Abstract
Replication of the giant RNA genome of severe acute respiratory syndrome (SARS) coronavirus (CoV) and synthesis of as many as eight subgenomic (sg) mRNAs are mediated by a viral replicase-transcriptase of outstanding complexity that includes an essential endoribonuclease activity. Here, we show that the CoV replicative machinery, unlike that of other RNA viruses, also uses an exoribonuclease (ExoN) activity, which is associated with nonstructural protein (nsp) 14. Bacterially expressed forms of SARS-CoV nsp14 were shown to act on both ssRNAs and dsRNAs in a 3′→5′ direction. The activity depended on residues that are conserved in the DEDD exonuclease superfamily. The protein did not hydrolyze DNA or ribose-2′-O-methylated RNA substrates and required divalent metal ions for activity. A range of 5′-labeled ssRNA substrates were processed to final products of ≈8–12 nucleotides. When part of dsRNA or in the presence of nonlabeled dsRNA, the 5′-labeled RNA substrates were processed to significantly smaller products, indicating that binding to dsRNA in cis or trans modulates the exonucleolytic activity of nsp14. Characterization of human CoV 229E ExoN active-site mutants revealed severe defects in viral RNA synthesis, and no viable virus could be recovered. Besides strongly reduced genome replication, specific defects in sg RNA synthesis, such as aberrant sizes of specific sg RNAs and changes in the molar ratios between individual sg RNA species, were observed. Taken together, the study identifies an RNA virus ExoN activity that is involved in the synthesis of multiple RNAs from the exceptionally large genomic RNA templates of CoVs.
Keywords: replication, ribonuclease, severe acute respiratory syndrome
Coronaviruses (CoVs) are important human pathogens that are mainly associated with respiratory disease, including the severe acute respiratory syndrome (SARS) (1). With genome sizes of ≈30,000 nt, CoVs feature the largest nonsegmented genomes among RNA viruses. The family Coronaviridae comprises two genera, Coronavirus and Torovirus, and is part of the order Nidovirales, which also contains the families Arteriviridae and Roniviridae (2, 3). Expression of the CoV plus-strand RNA genome starts with the translation of two large overlapping 5′-terminal ORFs, 1a and 1b (Fig. 1). The translation products are called polyprotein (pp) 1a (450–500 kDa) and pp1ab (750–800 kDa) (4). Expression of pp1ab requires ribosomal frameshifting into the −1 reading frame, occurring just upstream of the ORF 1a translation stop codon (5). The pps are autocatalytically processed by proteases to release at least 16 nonstructural proteins (nsps) (6) that, together with several cellular proteins, form the replicase complex directing the replication and transcription of the CoV genome (7). In the latter process, a virus-specific nested set of subgenome-length mRNAs is generated to express the ORFs downstream of ORF1b, which encode the structural and several (virus-specific) accessory proteins (3). All of the CoV subgenome-length plus-strand RNAs have a common leader sequence at their 5′-ends that is identical to the sequence present at the 5′-end of the genome (8). The subgenome-length mRNAs are synthesized by using subgenome-length minus strands as templates (9). Because of a process called discontinuous extension of minus strands, all these subgenome-length minus strands carry the complement of the leader sequence at their 3′-ends (10, 11).
Fig. 1.
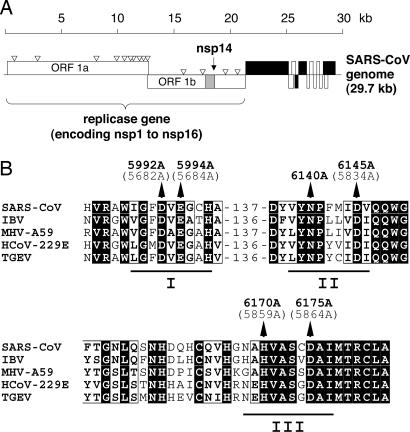
CoV replicase genes encode a putative ExoN. (A) Functional ORFs in the SARS-CoV genome are expressed from both genomic RNA and a set of eight sg mRNAs (17). ORFs encoding the four major structural proteins, S, E, M, and N, are shown in black. The ORF1a- and ORF1b-encoded proteins, pp1a and pp1ab, are cleaved by viral proteases to yield 16 processing end products called nsp1–16. The N-terminal part of nsp14 was predicted to harbor an ExoN domain (13). (B) Partial sequence alignment of representative CoV ExoN domains. ExoN residues (pp1ab numbering) that were targeted in this study by site-directed mutagenesis are indicated for SARS-CoV (denoted in bold type) and HCoV-229E (given in brackets). Also shown are the conserved exonuclease sequence motifs I–III (18).
CoV RNA synthesis is thought to involve an enzymology that is significantly more complex than that of most other RNA viruses. These enzymes include those that are also conserved in other nidoviruses and arranged in the respective replicase pps in the following order (from N to C terminus): chymotrypsin-fold main protease, superfamily 1 RNA-dependent RNA polymerase, zinc finger-containing superfamily 1 helicase, and endoribonuclease (12). In addition, CoVs encode one or two papain-like proteases, as well as putative exoribonuclease (ExoN), ribose-2′-O methyltransferase, and ADP-ribose-1′′-phosphatase (Appr-1′′-pase) activities (7, 13). Some members of the family Coronaviridae also encode putative cyclic nucleotide phosphodiesterase activities (13). Because of the large size of CoV genomes, which until recently has limited the use of reverse genetics approaches, the activities and functions of the various enzymes in the CoV life cycle are only slowly beginning to emerge. Recently, a CoV endoribonuclease activity has been identified and shown to have a critical role in CoV replication and transcription (14, 15). Here, we report that CoVs encode a second ribonucleolytic activity. The SARS-CoV nsp14 is demonstrated to mediate a metal ion-dependent 3′→5′ ExoN activity that acts on both ssRNA and dsRNA. The data suggest that binding of nsp14 to dsRNA (in cis or trans) significantly stimulates the ribonucleolytic activity of the enzyme. Both DNA and ribose-2′-O-methylated RNA proved to be resistant to SARS-CoV nsp14-mediated cleavage. Comparative sequence analysis and characterization of mutant versions of SARS-CoV nsp14 expressed in Escherichia coli suggest that the CoV ExoN belongs to the DEDD superfamily of nucleases (13). Transfection of full-length human CoV 229E (HCoV-229E) RNAs encoding mutant forms of ExoN into susceptible cells consistently revealed severe defects in viral RNA synthesis. The data we obtained implicate ExoN activity in both genomic and subgenomic (sg) RNA synthesis. That the related arteriviruses, featuring genomes approximately two times smaller, do not seem to encode an ExoN homolog leads us to speculate that CoVs have evolved ExoN to control RNA synthesis on exceptionally large RNA templates.
Results and Discussion
Identification of SARS-CoV nsp14-Associated Nucleolytic Activity.
SARS-CoV nsp14 was expressed as a maltose-binding protein (MBP) fusion protein in E. coli and purified by amylose-affinity chromatography, an approach that previously has proven suitable for the expression of several SARS-CoV enzymes (15–17). A significant portion of the overexpressed MBP-nsp14 fusion protein could be obtained in a soluble form and was purified by using a single-step purification protocol (Fig. 6A, which is published as supporting information on the PNAS web site). A range of mutant forms of MBP-nsp14 were expressed and purified in an identical manner as the wild-type protein (Figs. 1B and 6B). They carried substitutions of predicted active-site (including metal ion-coordinating) residues in the conserved exonuclease motifs, Exo I, II, and III (13, 18, 19). As Fig. 6C shows, MBP-nsp14 readily hydrolyzed a 5′-labeled ssRNA substrate to a final product of ≈12 nt. By contrast, the MBP-nsp14_D5992A/E5994A and MBP-nsp14_H6170A proteins had strongly reduced nucleolytic activities, whereas the MBP-nsp14_N6140A, MBP-nsp14_D6145A, and MBP-nsp14_D6175A proteins were nearly inactive under the conditions used in our assay. The data confirm that the activity is associated with nsp14 rather than potentially copurified E. coli nucleases. This leads us to conclude that, besides the recently identified uridylate-specific endoribonuclease (15), SARS-CoV encodes a second nuclease activity.
Metal Ion Requirements of SARS-CoV ExoN.
To further characterize the biochemical properties of SARS-CoV nsp14, we sought to obtain highly purified nsp14 in an essentially unmodified form. Because the MBP-nsp14 fusion protein turned out to be largely uncleavable by Factor Xa, we decided to express nsp14 as a C-terminally His-tagged fusion protein (nsp14-HC). This protein was purified by Ni-NTA affinity chromatography and gel filtration to apparent homogeneity (Fig. 7A, which is published as supporting information on the PNAS web site) and used in all subsequent experiments. Assessment of the enzyme's metal ion requirements revealed a strict dependence of the nucleolytic activity on divalent cations. The protein was most active in the presence of Mg2+ or Mn2+ ions but also retained significant (albeit lower) activity in the presence of Zn2+ (0.5 mM) (Fig. 7B). Like other DEDD family proteins (20–22), nsp14 was inactive in the presence of Ca2+ and at high concentrations of Zn2+ ions. Fig. 7B also shows that the final product obtained in the presence of Mn2+ ions was slightly smaller than that seen in the presence of Mg2+ or Zn2+, suggesting that binding to different metal ions might affect the activity of the SARS-CoV ExoN by inducing subtle structural differences in the active site, as reported for several other nucleases (21, 23). The data presented in Figs. 6 and 7B, together with sequence comparisons of CoV ExoN domains with DEDD family exonucleases (13), and the biochemical and structural information available for several of these cellular homologs (24–29), suggest that the ExoN activity critically depends on two divalent metal ions that are coordinated by the SARS-CoV pp1ab residues Asp-5992 and Glu-5994 (both from Exo I), Asp-6145 (Exo II), and Asp-6175 (Exo III). With few exceptions (30), the conserved Exo I–III Asp and Glu residues are known to be essential for DEDD nuclease activity, whereas replacements of the conserved Exo III Tyr/His residue appear to be slightly less detrimental (26, 31, 32). The Exo II and Exo III Asp residues (6145 and 6175) proved to be essential for the SARS-CoV ExoN activity, whereas the D5992A/E5994A and H6170A substitutions resulted in substantially diminished (but still detectable) activities (see above). Furthermore, the conserved Asn-6140 residue proved to be essential for activity. The available structure information (24, 33) leads us to suggest that this Asn residue might contribute to substrate binding by forming a hydrogen bond with the ribose oxygen of the penultimate nucleotide.
Given that the various DEDD family members use a similar but not identical set of residues to coordinate the two metal ions and to orient the attacking hydroxide ion and its nucleotide substrate (24, 25, 27, 30, 33), more experimentation and structure information will be required to understand the precise functional and structural role of individual ExoN residues and the other subdomains present in nidovirus nsp14 proteins. In this context, the role of the putative Zn-finger structure, which uniquely separates the Exo I and II motifs in CoV and torovirus ExoNs (13), will be of major interest.
SARS-CoV ExoN Acts with 3′→5′ Polarity on both ssRNA and dsRNA Substrates.
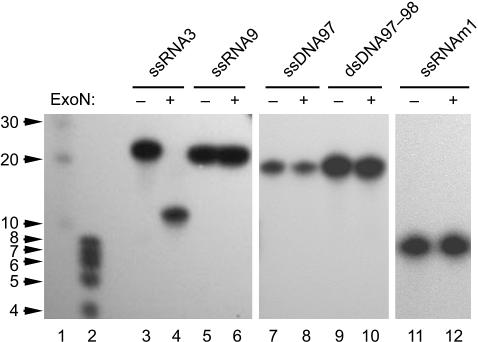
To gain insight into the specificity and directionality of the ExoN activity, we incubated a range of nucleic acid substrates with purified nsp14-HC and analyzed the reaction products by denaturing PAGE and autoradiography. Similar to MBP-nsp14 (see above and Fig. 6C), nsp14-HC readily hydrolyzed ssRNA in the presence of Mg2+ ions (Fig. 2, lanes 3 and 4). In our experiments, the protein was completely inactive on ssDNA and dsDNA (Fig. 2, lanes 7–10), suggesting that the ribose-2′-hydroxyl group is critically involved in the reaction. Accordingly, ribose-2′-O-methylated RNA substrates proved to be resistant to cleavage by nsp14 (Fig. 2, lanes 5 and 6, and data not shown). That, in all our experiments (see below), 5′-labeled RNA substrates were gradually hydrolyzed to increasingly smaller products that still carried their 5′-label led us to conclude that ExoN operates in a 3′-to-5′ direction. To further support this conclusion, we produced 5′- and 3′-labeled ssRNA substrates and compared their hydrolysis by nsp14-HC over time. Hydrolysis of the 3′-labeled RNA substrate by nsp14-HC did not lead to detectable reaction intermediates or stable end products but rather resulted in a signal at the bottom of the gel that, apparently, represented the released 3′-label, 5′-[32P]pCp (Fig. 8, which is published as supporting information on the PNAS web site, and data not shown). Taken together, the data strongly suggest that ExoN hydrolyzes its substrates in a 3′→5′ direction.
Fig. 2.
Substrate specificity of the SARS-CoV ExoN. SARS-CoV nsp14-HC was incubated with different 5′-[32P]-labeled substrates, and the reaction products were analyzed by denaturing PAGE and autoradiography. Lanes 1 and 2, 5′-[32P]-labeled RNA markers with sizes indicated to the left; lanes 3 and 4, reactions with ssRNA3; lanes 5 and 6, reactions with a ribose-2′-O-methylated form of ssRNA3 (designated ssRNA9); lanes 7 and 8, reactions with ssDNA97; lanes 9 and 10, reactions with dsDNA97/98; lanes 11 and 12, reactions with ssRNAm1. Reactions were done in the presence (lanes 4, 6, 8, 10, and 12) or absence (lanes 3, 5, 7, 9, and 11) of nsp14-HC (ExoN).
Using more than a dozen ssRNA substrates (see Materials and Methods and Table 1, which is published as supporting information on the PNAS web site), we confirmed that nsp14-HC is capable of hydrolyzing ssRNAs of varying length and sequence to final products of between 8 and 12 nt (Figs. 2 and 3 and data not shown). In all cases, these products remained stable even after substantially prolonged incubation times. Consistent with these data, short oligoribonucleotides, such as the 8-nt ssRNA m1 shown in Fig. 2 (lanes 11 and 12) and RNAs shorter than ssRNA m1 (data not shown) were not cleaved by nsp14-HC. The data suggest a minimal ExoN-binding site for ssRNA of ≈8–12 nt. Comparison of the data obtained with a range of ssRNA substrates revealed differences in the velocities of the nucleolytic reactions and also the sizes of the reaction intermediates, and final products varied slightly among the substrates tested. Also, we did not observe a regular laddering of products with equally distributed 1-nt increments in length, indicating that specific 3′-terminal nucleotides were hydrolyzed by nsp14-HC more effectively than others (Fig. 3 A–D and data not shown). The functional and structural basis of the ExoN preference for specific 3′-terminal nucleotides and/or RNA structures (see below) and its implications for possible biologically relevant substrates remain to be characterized.
Fig. 3.
Ribonucleolytic activities of SARS-CoV ExoN on ssRNA and dsRNA substrates. The various RNA substrates (for sequences, see Table 1) were incubated with nsp14-HC for the indicated periods of time.
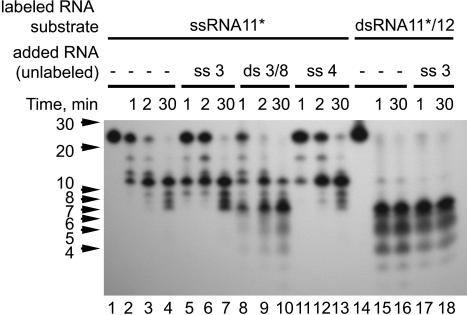
In the course of the study, we observed differences in the nsp14-HC-mediated processing of ssRNA versus dsRNA substrates. Whereas the hydrolysis of 5′-labeled RNA substrates proceeded to final products of 8–12 nt in the case of ssRNA (see above), it proceeded to only 4–7 nt if the labeled RNA was part of a duplex RNA molecule. Fig. 3 D and E illustrate this differential processing for single- and double-stranded versions of RNA11. Nsp14-HC hydrolyzed ssRNA11 almost completely within 5 min to yield a major product of ≈12 nt (lanes 4–6). However, if the same RNA11 was hybridized to unlabeled fully complementary ssRNA12 (resulting in dsRNA11/12), it was degraded to much smaller products of between 4 and 7 nt, suggesting that duplex or at least partial-duplex RNA substrates are much better ExoN substrates than ssRNA. To corroborate this result, we performed another set of experiments in which we supplemented the nuclease reactions with (noncomplementary) unlabeled ssRNA or dsRNA. The effects of these extra RNAs on the nsp14-HC-mediated hydrolysis of our labeled test substrates (5′-labeled ssRNA11 and its double-stranded derivative, dsRNA11/12, respectively) were studied by PAGE and autoradiography (Fig. 4). In reactions supplemented with unlabeled ssRNA, the major product had the same size of 12 nt as the nonsupplemented reaction, demonstrating that ssRNA had no significant effect on the hydrolysis of the test substrates by nsp14-HC (Fig. 4, lanes 2–4, 5–7, 11–13, and 17–18). By contrast, addition of dsRNA caused nsp14-HC to produce a major processing product of 7 nt (Fig. 4, compare lanes 9 and 10 with lanes 3, 4, 6, 7, 12, and 13). The 7-nt product and several minor products of even smaller sizes comigrated with the major products of the dsRNA11/12 reaction (Fig. 4, lanes 15 and 16), suggesting that dsRNA (irrespective of whether it is provided in cis or in trans) promotes or modulates the ribonucleolytic activity of nsp14. The observation that dsRNA (i) was much more completely processed than ssRNA and (ii) assisted in trans to process ssRNA to near completion leads us to believe that the biologically relevant substrate of nsp14 may be (partial) duplex rather than ssRNA. However, ExoN (like other nucleases) most likely requires at least a few unpaired 3′-terminal nucleotides to initiate its 3′-to-5′ exonuclease activity (34, 35). The questions of whether such a local melting of duplex RNA occurs and how this then is achieved remain to be investigated in future experiments.
Fig. 4.
dsRNA affects the exonucleolytic activity of the SARS-CoV ExoN. 5′-[32P]-labeled RNA11 (lanes 2–13) or a ds version of RNA11 (lanes 15–18), which was obtained by annealing 5′-[32P]-labeled RNA11 with the complementary unlabeled RNA12, were incubated with nsp14-HC for the indicated periods of time in the absence (lanes 2–4, 15, and 16) or presence (lanes 5–13, 17, and 18) of the indicated unlabeled RNAs (ssRNA3, dsRNA3/8, and ssRNA4, respectively). Positions of RNA size markers are given to the left.
Critical Role of ExoN Activity in CoV Replication and Transcription.
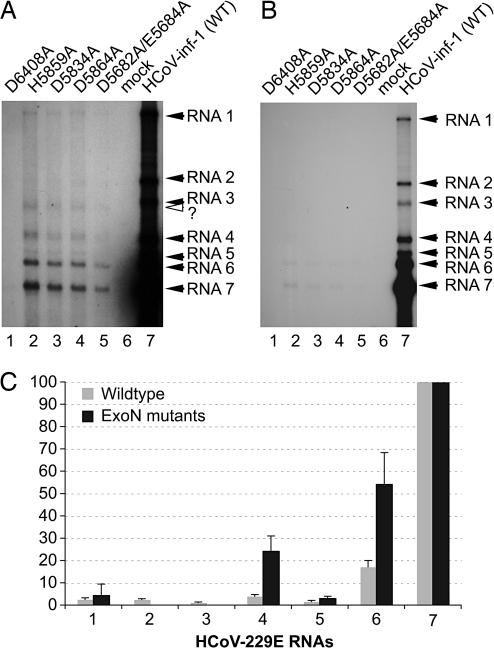
To gain insight into possible functions of the ExoN in the CoV life cycle, we constructed a series of HCoV-229E mutants with substitutions in the presumed ExoN active site. HCoV-229E was chosen for these experiments because of its superior biological safety compared to SARS-CoV. We introduced codon substitutions with Ala of five presumed catalytic residues, D5682/E5684 (double mutant in the Exo I motif), D5834 (Exo II), H5859, and D5864 (both in Exo III) that were shown for SARS-CoV nsp14 to interfere with ExoN activity (see above). Three days after transfection of BHK-HCoV-N cells with wild-type and mutated full-length HCoV-229E RNAs, respectively, accumulation of viral RNA was analyzed. A typical Northern blot experiment is shown in Fig. 5A and B. The data consistently revealed that accumulation of viral RNA was drastically reduced in each of the ExoN mutants tested. The essentially identical pattern of viral RNA species obtained for the four mutants suggests that the observed defects in viral RNA synthesis resulted from the lack of ExoN activity rather than from changes in nsp14 structure or genomic RNA structure. Quantitative analysis of the amount of the most abundant RNA7 was reduced ≈180-fold. We also observed other defects in the synthesis of subgenome-length RNAs. First, the common molar ratio of individual sg RNA species was altered, partially at the expense of mRNA7 (Fig. 5C). Second, the band representing mRNA4 was repeatedly found to be broadened or even appeared as a double band, indicating that an alternative (noncanonical?) leader-body junction might be used in this case. Third, in all of the mutants, mRNA3 appeared to migrate faster. Fourth, in the mutants, mRNA2 was nontypically less abundant than the genome RNA and, even after exposure of the blots for 4 weeks, barely detectable (Fig. 5A). The severe defects in viral RNA synthesis and/or accumulation are consistent with our failure to recover infectious virus progeny after transfection of full-length RNAs encoding ExoN-deficient replicases into susceptible cells. Taken together, the data lead us to suggest that, besides its role in genome replication, ExoN might have a distinct role in sg RNA synthesis. More specifically, the increased relative abundance of RNA4–6 (which, apparently, occurs at the expense of the more 3′-terminal RNA species) and the aberrant sizes observed for mRNA3 and (in part) mRNA4, appear to suggest that (one of the) central mechanisms involved in sg RNA synthesis might be dysregulated if the CoV replicase lacks ExoN activity. It remains to be investigated whether ExoN has a direct or indirect role in the discontinuous extension of minus strands, for example, by assisting the minus-strand polymerase in changing its template at transcription-regulating sequence (TRS) elements or whether it has other (RNA processing) functions in sg minus- and/or plus-strand synthesis. Complementary basepairing-assisted template switching at TRS elements is a key step in the production of antileader-containing sg minus strands and, in fact, causes the nonequimolar distribution of coronaviral sg RNAs, with the smaller RNAs being significantly more abundant than the larger ones (see wildtype in Fig. 5 B and C). Partial defects in template switching at TRS elements would likely shift the molar ratio of RNAs toward the larger RNA species or, with a more visible impact, cause a significant portion of the negative-strand polymerases (when stalled at TRS sequences but unable to perform the template switch) to dissociate from their template. In both scenarios, defects in genomic and sg RNA synthesis (similar to those we described for the ExoN mutants) could be expected. Future experiments are required to explore these and other possibilities. Mechanistically, the 3′→5′ ExoN could act to process (“trim”?) the nascent minus strand's 3′ end before or after its transfer to the leader TRS to produce a 3′-terminal sequence that is fully complementary to the (new) template.
Fig. 5.
Effects of ExoN active-site mutations on the intracellular accumulation of viral RNAs. (A and B) BHK-21-N cells (4 × 106) were mock-transfected or transfected with HCoV-229E full-length RNAs transcribed from purified genomic DNA of vHCoV-1ab_H5859A, vHCoV-1ab_D5834A, vHCoV-1ab_D5864A, vHCoV-1ab_D5682A/E5684A, and vHCoV-inf-1 (WT), respectively. Seventy-two hours after transfection, intracellular poly(A) RNA was isolated and analyzed by Northern hybridization, as described in Materials and Methods. The blot was exposed to x-ray film for 28 (A) or 2 days (B). Transfections were done with the following RNAs: lane 1, HCoV-1ab-D6408A RNA (negative control) (15); lane 2, HCoV-1ab-H5859A RNA; lane 3, HCoV-1ab-D5834A; lane 4, HCoV-1ab-D5864A; lane 5, HCoV-1ab-D5682A/E5684A; lane 6; no RNA (mock transfection); lane 7, HCoV-229E RNA (wild-type sequence). HCoV-229E-specific RNAs 1–7 are indicated by filled arrowheads and the position of a mutant-specific sg RNA migrating slightly faster than HCoV-229E RNA3 is indicated by an open arrowhead. (C) Analysis of ratios of viral RNA species. Quantitation of bands was done in the linear range of exposure by phosphorimaging by using advanced image data analysis software (Raytest, Straubenhardt, Germany). The relative abundance of HCoV-229E (wild-type) RNAs was determined from 10 independent transfections of full-length RNA derived from vHCoV-inf-1. Transfections of RNAs containing ExoN active-site substitutions were performed in triplicate. Because very similar results were obtained for each of the four mutants, the data are presented here as a single set of data. In each experiment, the signal obtained for RNA7 was taken to be 100%, and all other signals were normalized to this value. Mean values and standard deviations are given. Because of low abundance and aberrant size, RNA2 and -3 of the ExoN mutants could not be identified unambiguously and therefore were excluded from this analysis.
Concluding Remarks.
This study identifies an RNA virus 3′→5′ ExoN activity that is uniquely conserved in CoV, torovirus, and ronivirus replicases (13) and appears to be critically involved in viral RNA synthesis. As noted (7, 13), the apparently much more complex enzymology involved in CoV RNA synthesis that, besides polymerase, helicase, and protease activities, has been predicted (or recently proven) to use endoribonuclease, ADP-ribose-1′′-phosphatase, ribose-2′-O methyltransferase, and (in some cases) cyclic nucleotide phosphodiesterase activities, can probably be attributed to the specifics of the CoV (nidovirus) life cycle. The sequence relationships between virus-encoded and DNA polymerase-associated 3′→5′ ExoNs suggest a possible role of nsp14 in some sort of proofreading during CoV RNA synthesis, which might prevent the polymerase from crossing a postulated “error threshold” above which the survival of the viral quasispecies would become impossible (36). Although clearly this idea needs to be substantiated by more experiments, it receives support by the fact that the much smaller arteriviruses (despite an otherwise very similar domain organization of their replicase pps) lack this ExoN domain. Furthermore, the observed defects in the synthesis of genome- and subgenome-length RNAs are important findings that deserve further investigation. Thus, for example, in-depth analyses of the enzyme's substrate specificity, sequence analysis of leader-body junctions of sg RNAs with aberrant sizes and comparison of error frequencies between wild-type CoVs and viable ExoN mutants generated by reverse genetics should provide insight into the physiological role of this activity and the molecular basis of the observed functional defects of ExoN mutants. In this context, it should be noted that single amino acid substitutions in the largely uncharacterized C-terminal domain of nsp14 (outside ExoN) attenuated virulence of mouse hepatitis virus A59 in mice without affecting viral replication in cultured cells (37), indicating that nsp14 may be a multifunctional protein with diverse roles in RNA synthesis and pathogenesis.
Materials and Methods
Plasmid Construction.
To construct pMal-SARS-CoV-nsp14, the coding sequence of SARS-CoV (strain Frankfurt 1, GenBank accession no. AY291315) was amplified by RT-PCR from poly(A) RNA isolated from SARS-CoV-infected Vero cells (17) using the following oligonucleotides: 5′-GCAGAAAATGTAACTGGACTTTT-3′ and 5′-AAAGGATCCTTACTGTAACCTGGTAAATGTATT-3′. The purified PCR product was treated with T4 DNA polymerase, polynucleotide kinase, and BamHI and subsequently ligated with XmnI/BamHI-digested pMal-c2 plasmid DNA (New England Biolabs). Plasmid pMal-SARS-CoV-nsp14 encoded an E. coli maltose-binding protein-SARS-CoV nsp14 fusion protein. Mutations in the nsp14 coding sequence were introduced by using PCR-based methods. To construct pDEST14–75HC, the SARS-CoV nsp14 coding sequence (nucleotides 17970–19550), preceded by one Met codon and followed by six His codons and a translation stop codon, was inserted into plasmid pDEST14 (Invitrogen) according to the manufacturer's instructions. The pDEST14–75HC plasmid DNA was used to express a C-terminally His-tagged form of SARS-CoV nsp14.
Protein Expression and Purification.
To express MBP-nsp14 fusion proteins, E. coli Rosetta (DE3)pLysS cells (Novagen) were transformed with pMal-SARS-CoV-nsp14 plasmid DNA and its mutant derivatives, respectively. The cells were cultured at 37°C in LB medium supplemented with antibiotics until the culture density (A600) reached 0.6. After induction of expression with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG), the cells were grown for 4 h at 18°C. Cells were harvested, and MBP fusion proteins were purified by amylose affinity chromatography by using previously described protocols (17). Briefly, cells were suspended and lysed in APP buffer [20 mM Tris·HCl (pH 8.0)/300 mM NaCl/10% glycerol/2 mM DTT] containing 0.25 mg/ml lysozyme, 1 mM PMSF, 0.1% Triton X-100, and 1 μg/ml DNase. After sonication and centrifugation, the soluble fraction was loaded onto amylose columns preequilibrated in buffer APP. After washing with APP buffer, the protein was eluted by using APP buffer containing 10 mM maltose. The protein-containing fractions were pooled, dialyzed against buffer APP containing 50% glycerol, and stored at −20°C.
To express His-tagged SARS-CoV nsp14 protein, E. coli Rosetta (DE3)pLysS cells were transformed with pDEST14–75HC plasmid DNA and cultured as described above. In this case, protein expression was induced with 50 μM isopropyl-β-d-1-thiogalactopyranoside, and cells were grown for 16 h at 17°C. Cells were harvested, resuspended in buffer containing 50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 10 mM imidazole, 0.25 mg/ml lysozyme, 1 μg/ml DNase, and 20 mM MgSO4. After sonication and centrifugation, the soluble supernatant was loaded onto Ni-NTA affinity columns that were preequilibrated with buffer containing 50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 10 mM imidazole. After washing with buffers containing 10 and 50 mM imidazole, respectively, the His-tagged nsp14 was eluted with buffer containing 250 mM imidazole. The pooled peak fractions were loaded onto a Superdex 200 column (Amersham Pharmacia Biosciences), which was run under isocratic conditions by using buffer containing 10 mM Tris·HCl (pH 8.0), 300 mM NaCl, and 5 mM DTT. The purified protein was stored at −20°C.
Nucleic Acid Substrates and Size Markers.
The following oligoribonucleotides were used: RNA1 (5′-CGCAGUGAGCUCCUAUUCGCCC-3′); RNA2 (5′-CGCAGUGAGCUCCUAAUCGCCC-3′); RNA3 (5′-CGCAGUUAGCUCCUAAUCGCCC-3′); RNA4 (5′-GCGAGUGAGCCCCCUUCCCCGCUCACUCGC-3′); RNA5 (5′-GCGAGUGAGCCCCCAACCCCGCUCACUCGC-3′); RNA6 (5′-GCGUCACUCGAGGAUAAGCGGG-3′); RNA7 (5′-AUACAGGGCGAAUAGGAGCUCA-3′); RNA8 (5′-GGGCGAUUAGGAGCUAACUGCG-3′); RNA9 (2′-O-methyl RNA3); RNA10 (5′-AACAUCACUAACAUACAG-3′); RNA11 (5′-AACAUCACUAACAUACAGUGACCAA-3′); RNA12 (2′-O-methyl 5′-UUGGUCACUGUAUGUUAGUGAUGUU-3′); RNA13 (5′-GGGCGAUUAGGAGCUCACUGCG-3′); RNAm1 (5′-CGCAGUUA-3′), RNAm2 (5′-CGCAGUU-3′), RNAm3 (5′-CGCAGU-3′), RNAm4 (5′-CGCAG-3′), RNAm5 (5′-CGCA-3′) (Table 1). Oligonucleotides used in this study were: DNA97 (5′-CGCAGTTAGCTCCTAATCGCCC-3′) and DNA98 (5′-GGGCGATTAGGAGCTAACTGCG-3′). To produce a ds form of RNA11, the oligoribonucleotide was annealed with a 1.5-fold excess of RNA12. To produce a ds form of DNA97, the oligonucleotide was annealed with a 1.5-fold excess of DNA98. 5′-labeling of ssRNA or ssDNA was done by using T4 polynucleotide kinase and γ-[32P]-ATP. 3′-labeling of ssRNA was done by using T4 RNA ligase and 5′-[32P]-pCp, as described (15). As size markers, 5′-[32P]-labeled RNA Decade Marker (Ambion, Austin, TX) and a mixture of 5′-[32P]-labeled ssRNAs m1, m2, m3, m4, and m5 were used.
Nuclease Assay.
Standard reactions contained 200 nM nsp14-HC (or 200 nM MBP-nsp14 fusion protein or one of its mutant derivatives) and 40 nM (labeled) or 400 nM (unlabeled) oligo(ribo)nucleotide. Reactions were performed in 25 mM Hepes-KOH/50 mM NaCl/5 mM MgCl2/1 mM DTT. To determine the role of metal ions for SARS-CoV ExoN activity, MgCl2 was replaced with 5 mM MnCl2, 5 mM CaCl2, and 5 and 0.5 mM Zn-acetate, respectively. After incubation at 37°C for up to 60 min, the reactions were stopped by addition of an equal volume of loading buffer (formamide containing 10 mM EDTA). The products were then analyzed in 7 M urea-containing 20% polyacrylamide gels (acrylamide/bisacrylamide ratio 19:1) buffered with 0.5× Tris-borate-EDTA.
Mutagenesis of the HCoV-229E Full-Length cDNA Clone.
The recombinant vaccinia virus (VV) vHCoV-inf-1 containing the full-length cDNA of HCoV-229E (38) was used to construct mutant derivatives in which the codons of the pp1ab residues Asp-5682, Glu-5684, Asp-5834, His-5859, and Asp-5864, respectively, were substituted with Ala codons (Fig. 1B). The respective vHCoV-inf-1 derivatives were called vHCoV-1ab-D5682A/E5684A, vHCoV-1ab-D5834A, vHCoV-1ab-H5859A, and vHCoV-1ab-D5864A. Mutagenesis of vHCoV-inf-1 involved several steps of VV-mediated recombination using the protocols described previously (15). Genome-length RNAs were prepared by in vitro transcription by using purified genomic DNA from vHCoV-inf-1 and its mutant derivatives (38). Fifteen micrograms of HCoV-inf-1 RNA, HCoV-1ab-D5682A/E5684A RNA, HCoV-1ab-D5834A RNA, HCoV-1ab-H5859A RNA, and HCoV-1ab-D5864A RNA, respectively, were used for electroporation of 4 × 106 BHK-21 cells expressing the HCoV-229E nucleocapsid (N) protein (BHK-HCoV-N cells) (39). Seventy-two hours after transfection, poly(A) RNA was isolated, separated by agarose-formaldehyde gel electrophoresis, and transferred to nylon membrane. Viral RNAs were analyzed by Northern hybridization by using an [α-32P] multiprime-labeled DNA probe specific for HCoV-229E nucleotides 26857–27277 and the negative-strand complement of this sequence.
Supplementary Material
Acknowledgments
We thank Volker Thiel (Cantonal Hospital, St. Gallen, Switzerland) and Stuart Siddell (University of Bristol, Bristol, U.K.) for the recombinant vaccinia virus vHCoV-inf-1. This work was supported by the Deutsche Forschungsgemeinschaft (Grants SFB 479, TP A8, and ZI 618/4) and the Euro-Asian SARS-DTV Network (Grant SP22-CT-2004-511064).
Abbreviations
- SARS
severe acute respiratory syndrome
- CoV
coronavirus
- ExoN
exoribonuclease
- nsp
nonstructural protein
- sg
subgenomic
- pp
polyprotein
- MBP
maltose-binding protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Peiris J. S., Yuen K. Y., Osterhaus A. D., Stohr K. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez J. M., Gomez-Puertas P., Cavanagh D., Gorbalenya A. E., Enjuanes L. Arch. Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddell S. G., Ziebuhr J., Snijder E. J. In: Topley and Wilson's Microbiology and Microbial Infections. Mahy B. W. J., ter Meulen V., editors. London: Hodder Arnold; 2005. pp. 823–856. [Google Scholar]
- 4.Ziebuhr J. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziebuhr J., Snijder E. J., Gorbalenya A. E. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 7.Ziebuhr J. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B. A., Siddell S. G. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawicki D., Wang T., Sawicki S. J. Gen. Virol. 2001;82:385–396. doi: 10.1099/0022-1317-82-2-385. [DOI] [PubMed] [Google Scholar]
- 10.Sawicki S. G., Sawicki D. L. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 11.Zúñiga S., Sola I., Alonso S., Enjuanes L. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya A. E. Adv. Exp. Med. Biol. 2001;494:1–17. [PubMed] [Google Scholar]
- 13.Snijder E. J., Bredenbeek P. J., Dobbe J. C., Thiel V., Ziebuhr J., Poon L. L., Guan Y., Rozanov M., Spaan W. J., Gorbalenya A. E. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj K., Guarino L., Kao C. C. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov K. A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A. E., Ziebuhr J. Proc. Natl. Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov K. A., Thiel V., Dobbe J. C., van der Meer Y., Snijder E. J., Ziebuhr J. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiel V., Ivanov K. A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E. J., Rabenau H., Doerr H. W., et al. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 18.Moser M. J., Holley W. R., Chatterjee A., Mian I. S. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo Y., Deutscher M. P. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 21.Deutscher M. P., Marlor C. W. J. Biol. Chem. 1985;260:7067–7071. [PubMed] [Google Scholar]
- 22.Lehman I. R., Richardson C. C. J. Biol. Chem. 1964;239:233–241. [PubMed] [Google Scholar]
- 23.Ren Y. G., Kirsebom L. A., Virtanen A. J. Biol. Chem. 2004;279:48702–48706. doi: 10.1074/jbc.M403858200. [DOI] [PubMed] [Google Scholar]
- 24.Beese L. S., Steitz T. A. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y., Patel D. J. J. Mol. Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derbyshire V., Grindley N. D., Joyce C. M. EMBO J. 1991;10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamdan S., Carr P. D., Brown S. E., Ollis D. L., Dixon N. E. Structure (London) 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 28.Joyce C. M., Steitz T. A. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 29.Steitz T. A., Steitz J. A. Proc. Natl. Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thore S., Mauxion F., Seraphin B., Suck D. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elisseeva E., Mandal S. S., Reha-Krantz L. J. J. Biol. Chem. 1999;274:25151–25158. doi: 10.1074/jbc.274.35.25151. [DOI] [PubMed] [Google Scholar]
- 32.Taft-Benz S. A., Schaaper R. M. Nucleic Acids Res. 1998;26:4005–4011. doi: 10.1093/nar/26.17.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrino F. W., Harvey S., McMillin S., Hollis T. J. Biol. Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- 34.Cowart M., Gibson K. J., Allen D. J., Benkovic S. J. Biochemistry. 1989;28:1975–1983. doi: 10.1021/bi00431a004. [DOI] [PubMed] [Google Scholar]
- 35.Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Proc. Natl. Acad. Sci. USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domingo E., Escarmis C., Lazaro E., Manrubia S. C. Virus Res. 2005;107:129–139. doi: 10.1016/j.virusres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Sperry S. M., Kazi L., Graham R. L., Baric R. S., Weiss S. R., Denison M. R. J. Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel V., Herold J., Schelle B., Siddell S. G. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- 39.Schelle B., Karl N., Ludewig B., Siddell S. G., Thiel V. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.