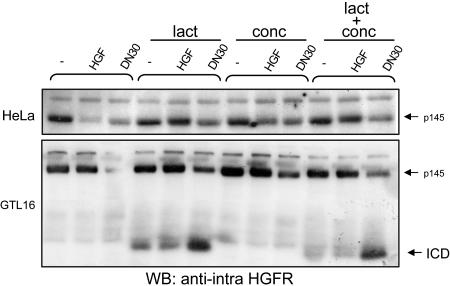
Ab-induced and ligand-dependent down-regulation exploit different pathways. HeLa (Upper) and GTL16 (Lower) cells were pretreated with lactacystine (lact), concanamycin (conc), or both for 2 h before treatment with HGF or DN30. HGFR down-regulation was evaluated on total cell lysates. In the presence of the proteasome inhibitor (lact), ligand-induced HGFR down-regulation was impaired, whereas Ab-induced was not. In this condition, a 60-kDa fragment [intracellular domain (ICD)], detectable by an Ab directed against the intracellular portion, accumulated in cells.
We then verified whether the DN30 mAb could trigger receptor down-regulation in cells expressing normal levels of HGFR (MDA-MB-435 β4). As shown in Fig. 5B, also in these cells, DN30 efficiently down-regulated HGFR. MAb-induced reduction of HGFR exposed at the cell membrane was also evaluated by cytofluorimetric analysis, which showed that mAb treatment reduced the amount of HGFR expressed at the cell surface with an efficiency higher than the cognate ligand HGF (see Fig. 13, which is published as supporting information on the PNAS web site). A similar reduction, in the same assay, was observed also in GTL16 cells (data not shown).
Molecular Mechanism of DN30-Induced HGFR Down-Regulation.
Ligand-dependent and Ab-induced down-regulation may follow different pathways. Ligand-dependent down-regulation of RTKs is a multistep process including internalization, ubiquitinylation, endosomal sorting, and, finally, lysosomal or proteasomal degradation (24). To assess which degradation pathway is involved in mAb-induced HGFR down-regulation, we blocked the activity of either the lysosome or the proteasome by using specific inhibitors (concanamycin and lactacystin/MG132, respectively) before mAb stimulation. Surprisingly, although inhibition of the proteasomal pathway severely impaired ligand-induced HGFR degradation, it did not affect receptor down-regulation due to DN30 treatment (Fig. 6; and see Fig. 14A, which is published as supporting information on the PNAS web site), thus indicating that this mAb and HGF promote HGFR down-regulation through different mechanisms. When proteasome activity was impaired, a fragment of 60 kDa, barely detectable in basal condition, was heavily accumulated in cells upon DN30 treatment (Figs. 6 and 14 A and B). This fragment was detectable on Western blots with an anti-intracellular HGFR Ab and consisted in the cytoplasmic domain of the receptor [intracellular domain (ICD)]. As expected for molecules committed to proteasomal degradation, the 60-kDa fragment was tagged with ubiquitin moieties (Fig. 14C).
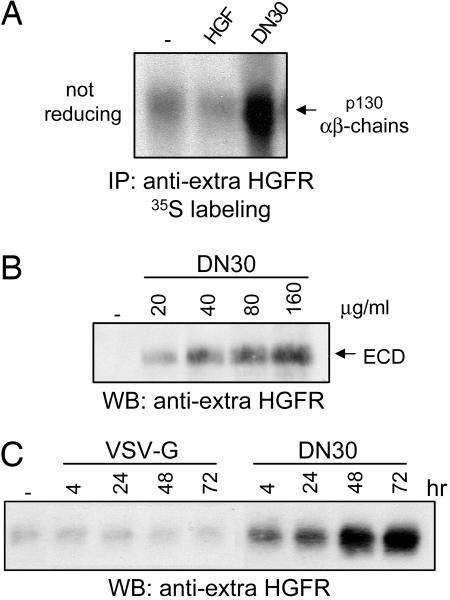
Because the extracellular domain (ectodomain) of the receptor was not detectable in cell lysates upon DN30 treatment, we verified whether it was released outside the cells upon cleavage, a process known as shedding (25). To test this hypothesis, we looked for the presence of HGFR ectodomain in cell culture medium. As shown in Fig. 7A, from culture media of metabolically labeled cells, we immunoprecipitated a band showing, under nonreducing conditions, an apparent molecular mass of 130 kDa (consistent with the complex of the extracellular αβ-chains); when the samples were analyzed under reducing conditions, the complex was resolved in the two bands of 80 kDa (β-chain) and 45 kDa (α-chain) (see Fig. 15A, which is published as supporting information on the PNAS web site). Notably, whereas HGF stimulation did not enhance receptor shedding, this process was dramatically increased upon DN30 treatment. According to previous data (26, 27), a slight amount of HGFR ectodomain was basally present in the cell-conditioned media. DN30 was able to promote shedding of the HGFR extracellular domain not only in GTL16 and HeLa cells but in all of the cell lines tested expressing the endogenous receptor (Fig. 15B) as well as in those where we exogenously expressed it (Fig. 15C).
Fig. 7.
DN30 induces proteolytic cleavage of HGFR and shedding of the extracellular domain (ECD). (A) Supernatants obtained from metabolically labeled GTL16 cells were collected and immunoprecipitated with an anti-HGFR Ab directed against the extracellular domain. As shown, DN30, but not HGF, induced shedding of HGFR ectodomain. (B and C) HGFR shedding is dose- and time-dependent. Cells were stimulated either with increasing amounts of DN30 or for different times.
By treating cells with increasing amounts of Ab for 4 h or for different lengths of time, we showed that mAb-mediated HGFR shedding was specific (not observed with VSV-G) and dose- (Fig. 7B) and time-dependent (Fig. 7C). Unlike ligand-induced HGFR down-regulation, ectodomain shedding did not require clathrin-dependent endocytosis (Fig. 15D).
It is interesting to note that the ability to induce HGFR ectodomain shedding is not shared by all HGFR-specific mAbs. DO24, a HGFR mAb characterized in ref. 13, is able to induce receptor down-regulation but not shedding (Fig. 15E). Notably, DO24 is a full agonist mAb that does not impair HGFR activation.
HGFR Activation Is Not Required for Ab-Induced Shedding.
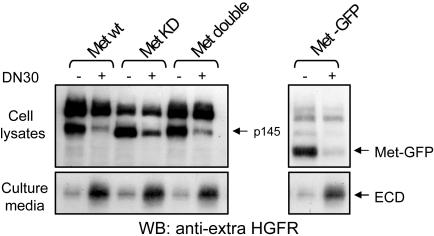
As we have reported, a complex containing endophilin, CIN85, and Cbl mediates ligand-dependent down-regulation of HGFR (28). This complex is recruited to the receptor upon HGF-induced activation and promotes receptor endocytosis, ubiquitinylation, and degradation. For the accomplishment of this process, both the kinase activity of the receptor and its ability to recruit intracellular transducers are required (29). To verify whether this is required also for mAb-induced down-modulation and shedding, we prompted the ability of DN30 to down-regulate various HGFR mutants. We expressed in COS-7 cells either WT HGFR or the following mutants: MET KD, encoding a receptor devoid of tyrosine kinase activity due to a Lys-Ala substitution in the ATP binding pocket (30); MET “double,” encoding a HGFR lacking the docking tyrosines Y1349 and Y1356 (30); and MET-GFP, a dominant-negative mutant, where the sequence encoding the whole intracellular domain of the receptor was replaced by the GFP sequence (31). Forty-eight hours after transfection, cells were treated with DN30. Unexpectedly, DN30 was able to trigger down-regulation and induce HGFR shedding in all of the mutants (Fig. 8). This experiment suggests that DN30-induced HGFR down-regulation does not require receptor kinase activity or the recruitment of cytoplasmic transducers and that the whole intracellular domain is dispensable for the process. This further confirms that the Ab and the ligand activate different down-regulatory mechanisms.
Fig. 8.
Activation of signal transduction is not required for HGFR shedding. COS-7 cells were transfected with the indicated HGFR mutants and, 48 h later, were treated with DN30 for 4 h. Equal amounts of total cell lysates and conditioned media were processed for Western blotting. As shown, DN30 was able to induce down-regulation and ectodomain (ECD) shedding of all HGFR mutants. HGFR mutants: Met KD, HGFR kinase dead; Met Double, HGFR mutant lacking the docking tyrosines 1349/1356; Met-GFP, HGFR mutant where the whole intracellular portion was replaced by the GFP sequence.
The HGFR Shed Ectodomain Behaves as a Decoy Receptor.
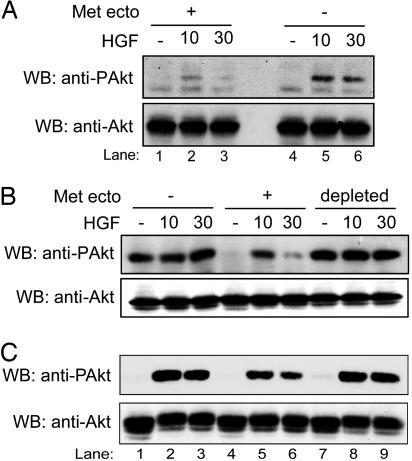
Because we have shown that an engineered extracellular domain of the HGFR can effectively function as a dominant negative decoy molecule (32), we tested the ability of the ectodomain shed upon DN30 treatment to inhibit HGFR signaling. Cells were stimulated for different times with HGF either in the presence (Fig. 9A, lanes 1–3) or in the absence (Fig. 9A, lanes 4–6) of HGFR ectodomain (obtained by pretreating cells with DN30 for 72 h), and Akt activation was assessed. As shown, in the presence of HGFR ectodomain, HGF-triggered Akt phosphorylation was strongly impaired. To prove that this impairment was, indeed, due to a decoy effect, we cleared the ectodomain out of the medium by multiple immunoprecipitation cycles before stimulating the cells with HGF. The depleted medium was no longer able to prevent HGFR activation (Fig. 9 B and C), thus supporting the idea that the shed fragment acts like a decoy.
Fig. 9.
The HGFR shed ectodomain behaves as a dominant negative molecule. (A) Cells pretreated for 72 h with DN30 were stimulated with HGF in either the presence (lanes 1–3) or absence (lanes 4–6) of the shed HGFR ectodomain in the culture medium. As shown, shed HGFR ectodomain impaired Akt activation. (B and C) GTL16 (B) and HeLa (C) cells were stimulated with HGF in the presence of control medium (lanes 1–3), medium containing HGFR ectodomain (lanes 4–6), or the same medium depleted of the shed ectodomain (lanes 7–9). As shown, the depleted medium was no longer able to prevent Akt activation.
Discussion
The HGFR encoded by the MET protooncogene is a RTK that, upon activation, elicits a complex spectrum of biological responses known as “invasive growth,” implying induction and coordination of cell proliferation, migration, differentiation, and survival. Under physiological conditions, this invasive growth program plays a pivotal role during embryo development, but, when unleashed in cancer, contributes to tumor progression and metastasis (33). The involvement of HGFR in human tumors is now firmly established, as germ-line missense mutations of the MET gene are responsible for some hereditary forms of cancer (9, 10), and inappropriate HGFR activation has been shown in most types of solid tumors, often correlating with poor prognosis (reviewed in ref. 34). The most frequent alteration in human cancers is receptor overexpression (33) that leads to constitutive dimerization and activation of the receptor, even in the absence of ligand (35). Increased HGFR expression can be due to (i) gene amplification, as in colorectal tumors, where MET confers to neoplastic cells a selective advantage for liver metastasis (11); (ii) enhanced transcription, induced by other oncogenes, such as Ras, Ret, and Ets (36–39); or (iii) hypoxia-activated transcription, leading to higher amounts of receptor that hypersensitize the cells to HGF and promote tumor invasion (40).
Two strategies are currently used in the clinical setting to interfere with RTKs: (i) treatment with small molecules inhibiting the tyrosine kinase activity; (ii) treatment with antibodies interfering with receptor activation. Very few HGFR tyrosine kinase inhibitors are currently available, and they are not highly specific for this kinase (41–43). Here, we describe an anti-HGFR mAb (DN30) inducing receptor down-regulation. As in the case of the HER2-specific mAb Trastuzumab (7), DN30 is very efficient in reducing receptor levels in cells where HGFR is overexpressed and, consequently, constitutively activated. Because overexpression is the most frequent alteration of MET in human tumors (34), our observations might have an impact for antineoplastic therapy. DN30-induced HGFR down-regulation leads to inhibition of receptor-mediated signal transduction and, in particular, of the Akt pathway, known to be involved in the antiapoptotic response. This finding is consistent with our observations, because we have shown that in vitro treatment with DN30 resulted in impairment of anchorage-independent growth, a property that requires the escape from apoptosis due to lack of anchorage. In vivo, we observed that tumors in animals treated with DN30 displayed an increased rate of apoptosis. On the other hand, we did not observe modification of cellular growth properties in response to the mAb, in agreement with the lack of inhibitory effect of DN30 on the activation of MAPK pathway (data not shown).
DN30-induced HGFR down-regulation is due to a mechanism different from that promoted by HGF, because it involves shedding of the extracellular portion of the receptor. The ectodomain of many membrane proteins, including growth factor receptors, can be released from the surface by a general shedding system activatable by protein kinase C (25, 44, 45). The proteases most commonly involved in this process are the α-secretases of the ADAM family (25). In an attempt to identify the enzyme responsible for HGFR shedding, we inhibited ADAMs and other Zn-dependent proteases, urokinase, acidic proteases, serine and cysteine proteases, and PKC, but, in all cases, receptor shedding was unaffected (data not shown; and see Supporting Text, which is published as supporting information on the PNAS web site), indicating that the enzyme responsible for HGFR ectodomain release is outside the list of the proteases usually involved in receptor shedding.
In this article, we provide evidence that DN30 is active in vivo, where it impairs tumor growth and formation of spontaneous metastases from cancer cells engrafted into nude mice. Our experiments suggest that these effects are HGFR-dependent and are mediated by the action of the Ab on cancer cells. We observed a significant reduction of intratumor neovascularization due to a decrease of the number of sprouting vessels of the microenvironment. Because DN30 does not bind to mHGFR, the effect on tumor vascularization is indirect and is likely due to the loss of release of angiogenic factors that usually follows HGFR activation in tumor cells. The HGFR extracellular domain shed from cancer cells can sequester active HGF, thus preventing activation of HGFR exposed on endothelial cells. Interestingly, Michieli et al. (32) obtained similar findings targeting HGFR by using a soluble receptor form (decoy Met) corresponding to the shed ectodomain produced upon Ab treatment.
It is worth noting that treatment with DN30 did not impact the functionality of different organs such as spleen, bone marrow, liver, heart, bone, and kidney, which did not show evident pathological alterations (data not shown) after long-term exposure to the Ab.
In conclusion, our results suggest Ab-induced down-regulation of HGFR as a candidate tool for immunotherapy, because down-regulation of growth factor receptors is considered a critical mechanism of signal attenuation (46, 47). This specific Ab exploits its effect in inhibiting HGFR signaling by a dual mechanism: On one hand it reduces the number of receptor molecules on the cell surface; on the other hand it promotes the release of a decoy HGFR which, according to our past (32) and present observations, is endowed with a dominant negative activity. Another important observation is that the inhibitory mechanism activated by the Ab does not require HGFR tyrosine kinase activity. This feature represents a relevant advantage in the perspective of a therapeutic approach, because, in clinical practice, it is frequent to combine different drugs to improve the effect on the target molecule. In the case of HGFR, it would thus be possible to combine kinase inhibitors with the Ab, allowing the contemporary action on both HGFR activation and levels that is likely to enhance the therapeutic efficacy of target therapy in HGFR-overexpressing tumors, with the aim of interfering with both tumor growth and the acquisition of an invasive–metastatic phenotype.
Materials and Methods
Reagents.
Anti-HGFR mAbs DN30, DO24, and DL21 were characterized in ref. 13. Other used Abs are anti-HGFR C12 (Santa Cruz Biotechnology), anti-human phospho-HGFR (Cell Signaling Technology), anti-ptyr PY20 (Transduction Laboratories), anti-ubiquitin (Babco), anti-Hsp70 (Stressgen), anti-phospho Akt (Ser-473, Cell Signaling Technology), anti-Akt (Santa Cruz Biotechnology), anti-mouse CD31 (Pharmingen), and anti-vesicular stomatitis virus (VSV-G, Sigma). Lactacystin, concanamycin, and MG132 were purchased from Calbiochem.
Down-Regulation Assay.
DN30 (80 μg/ml) and HGF (80 ng/ml) were added to serum-free DMEM. Where indicated, cells were preincubated for 2 h with either 10 μM lactacystin or 100 nM concanamycin. HGFR degradation was studied as described in ref. 28.
Metabolic Labeling and Analysis of HGFR Shedding.
Serum-starved cells were pulse-labeled with [35S]methionine and [35S]cysteine [100μCi/ml (1 Ci = 37 GBq)], Amersham Pharmacia) for 30 min and treated with DN30, VSV-G, or HGF for 4 h. Cell-conditioned media were collected and subjected to immunoprecipitation with anti-HGFR extracellular Ab.
In Vitro Biological Assays.
For evaluation of anchorage-independent growth, GTL16 was pretreated with either DN30 or VSV-G for 48 h. Then 1,500 cells per well were seeded in DMEM 2%/FBS 0.5% soft agar and maintained in the presence of the indicated amounts of Abs or HGF for 10 days. Grown colonies were visualized by staining with tetrazolium salt (48). As described in ref. 31, the invasion assay was performed in Transwell chambers (Corning) with 5 × 104 cells pretreated with either DN30 or VSV-G.
In Vivo Experiments.
The in vivo experiments were performed by inoculating s.c. either 1.5 × 106 GTL16 or 2.5 × 106 MDA-MB-435 β4 into the posterior flank of immunodeficient nu/nu female mice on Swiss CD1 background (Charles River Breeding Laboratories). Upon appearance of the tumor, mice bearing masses of comparable size were selected and inoculated either i.p. or in situ twice a week with the indicated amounts of mAbs. GTL16 and MDA-MB-435 β4-injected mice were killed after 4 or 8 weeks of treatment, respectively, and tumor weight was evaluated. HGFR phosphorylation in primary tumors was analyzed by immunohistochemical staining by an anti-phospho-HGFR Ab (Cell Signaling Technology). In mice injected with MDA-MB-435 β4, the lungs were analyzed for the presence of metastasis by means of microscopic observation. Tumor vascularization was evaluated as described in ref. 32.
Supplementary Material
Acknowledgments
We thank R. Albano for mAb production and purification; A. Sottile for experimental help; R. Lo Noce for mouse care; L. Palmas for technical assistance; and S. Corso, L. Tamagnone, T. Crepaldi, E. Vigna, and our colleagues for helpful discussions. This work was supported by Associazione Italiana per la Ricerca sul Cancro (Italy) (AIRC) grants (to S.G. and P.M.C.) and a Ministero dell’ Istruzione, dell’ Università e della Ricerca grant (to S.G.). A. Petrelli, P.C., and L.G. are recipients of AIRC fellowships.
Abbreviations
- HGF
hepatocyte growth factor
- HGFR
HGF receptor
- RTK
tyrosine kinase receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hudson P. J. Curr. Opin. Immunol. 1999;11:548–557. doi: 10.1016/s0952-7915(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 2.Hudson P. J., Souriau C. Nat. Med. 2003;9:129–134. doi: 10.1038/nm0103-129. [DOI] [PubMed] [Google Scholar]
- 3.Gschwind A., Fischer O. M., Ullrich A. Nat. Rev. Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 4.Cragg M. S., French R. R., Glennie M. J. Curr. Opin. Immunol. 1999;11:541–547. doi: 10.1016/s0952-7915(99)00010-2. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara N., Hillan K. J., Gerber H. P., Novotny W. Nat. Rev. Drug Discovery. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 6.Li S., Schmitz K. R., Jeffrey P. D., Wiltzius J. J., Kussie P., Ferguson K. M. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Hynes N. E., Lane H. A. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 8.Trusolino L., Comoglio P. M. Nat. Rev. Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt L., Duh F. M., Chen F., Kishida T., Glenn G., Choyke P., Scherer S. W., Zhuang Z., Lubensky I., Dean M., et al. Nat. Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 10.Kim I. J., Park J. H., Kang H. C., Shin Y., Lim S. B., Ku J. L., Yang H. K., Lee K. U., Park J. G. J. Med. Genet. 2003;40:e97. doi: 10.1136/jmg.40.8.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Renzo M. F., Olivero M., Giacomini A., Porte H., Chastre E., Mirossay L., Nordlinger B., Bretti S., Bottardi S., Giordano S., et al. Clin. Cancer Res. 1995;1:147–154. [PubMed] [Google Scholar]
- 12.Corso S., Comoglio P. M., Giordano S. Trends Mol. Med. 2005;11:284–292. doi: 10.1016/j.molmed.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Prat M., Crepaldi T., Pennacchietti S., Bussolino F., Comoglio P. M. J. Cell Sci. 1998;111:237–247. doi: 10.1242/jcs.111.2.237. [DOI] [PubMed] [Google Scholar]
- 14.Giordano S., Ponzetto C., Di Renzo M. F., Cooper C. S., Comoglio P. M. Nature. 1989;339:155–156. doi: 10.1038/339155a0. [DOI] [PubMed] [Google Scholar]
- 15.Frisch S. M., Francis H. J. Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Q., Chen S., You Z., Yang F., Carey T. E., Saims D., Wang C. Y. J. Biol. Chem. 2002;277:25203–25208. doi: 10.1074/jbc.M201598200. [DOI] [PubMed] [Google Scholar]
- 17.Qiao H., Hung W., Tremblay E., Wojcik J., Gui J., Ho J., Klassen J., Campling B., Elliott B. J. Cell. Biochem. 2002;86:665–677. doi: 10.1002/jcb.10239. [DOI] [PubMed] [Google Scholar]
- 18.Qiao H., Saulnier R., Patryzkat A., Rahimi N., Raptis L., Rossiter J., Tremblay E., Elliott B. Cell Growth Differ. 2000;11:123–133. [PubMed] [Google Scholar]
- 19.Trusolino L., Bertotti A., Comoglio P. M. Cell. 2001;107:643–654. doi: 10.1016/s0092-8674(01)00567-0. [DOI] [PubMed] [Google Scholar]
- 20.Bussolino F., Di Renzo M. F., Ziche M., Bocchietto E., Olivero M., Naldini L., Gaudino G., Tamagnone L., Coffer A., Comoglio P. M. J. Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sengupta S., Gherardi E., Sellers L. A., Wood J. M., Sasisekharan R., Fan T. P. Arterioscler. Thromb. Vasc. Biol. 2003;23:69–75. doi: 10.1161/01.atv.0000048701.86621.d0. [DOI] [PubMed] [Google Scholar]
- 22.Worden B., Yang X. P., Lee T. L., Bagain L., Yeh N. T., Cohen J. G., Van W. C., Chen Z. Cancer Res. 2005;65:7071–7080. doi: 10.1158/0008-5472.CAN-04-0989. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y. W., Su Y., Volpert O. V., Vande Woude G. F. Proc. Natl. Acad. Sci. USA. 2003;100:12718–12723. doi: 10.1073/pnas.2135113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterman H., Yarden Y. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 25.Arribas J., Borroto A. Chem. Rev. 2002;102:4627–4638. doi: 10.1021/cr010202t. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers M., Taylor G. A., Weidner K. M., Omura S., Vande Woude G. F. Mol. Cell. Biol. 1997;17:799–808. doi: 10.1128/mcb.17.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prat M., Crepaldi T., Gandino L., Giordano S., Longati P., Comoglio P. Mol. Cell. Biol. 1991;11:5954–5962. doi: 10.1128/mcb.11.12.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrelli A., Gilestro G. F., Lanzardo S., Comoglio P. M., Migone N., Giordano S. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 29.Peschard P., Fournier T. M., Lamorte L., Naujokas M. A., Band H., Langdon W. Y., Park M. Mol. Cell. 2001;8:995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 30.Ponzetto C., Bardelli A., Zhen Z., Maina F., Dalla Z. P., Giordano S., Graziani A., Panayotou G., Comoglio P. M. Cell. 1994;77:261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 31.Giordano S., Corso S., Conrotto P., Artigiani S., Gilestro G., Barberis D., Tamagnone L., Comoglio P. M. Nat. Cell Biol. 2002;4:720–724. doi: 10.1038/ncb843. [DOI] [PubMed] [Google Scholar]
- 32.Michieli P., Mazzone M., Basilico C., Cavassa S., Sottile A., Naldini L., Comoglio P. M. Cancer Cell. 2004;6:61–73. doi: 10.1016/j.ccr.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. Nat. Rev. Mol. Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 34.Maulik G., Shrikhande A., Kijima T., Ma P. C., Morrison P. T., Salgia R. Cytokine Growth Factor Rev. 2002;13:41–59. doi: 10.1016/s1359-6101(01)00029-6. [DOI] [PubMed] [Google Scholar]
- 35.Kong-Beltran M., Stamos J., Wickramasinghe D. Cancer Cell. 2004;6:75–84. doi: 10.1016/j.ccr.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 36.Ivan M., Bond J. A., Prat M., Comoglio P. M., Wynford-Thomas D. Oncogene. 1997;14:2417–2423. doi: 10.1038/sj.onc.1201083. [DOI] [PubMed] [Google Scholar]
- 37.Gambarotta G., Boccaccio C., Giordano S., Ando M., Stella M. C., Comoglio P. M. Oncogene. 1996;13:1911–1917. [PubMed] [Google Scholar]
- 38.Furge K. A., Kiewlich D., Le P., Vo M. N., Faure M., Howlett A. R., Lipson K. E., Woude G. F., Webb C. P. Proc. Natl. Acad. Sci. USA. 2001;98:10722–10727. doi: 10.1073/pnas.191067898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sariola H., Saarma M. J. Cell Sci. 2003;116:3855–3862. doi: 10.1242/jcs.00786. [DOI] [PubMed] [Google Scholar]
- 40.Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P. M. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 41.Morotti A., Mila S., Accornero P., Tagliabue E., Ponzetto C. Oncogene. 2002;21:4885–4893. doi: 10.1038/sj.onc.1205622. [DOI] [PubMed] [Google Scholar]
- 42.Christensen J. G., Schreck R., Burrows J., Kuruganti P., Chan E., Le P., Chen J., Wang X., Ruslim L., Blake R., et al. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 43.Berthou S., Aebersold D. M., Schmidt L. S., Stroka D., Heigl C., Streit B., Stalder D., Gruber G., Liang C., Howlett A. R., et al. Oncogene. 2004;23:5387–5393. doi: 10.1038/sj.onc.1207691. [DOI] [PubMed] [Google Scholar]
- 44.Arribas J., Coodly L., Vollmer P., Kishimoto T. K., Rose-John S., Massague J. J. Biol. Chem. 1996;271:11376–11382. doi: 10.1074/jbc.271.19.11376. [DOI] [PubMed] [Google Scholar]
- 45.Blobel C. P. Curr. Opin. Cell Biol. 2000;12:606–612. doi: 10.1016/s0955-0674(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 46.Di Fiore P. P., De C. P. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 47.Sorkin A., Von Z. M. Nat. Rev. Mol. Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 48.Schaeffer W. I., Friend K. Cancer Lett. 1976;1:259–262. doi: 10.1016/s0304-3835(75)97506-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.