Abstract
The development of vertebrate basal forebrain dopaminergic (DA) neurons requires the conserved zinc finger protein Too Few (Tof/Fezl) in zebrafish. However, how Tof/Fezl regulates the commitment and differentiation of these DA neurons is not known. Proneural genes encoding basic helix–loop–helix transcription factors regulate the development of multiple neuronal lineages, but their involvement in vertebrate DA neuron determination is unclear. Here we show that neurogenin 1 (ngn1), a vertebrate proneural gene related to the Drosophila atonal, is expressed in and required for specification of DA progenitor cells, and when overexpressed leads to supernumerary DA neurons in the forebrain of zebrafish. Overexpression of ngn1 is also sufficient to induce tyrosine hydroxylase expression in addition to the pan-neuronal marker Hu in nonneural ectoderm. We further show that Tof/Fezl is required to establish basal forebrain ngn1-expressing DA progenitor domains. These findings identify Ngn1 as a determinant of brain DA neurons and provide insights into how Tof/Fezl regulates the development of these clinically important neuronal types.
Keywords: neurogenin 1, pluripotent neural stem cell, neurotransmitter phenotype, commitment and differentiation
The determination of neurotransmitter phenotype is an important aspect of neuronal differentiation, and in this regard, dopaminergic (DA) neurons have attracted considerable attention because of their functional and medical importance (1). Degeneration of substantia nigra DA neurons in humans is a hallmark of Parkinson's disease, and the malfunction of DA neurons in other brain regions is implicated in psychiatric disorders and neuroendocrine dysregulation. Therefore, understanding the determination of DA phenotype and the specification of DA neuronal circuitry may provide mechanistic and therapeutic insights into these disorders. To date, only limited number of known or putative transcriptional regulators, including Pax6, Dlx, Nurr1, Lmx1a, Lmx1b, Msx1, Foggy, and Too Few (Tof/Fez1), have been implicated in the specification of DA phenotype in vertebrates (2–7, 35). Despite this knowledge, the mechanisms leading to the early commitment of pluripotent neural stem cells to DA lineage remain elusive.
The earliest DA neurons in zebrafish are detected at ≈24 h postfertilization (hpf) in the basal forebrain (8). They express tyrosine hydroxylase (TH), the rate-limiting enzyme in dopamine synthesis, and dopamine transporter (DAT), a protein involved in dopamine reuptake (9). Later during development, these DA neurons have both ascending and descending projections, and are believed to be homologous to mammalian DA neurons of both the basal forebrain and midbrain (10, 11).
Through forward genetic analysis, an adult viable zebrafish mutant named too few (tofm808) has been isolated that displays selective deficits of basal forebrain DA as well as adjacent serotonergic (5HT) neurons (8). Molecular characterizations have revealed that the too few mutant carries a point mutation that changes Cys-287 to Ser in the second of the six zinc finger motifs of the conserved zinc finger protein Fezl (Tof/Fezl) (5). Whereas these studies establish an important role of Tof/Fezl in DA neuron development, the mechanism by which Tof/Fezl acts in DA neuron specification is not clear.
The role of basic helix–loop–helix (bHLH) proteins in neural development has been initially discovered in Drosophila (12) and later studied in multiple vertebrate neuronal lineages (13, 14). Neurogenins are a family of bHLH proteins related to Drosophila Atonal (15). Multiple members of the Neurogenin family have been discovered in mice (16, 17), and they play partially overlapping roles in regulating neuronal specification. For example, Neurogenin1 and Neurogenin2 are involved in the development of dorsal root ganglia (18, 19), and Neurogenin2 is a determination factor for placodal sensory neuron development (20). In zebrafish, only one neurogenin-like gene, named neurogenin 1 (ngn1), has been identified so far (21). Ngn1 is required for the development of spinal, cranial sensory, and epiphysial neurons (22–24).
Here we report that early-born basal forebrain DA neurons are derived from Ngn1-expressing progenitor cells. We demonstrate that Ngn1 is necessary for the development of these DA neurons, and overexpression of Ngn1 leads to supernumerary DA neurons in the zebrafish forebrain, and induces TH+ cells with an apparent neuronal morphology on the yolk surface ectoderm. Furthermore, we show that Tof/Fezl is expressed in an overlapping fashion with ngn1, and is required to specify ngn1-expressing DA progenitor domains in the basal forebrain. Together, our data identify early regulatory steps that lead to the commitment of pluripotent neural stem cells to dopaminergic phenotype.
Results
Ngn1 Is Expressed in Dopaminergic Progenitor Cells.
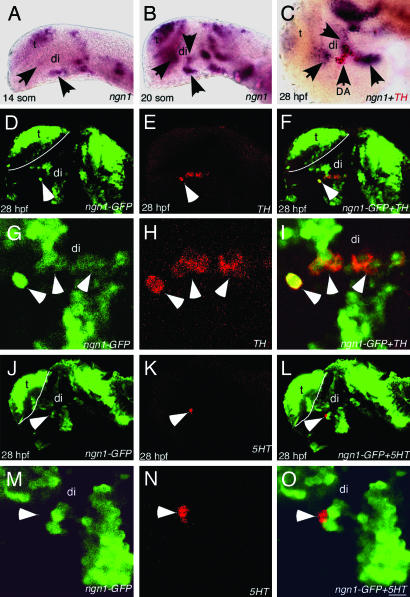
While examining genes that are expressed in the vicinity of DA neurons in the basal forebrain of zebrafish, we noted that ngn1 is expressed in the basal forebrain as distinct clusters before DA neuron appearance (Fig. 1 A and B), and later in close proximity to the appearing nascent TH+ DA neurons (Fig. 1C). At 28 hpf, between approximately three and six DA neurons were detected in the basal forebrain on each side of the midline, and they were the earliest born DA neurons in the entire zebrafish embryo (Fig. 1E). To determine whether these early-born basal forebrain DA neurons are derived from ngn1-expressing progenitor cells, a GFP transgene driven by the ngn1 promoter (25) was used to follow the fates of ngn1+ cells after they stop expressing ngn1, because GFP protein is more stable than ngn1 mRNA, and usually lasts >6 h after the cessation of gfp transcript synthesis (26). Because the first TH-immunoreactive (ir) DA neurons are detected ≈24 hpf (8), we would be able to observe that DA neurons were GFP+ at 28 hpf, should they be derived from ngn1-expressing progenitor cells. Therefore, double immunostaining with GFP and TH antibodies were carried out in 28 hpf embryos, and the results revealed that all TH+ DA neurons were positive for GFP at 28 hpf (Fig. 1 D–I, and Fig. 7 A–F, which is published as supporting information on the PNAS web site). Among GFP+TH+ neurons, some displayed strong GFP signal that might correspond to newly born cells, whereas faint GFP signals were detected in all other TH+ DA neurons that were likely born at earlier times (Fig. 1I).
Fig. 1.
ngn1 is expressed in DA progenitors. All images are lateral views of anterior brain regions. Anterior is to the left, and dorsal is up. (A and B) ngn1 expression at 14- and 20-somite stages, respectively (arrows point to several clusters in the ventral forebrain). (C) A 28-hpf embryo showing ngn1 expression (purple) in close proximity to TH+ DA neurons (red). (D–F) Confocal images of 28 hpf ngn1-GFP transgenic embryos immunostained with GFP antibody (green, D), TH antibody (red, E), and the merged image (F), showing that GFP is detected in TH+ DA neurons. (G–I) High-magnification views of D–F. (J–L) Confocal images of 28-hpf ngn1-GFP transgenic embryos immunostained with GFP antibody (green, J), 5HT antibody (red, K), and the merged image (L), showing that GFP is not detected in 5HT neurons. (M–O) High-magnification views of J–L. di, diencephalon, t, telencephalon. (Scale bars, 64 μm in A and B, 60 μm in C–F and J–L, and 3 μm in G–I and M–O.)
A group of serotonin (5HT) neurons (usually about one to two neurons at ≈28 hpf) develop in close proximity to DA neurons in the basal forebrain (4). To determine whether ngn1-GFP is also detectable in these neighboring 5HT neurons, we carried out double immunostaining with GFP and 5HT antibodies. These results indicate that these neighboring serotonergic (5HT) neurons were always GFP negative (Figs. 1 J–O and 7 G–L). This observation indicates that the early-born basal forebrain DA neurons are derived from ngn1-expressing progenitor cells, but 5HT neurons are probably not.
Ngn1 Is Required for the Development of Dopaminergic Neurons in the Basal Forebrain.
To determine whether ngn1 is required for DA neuron development, we injected a ngn1 morpholino (MO) that has been shown to effectively and specifically knockdown Ngn1 protein expression (22, 23). Abrogation of Ngn1 activity by MO injection led to a severe loss of DA neurons in the ventral forebrain (95% of embryos showing none or few TH or DAT-positive cells by 48 hpf, n = 54) (Fig. 2 A–D). Some residual TH staining was detected in 48 hpf ngn1 MO embryos, but the intensity was much reduced as compared to control embryos (Fig. 2 A and B). DAT expression appeared to be completely abolished in these embryos (Fig. 2 C and D). This phenotype was confirmed in ngn1 mutants (see Fig. 6 G and H) isolated from mutagenesis screens (24, 27). Examination of an earlier developmental stage revealed that, at 32 hpf, DA neurons were completely absent in Ngn1-defective animals (Fig. 2 E and F). Examination of a later postembryonic larval stage show that, at 72 hpf, DA neurons appeared to recover somewhat but remained significantly defective in the basal forebrain (estimated to be ≈80–90% deficiency compared to control) (Fig. 2 G and H). The overall brain patterning of ngn1 morphants was normal, and TH+ DA neurons in the retina (Fig. 2 I and J), TH+ noradrenergic neurons of locus coeruleus (Fig. 2 A and B) and the sympathetic ganglia (Fig. 2 K and L) appeared not significantly affected in the ngn1 morphants. Moreover, the neighboring basal forebrain 5HT neurons were not obviously reduced (Fig. 2 M, N, and Insets). The lack of 5HT defects in the absence of ngn1 activity is consistent with the lineage study showing that these 5HT neurons were not derived from ngn1-expressing progenitor cells (Fig. 1 J–O). These data suggest that ngn1 is required for the development of basal forebrain DA neurons, but not for the development of neighboring 5HT neurons and several TH+ neuronal groups in other regions of the nervous system.
Fig. 2.
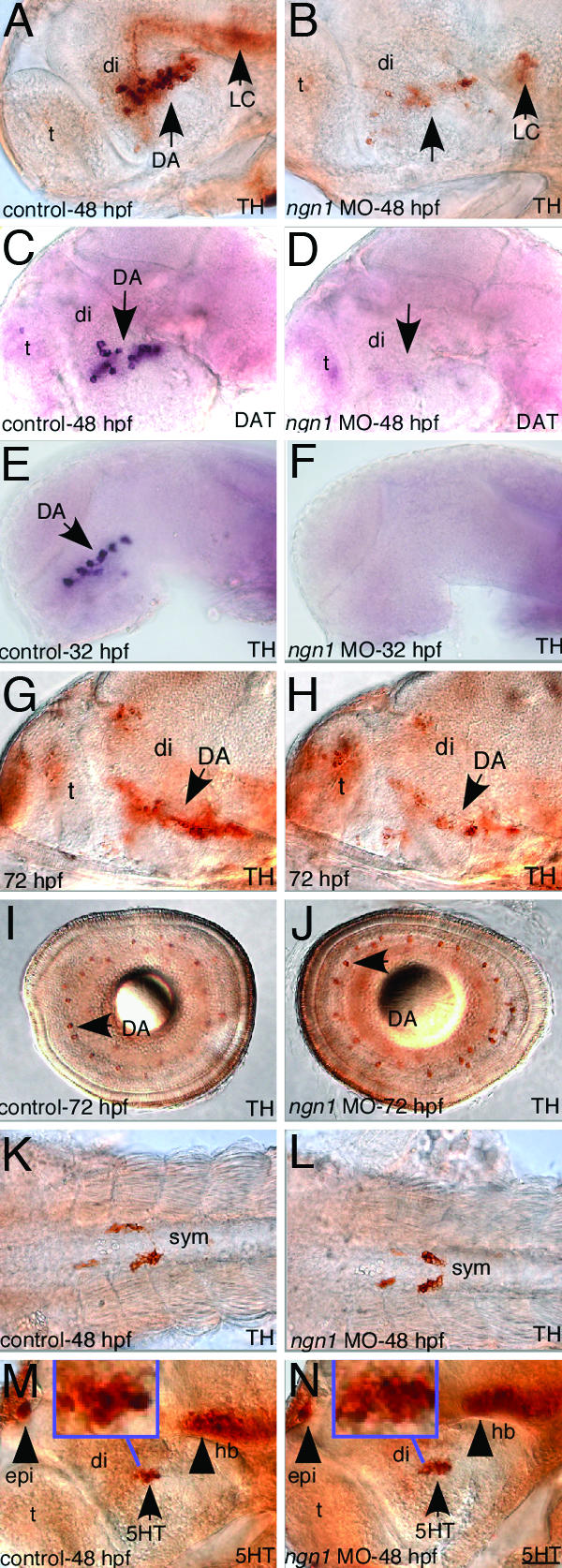
ngn1 is required for DA neuron development in the basal forebrain. All images are lateral views of anterior brain regions except I–L. Anterior is to the left, and dorsal is up. (A–L) Immunostaining with TH antibody (A, B, G, and L), in situ with TH (E and F), or DAT antisense probe (C and D), or immunostaining with 5HT antibody (M and N), shows that DA neurons in the ventral forebrain is completely absent at 32 hpf, largely absent at 48 hpf, and remain significantly defective at 72 hpf in ngn1-morphants, whereas TH+ DA neurons appear normal in the retina, TH+ gut sympathetic neurons are normal in the trunk region of ngn1 morphants, and 5HT neurons are not affected. (Insets) Magnified views of basal forebrain 5HT neurons. DA, dopaminergic neurons; DAT, dopamine transporter; di, diencephalon; epi, epiphysis; hb, hindbrain; LC, locus coeruleus; mhb, midhindbrain boundary; t, telencephalon. (Scale bar, 32 μm.)
Fig. 6.
The requirement of tof/fezl in establishing ngn1-expressing DA progenitor domains. (A–D) ngn1 expression in 20-somite wild-type sibling and the tof mutant embryos shows that ngn1 expression in the basal forebrain (arrows) is reduced at 20 somites (A and B) but appears normal at 28 hpf (C and D). (E and F) ngn1 expression is largely absent in the basal forebrain of tof/fezl morphant. (G and H) TH and tof/fezl in situ (both purple) shows that, although TH+ DA neurons are largely absent, the fezl expression appears normal in the ngn1 mutant. di, diencephalon; t, telencephalon. (Scale bars, 32 μm in G and H and 64 μm in A–F.)
To determine the state of DA progenitor domains in the absence of ngn1 activity, we analyzed ngn1 expression in the ngn1 morphants. We observed strongly enhanced ngn1 RNA labeling in the morphant embryos, possibly due to the fact that the ngn1 MO inhibits the translation of ngn1 transcript, and somehow leads to an enhanced transcript stability, although a negative feedback regulation of ngn1 transcription by Ngn1 protein is also a possibility. Nevertheless, these analyses revealed that the basal forebrain ngn1-expressing progenitor domains were intact in the ngn1 morphants at multiple developmental stages examined (Fig. 3), suggesting that, in the absence of ngn1 activity, progenitor domains are formed but fail to commit and differentiate into DA fate.
Fig. 3.
ngn1 expression in ngn1 morphants. Anterior is to the left, and dorsal is up. ngn1 expression is higher in the ngn1 morphants, and Ngn1-expressing progenitor domains are intact in the basal forebrain of ngn1 morphants at various developmental stages as indicated.
Overexpression of Ngn1 Leads to Supernumerary DA Neurons in the Basal Forebrain and TH+ Cells with an Apparent Neuronal Morphology on the Yolk Surface Ectoderm.
Given that ngn1 is essential for DA neuron development, we next determined whether overexpressing ngn1 has an impact on DA phenotype by microinjecting ngn1 mRNA into one- to eight-cell-stage zebrafish embryos. A proportion of ngn1 mRNA injected embryos (usually <15%) displayed morphological deformity including malformed eyes and brain (data not shown), as has been reported (21). Therefore, in all mRNA injection experiments carried out in our study, only the injected embryos that display grossly normal brain morphology were used to determine the impact of ngn1 misexpression on DA neuron development. We found that misexpression of ngn1 at 100 ng/ml led to a significant increase of DA neurons in the basal forebrain (77% embryos showing a significant increase of DA neurons, n = 64) (Fig. 4 A–H) and in some instances a few ectopic DA neurons were detected in the telencephalon (13%, n = 220, Fig. 4 E–H). Using the pan-neuronal marker HuC, we observed that, as previously reported (21), the expression of pan-neuronal marker HuC in the CNS was not grossly increased by Ngn1 overexpression (data not shown). These observations suggest that Ngn1's ability in increasing DA neuronal subtypes is not a consequence of an overall increased neuronal production in CNS. Interestingly, we found that misexpression of ngn1 also induced TH+ cells on the yolk surface ectoderm, albeit at a low number and a low frequency (≈10% of injected embryos have about one to three TH+ cells on the yolk surface ectoderm, n = 50) (Fig. 4 J and L), despite this, such ectopic TH+ cells were never observed in control embryos (0% of control embryos have TH+ cells on the yolk surface ectoderm, n = 211) (Fig. 4 I and K). DAT-positive cells appeared not induced on the yolk surface ectoderm by ngn1 misexpression at the concentration that we tested (data not shown). These TH+ cells appeared to have a neuronal morphology (Fig. 4 L and Inset). This observation is consistent with the fact that ngn1 is shown to be capable of inducing the pan-neuronal marker Hu on the yolk surface ectoderm (21). Taken together, we conclude that overexpression of ngn1 is able to increase the production of DA neurons in the forebrain and induce TH+ neurons in nonneural ectoderm. It is worth noting that increased production of DA neurons was largely detected in their endogenous location of the basal forebrain, suggesting that Ngn1 requires additional factors in this region to induce DA neuron production.
Fig. 4.
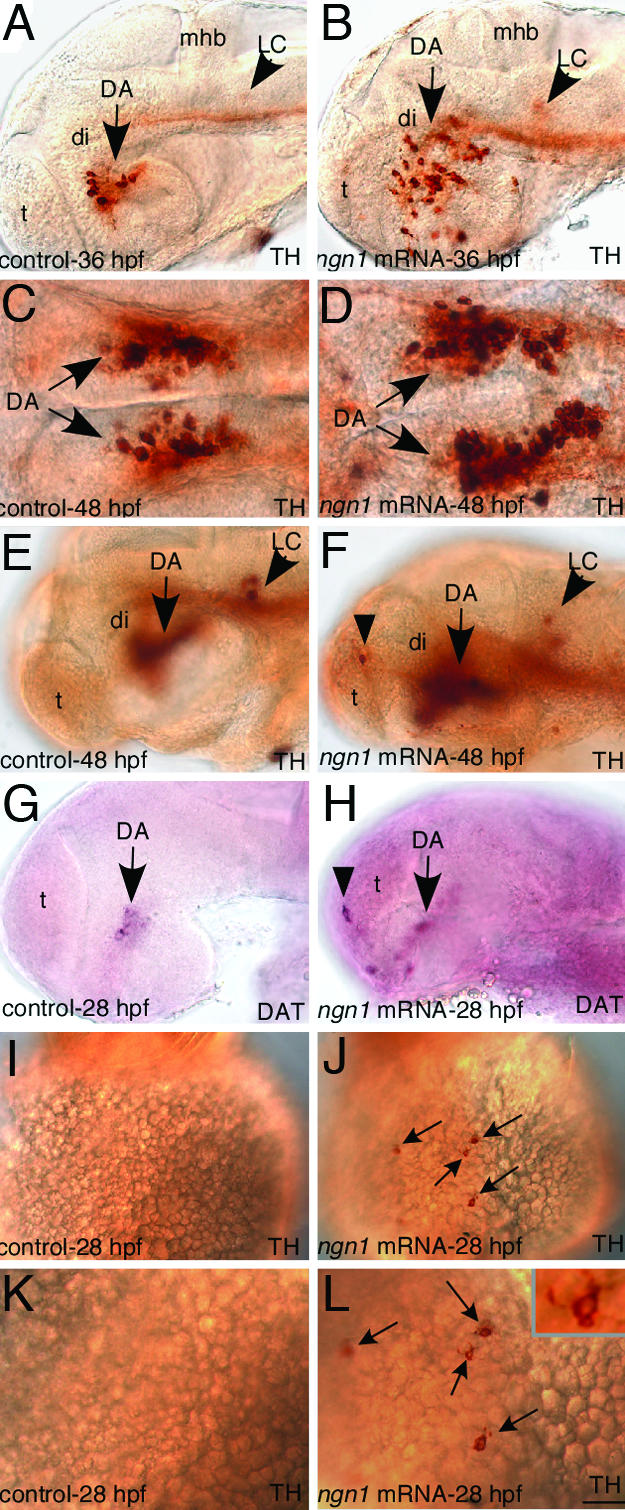
Misexpression of ngn1 leads to supernumerary DA neurons in the forebrain and induces TH+ cells with an apparent neuronal morphology on the yolk surface ectoderm. (A–H) Immunostaining with TH antibody (A–F) and in situ with DAT antisense probe (G and H) shows that DA neurons are significantly increased in the ventral forebrain and ectopically induced in the telencephalon (Di DA neurons are out of focal planes in E–H) in ngn1-mRNA injected embryos. (I and J) Immunostaining with TH antibody showing an ectopic TH+ cell on the yolk surface ectoderm of ngn1-injected embryo. (K and L) A high magnification view of (I and J), and Inset showing an ectopic TH+ cell with an apparent neuronal morphology on the yolk surface ectoderm. DA, dopaminergic neurons; DAT, dopamine transporter; di, diencephalon; LC, locus coeruleus; mhb, midhindbrain boundary; t, telencephalon. (Scale bar, 15 μm in K and L, 32 μm in A–H, and 64 μm in I and J.)
Tof/Fezl Expression Overlaps with ngn1-Expressing DA Progenitor Domains in the Basal Forebrain.
Our loss- and gain-of-function analyses indicate that Ngn1 is a crucial factor for basal forebrain DA neurons. To explore how Ngn1 might be regulated in the development of DA neurons, we investigated the relationship between tof/fezl and ngn1. Tof/Fezl was previously identified through zebrafish forward genetics as an essential factor for the development of basal forebrain DA neurons; however, its mechanism of action is unknown. In 10-somite (≈14 hpf) embryos, tof/fezl expression was detected broadly in the forebrain (Fig. 5A), preceding the detection of ngn1 expression in this region (Fig. 5B). At ≈18 hpf, whereas tof/fezl expression remained broad in the forebrain (Fig. 5C), ngn1 expression became detectable in distinct clusters of progenitor cells in the forebrain as well as other brain regions (Fig. 5D). During the period when DA neurons are specified, tof/Fezl expression became less broadly expressed and was detected in telencephalon, as well as, in diencephalon (the basal forebrain), close to but not within DA neurons (Fig. 5E). Double labeling experiments showed that tof/fezl expression domains encompassed the ngn1-expressing domains in the basal forebrain (Fig. 5F). Both tof/fezl and ngn1 expression in these domains were down-regulated by 48 hpf (Fig. 5 G and H). These analyses suggest that ngn1 and tof/fezl are coexpressed in progenitor domains that are in close proximity to basal forebrain DA neurons.
Fig. 5.
tof/fezl is expressed earlier than ngn1 and later in overlapping domains with ngn1 in the basal forebrain All images are lateral views of anterior brain regions. (A–D) In situ hybridization with tof/fezl cRNA probe (A and C) and ngn1 cRNA probe (B and D). (E–H) Double in situ hybridization of tof/fezl (red)+TH (purple) (E and G), tof/fezl (red) + ngn1 (purple) (F), and TH (red) + ngn1 (pruple) (H). DA, dopaminergic neurons; di, diencephalon; t, telencephalon. (Scale bar, 32 μm in E–H and 64 μm in A and D.)
The Basal Forebrain Expression of Ngn1 Is Transiently Reduced in the too few Mutant and Is Abrogated in the tof/fezl Morphant.
Because tof/fezl expression is detected earlier than ngn1 in the forebrain region and later overlaps with basal forebrain ngn1-expressing progenitor domains, we determined whether tof/fezl acts upstream of ngn1 in regulating DA neuron development. We first examined ngn1 expression in the tofm808 mutant, which results from a single amino acid change in the Tof/Fezl protein (5). The results showed that ngn1 expression was reduced in the basal forebrain at ≈20-somite stage (Fig. 6 A and B), but was later found comparable to the wild-type siblings at 28 hpf (Fig. 6 C and D). Because it is possible that tofm808 mutation might be a hypomorphic allele, we sought to determine whether the basal forebrain ngn1 expression domains would be more severely affected in a case of stronger loss of Tof/Fezl activity. Therefore, we designed two splicing morpholinos that target different exon/intron boundaries to knockdown Tof/Fezl activity (Fig. 8, which is published as supporting information on the PNAS web site). Knockdown of Tof/Fezl with either morpholino led to a severe defect of ngn1-expressing domains in the basal forebrain, whereas ngn1 expressing domains in other brain regions were largely unperturbed (Fig. 6 E and F). These analyses suggest that tof/fezl is required to establish ngn1-expressing progenitor domains in the basal forebrain. tof/fezl expression in the ngn1 mutant was largely normal as compared to the wild-type siblings (Fig. 6 G and H), suggesting that ngn1 is not required to regulate tof/fezl expression.
Discussion
Compared to some other neuronal types, our knowledge on the development of vertebrate DA neurons is rather limited. The involvement of these neurons in multiple human neurological disorders including Parkinson's disease, addiction, and schizophrenia makes it a worthwhile endeavor to understand the mechanisms underlying their development. Here we provide in vivo evidence in zebrafish for an important role of Ngn1 in basal forebrain DA neuron development: Ngn1 is required for the development of DA neurons, and when overexpressed, can lead to supernumerary DA neurons in the forebrain and TH+ neurons on the yolk surface ectoderm. Furthermore, we show that Tof/Fezl, an evolutionarily conserved zinc finger protein, is required to establish basal forebrain ngn1-expressing progenitor domains. These analyses identify crucial early regulatory steps in the commitment of multipotent neural stem cells to a dopaminergic lineage.
The Requirement of Ngn1 in DA Neuron Development.
Expression analysis, lineage study, as well as loss-of-function genetic analysis establishes that ngn1 is expressed in the basal forebrain DA neuronal progenitor cells and is required for the specification of DA neurons. Because ngn1-expressing progenitor domains appear intact in the absence of Ngn1 activity, the cellular role of Ngn1 is likely required within the progenitor cells. Because the other neuronal types that the basal forebrain ngn1-expressing progenitor cells may give rise to (we showed that the adjacent 5HT neurons are unlikely to be derived from these ngn1-expressing progenitor cells and are not obviously increased in ngn1-defective embryos) are unlikely to be known, the fate of these basal forebrain ngn1-expressing progenitors in the absence of Ngn1 activity remains to be determined. In addition, although basal forebrain DA neuron defects remain in the postembryonic larval brain, they appear to be less severe than earlier stages, suggesting that additional mechanisms likely exist to regulate the generation of DA neurons at later stages.
Ngn1 may play one or multiple of the following roles in DA progenitor cells: it may regulate their proliferation or cell cycle exit, regulate their acquisition of general neuronal properties, and/or regulate their acquisition of subtype identity, as proposed based on studies of proneural genes in multiple other neuronal lineages (14). Understanding the precise role of Ngn1 in DA neuron development requires the understanding of Ngn1's function at the molecular level, for instance, the identification of Ngn1 target genes in the DA neuronal lineage. The well known direct target of Ngn1, NeuroD, is surprisingly not detected in the basal forebrain region in zebrafish (J.-Y.J. and S.G., unpublished data), although the possibility exists that perhaps other yet unidentified NeuroD-like genes are expressed in this region in zebrafish. Nevertheless, the requirement and sufficiency of Ngn1 for the appearance of TH indicates that Ngn1 may directly regulate DA neuron subtype-related genes. At present, few genes are known to be involved in DA neuron development in zebrafish. Although the differentiation of basal forebrain DA neurons in mice is shown to require Pax6 and Dlx (6, 7), and the differentiation of midbrain DA neurons in mice is shown to require the nuclear receptor Nurr1 (2, 28) and the LIM homeodomain transcription factor Lmx1b (3), the involvement of these factors in the development of basal forebrain DA neurons in zebrafish remains to be determined. Taken together, it will be of great interest to test whether Ngn1 regulate these factors and/or other unidentified pathways during DA neuron determination.
The Role of Ngn1 in Inducing DA Neurons.
Our gain-of-function analysis demonstrates that Ngn1 is capable of inducing supernumerary DA neurons and a DA subtype-related gene in nonneural ectodermal cells. These data strengthen the idea that Ngn1 may be involved not only in specifying general neuronal property, but also in specifying subtype identity in the DA neuronal lineage. However, a definitive role of Ngn1 in DA neuron subtype specification awaits future analysis assessing whether Ngn1 can directly regulate DA subtype-related genes.
Although a few ectopic DA neurons are detected in the telencephalon, the most striking increase of DA neurons is restricted within its endogenous location of the basal forebrain. In our view, this observation has two implications. First, because more Ngn1 leads to more DA neurons in the basal forebrain, it suggests that Ngn1 is a limiting factor in the development of basal forebrain DA neurons. The nature of this limiting effect remains to be understood: Ngn1 may be a limiting factor in determining how many progenitors are initially specified toward DA lineage, or it may be a limiting factor in determining how many rounds of proliferation can occur before DA progenitors can exit cell cycle and embark on a differentiation pathway. Second, because DA neuron induction by Ngn1 is largely restricted to the basal forebrain, it suggests that Ngn1 requires additional factors that are present in the basal forebrain to determine DA neurons, or its pro-DA activity is inhibited in other regions of the nervous system.
Regulation of Ngn1 by the Conserved Zinc Finger Protein Tof/Fezl.
Previous molecular genetic analysis using the too few mutant zebrafish has revealed the importance of Tof/Fezl, a conserved zinc finger protein, in DA neuron specification (5, 8), but its mechanism of action is unknown. The analyses carried out here reveal that Tof/Fezl controls DA neuron development in part by establishing ngn1-expressing DA progenitor domains in the basal forebrain. How does Tof/Fezl specify ngn1-expressing progenitor domains? The fact that Tof/Fezl protein contains six C2H2 zinc fingers suggests that it is a putative transcription regulator and may directly or indirectly regulate ngn1 expression. Therefore, the identification of downstream target genes of Tof/Fezl will provide crucial insights into understanding its role in specifying ngn1-expressing progenitor domains. In addition to the zinc finger domain, Tof/Fezl possesses a Groucho-TLE-like repressor domain, which is found in transcription factors such as TCF that can serve as both transcriptional activators and repressors. Thus, Tof/Fezl may repress a factor that normally inhibits the formation of ngn1-expressing progenitor domains; alternatively, Tof/Fezl may be involved in promoting progenitor domains by activating the expression of ngn1.
A Homologous Role of Neurogenins in the Development of Zebrafish Basal Forebrain DA and Mouse Mesencephalic DA Neurons.
While our work was being prepared for publication, it was reported that loss of neurogenin 2 (ngn2) function in mice impairs the development of mesencephalic DA neurons, whereas non-DA neurons in the midbrain are unaffected (29). The mouse study and our work presented here have two important implications: first, they indicate that the role of neurogenins in DA neuron development is evolutionarily conserved. Second, they show that basal forebrain DA neurons in zebrafish share common developmental mechanisms with the mammalian midbrain DA neurons. Together with the dye-tracing experiments (10) and a functional study (30), our work suggests that some basal forebrain DA neurons in zebrafish are developmentally and functionally homologous to mammalian midbrain DA neurons.
One difference between the mouse study and our work is that, although ngn2 is not sufficient to induce DA neurons in mice (29), we show that ngn1 is capable of inducing DA neurons in zebrafish. Thus, whereas ngn2 appears to have a permissive role in mouse midbrain DA neuron development, ngn1 may have an instructive role in basal forebrain DA neuron development in zebrafish. These differences may be gene- and/or species-dependent.
In conclusion, our study demonstrates an important role of ngn1 in basal forebrain DA neuron development in zebrafish, and moreover, reveals that the establishment of ngn1-expressing DA progenitor domains requires the conserved zinc finger protein Tof/Fezl. Future analyses of these transcription regulators promise to unravel further mechanisms governing the commitment and differentiation of multipotent neural stem cells to a dopaminergic fate.
Materials and Methods
Fish Stocks and Maintenance.
Fish breeding and maintenance were performed as described (31). Embryos were raised at 28.5°C and staged according to Kimmel et al. (32). Fish heterozygous for the ngn1hi1059 and tofm808 mutations were bred to obtain homozygous embryos for analysis: ngn1 homozygous embryos were identified by applying the ratio of 25% to a population (>50) of stained embryos; tof mutant embryos were identified by genotyping for the missense mutation (5).
Analysis of Neuronal Phenotypes in Transgenic ngn1:GFP Embryos.
Twenty-eight-hpf Tg(-8.4ngn1:GFP) embryos were fixed overnight in 4% PFA in 0.1 M phosphate buffer (pH 7.4) and stored in 100% methanol at −20°C. Whole mount immunohistochemistry was performed as described (33). The following primary antibodies were used: anti-GFP (Chemicon, monoclonal, 1:1,000), anti-TH (Chemicon, rabbit polyclonal, 1:1,000) and anti-5HT (DiaSorin, rabbit polyclonal, 1:4,000). The following secondary antibodies were used: anti-rabbit Alexa 633 (Molecular Probes, 1:200) and anti-mouse Alexa 488 (Molecular Probes, 1:200). Fluorescent labeling was analyzed by laser scanning confocal microscopy.
MO and mRNA Injections and Analysis.
The ngn1 MO was synthesized and injected as described (22, 23). For misexpression experiments, capped RNAs from pCS2-β-gal and pCS2-ngn1 plasmids were synthesized and injected at 100–800 ng/μl with 2–3 nl into the yolk of one- to eight-cell-stage embryos as described (34). For details, see Supporting Text, which is published as supporting information on the PNAS web site.
In Situ Hybridization and Immunohistochemistry.
RNA In situ hybridization and immunohistochemistry were performed as described (8).
Supplementary Material
Acknowledgments
We thank Drs. Frances Brodsky, Yuh Nung Jan, Bingwei Lu, and John Rubenstein for their helpful comments on the manuscript; Drs. Adam Amsterdam and Nancy Hopkins (Massachusetts Institute of Technology, Cambridge) for the ngn1 mutant; Uwe Strähle and Patrick Blader (University of Heidelberg, Heidelberg) for ngn1:GFP transgenic fish; and Drs. Say-Yeob Yeo (National Institutes of Health, Bethesda), Ajay Chitnis (National Institutes of Health, Bethesda), and Masahiko Hibi (RIKEN, Tokyo) for plasmids. This work was supported by Searle Scholars Award, Burroughs Wellcome Fund, and National Institutes of Health grants (to S.G.), and by grants from the Wellcome Trust, Biotechnology and Biological Sciences Research Council and European Community (to S.W.W.). The financial support of Telethon–Italy Fellowship GFP03011 (to S.M.) is gratefully acknowledged.
Abbreviations
- DA
dopaminergic
- TH
tyrosine hydroxylase
- hpf
hours postfertilization
- DAT
dopamine transporter
- 5HT
serotonin
- MO
morpholino
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Goridis C., Rohrer H. Nat. Rev. Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- 2.Zetterström R. H., Solomin L., Jansson L., Hoffer B. J., Olson L., Perlmann T. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 3.Smidt M. P., Asbreuk C. H. J., Cox J. J., Chen H., Johnson R. L., Burbach J. P. H. Nat. Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- 4.Guo S., Yamaguchi Y., Schilbach S., Wada T., Goddard A., Lee J., French D., Handa H., Rosenthal A. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- 5.Levkowitz G., Zeller J., Sirotkin H. I., French D., Schilbach S., Hashimoto H., Hibi M., Talbot W. S., Rosenthal A. Nat. Neurosci. 2003;6:28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- 6.Vitalis T., Cases O., Engelkamp D., Verney C., Price D. J. J. Neurosci. 2000;20:6501–6516. doi: 10.1523/JNEUROSCI.20-17-06501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews G. L., Yun K., Rubenstein J., Mastick G. S. Mol. Cell. Neurosci. 2003;23:107–120. doi: 10.1016/s1044-7431(03)00016-2. [DOI] [PubMed] [Google Scholar]
- 8.Guo S., Wilson S. W., Cooke S., Chitnis A. B., Driever W., Rosenthal A. Dev. Biol. 1999;208:473–487. doi: 10.1006/dbio.1999.9204. [DOI] [PubMed] [Google Scholar]
- 9.Holzschuh J., Ryu S., Aberger F., Driever W. Mech. Dev. 2001;101:237–243. doi: 10.1016/s0925-4773(01)00287-8. [DOI] [PubMed] [Google Scholar]
- 10.Rink E., Wullimann M. F. Brain Res. 2001;889:316–330. doi: 10.1016/s0006-8993(00)03174-7. [DOI] [PubMed] [Google Scholar]
- 11.Kapsimali M., Bourrat F., Vernier P. J. Comp. Neurol. 2001;431:276–292. doi: 10.1002/1096-9861(20010312)431:3<276::aid-cne1070>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.Jan Y. N., Jan L. Y. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 13.Chitnis A. B. Curr. Opin. Neurobiol. 1999;9:18–25. doi: 10.1016/s0959-4388(99)80003-8. [DOI] [PubMed] [Google Scholar]
- 14.Bertrand N., Castro D. S., Guillemot F. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 15.Jarman A. P., Grau Y., Jan L. Y., Jan Y. N. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 16.Ma Q., Kintner C., Anderson D. J. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 17.Sommer L., Ma Q., Anderson D. J. Mol. Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 18.Ma Q., Fode C., Guillemot F., Anderson D. J. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Q., Chen Z., Barrantes I., Pompa J., Anderson D. J. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 20.Fode C., Gradwohl G., Morin X., Dierich A., LeMeur M., Goridis C., Guillemot F. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 21.Blader P., Fischer N., Gradwohl G., Guillemot F., Strahle U. Development (Cambridge, U.K.) 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- 22.Andermann P., Ungos J., Raible D. W. Dev. Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- 23.Cornell R. A., Eisen J. S. Development (Cambridge, U.K.) 2002;129:2639–2648. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- 24.Cau E., Wilson S. W. Development (Cambridge, U.K.) 2003;130:2455–2466. doi: 10.1242/dev.00452. [DOI] [PubMed] [Google Scholar]
- 25.Blader P., Plessy C., Strahle U. Mech. Dev. 2003;120:211–218. doi: 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- 26.Halloran M. C., Sato-Maeda M., Warren J. T., Su F., Lele Z., Krone P. H., Kuwada J. Y., Shoji W. Development (Cambridge, U.K.) 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 27.Amsterdam A., Nissen R. M., Sun Z., Swindell E. C., Farrington S., Hopkins N. Proc. Natl. Acad. Sci. USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakurada K., Ohshima-Sakurada M., Palmer T. D., Gage F. H. Development (Cambridge, U.K.) 1999;126:4017–4026. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- 29.Andersson E., Jensen J. B., Parmar M., Guillemot F., Bjorklund A. Development (Cambridge, U.K.) 2006;133:507–516. doi: 10.1242/dev.02224. [DOI] [PubMed] [Google Scholar]
- 30.Lau B., Bretaud S., Huang Y., Lin E., Guo S. Genes Brain Behav. 2005 doi: 10.1111/j.1601-183X.2005.00185.x. in press. [DOI] [PubMed] [Google Scholar]
- 31.Guo S., Driever W., Rosenthal A. Handbook of Molecular-Genetic Techniques for Brain and Behavior Research. Amsterdam: Elsevier; 1999. pp. 166–176. [Google Scholar]
- 32.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 33.Concha M. L., Russell C., Regan J. C., Tawk M., Sidi S., Gilmour D. T., Kapsimali M., Sumoy L., Goldstone K., Amaya E., et al. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 34.Guo S., Brush J., Teraoka H., Goddard A., Wilson S. W., Mullins M. C., Rosenthal A. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- 35.Andersson E., Tryggvason U., Deng Q., Friling S., Alekseenko Z., Robert B., Perlmann T., Ericson J. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.