Abstract
The nuclear transcription factor E-26-like protein 1 (Elk-1) is thought to impact neuronal differentiation [Sharrocks, A. D. (2001) Nat. Rev. Mol. Cell Biol. 2, 827–837], cell proliferation [Sharrocks, A. D. (2002) Biochem. Soc. Trans. 30, 1–9], tumorigenesis [Chai, Y. L., Chipitsyna, G., Cui, J., Liao, B., Liu, S., Aysola, K., Yezdani, M., Reddy, E. S. P. & Rao, V. N. (2001) Oncogene 20, 1357–1367], and apoptosis [Shao, N., Chai, Y., Cui, J., Wang, N., Aysola, K., Reddy, E. S. P. & Rao, V. N. (1998) Oncogene 17, 527–532]. In addition to its nuclear localization, Elk-1 is found throughout the cytoplasm, including localization in neuronal dendrites [Sgambato, V., Vanhoutte, P., Pages, C., Rogard, M., Hipskind, R., Besson, M. J. & Caboche, J. (1998) J. Neurosci. 18, 214–226], raising the possibility that Elk-1 may have alternative extranuclear functions in neurons. Using coimmunoprecipitation and reciprocal coimmunoprecipitation from adult rat brain, we found an association between Elk-1 protein and the mitochondrial permeability transition pore complex (PTP), a structure involved in both apoptotic and necrotic cell death. Electron microscopy in adult rat brain sections confirmed this association with mitochondria. Elk-1 was also identified from purified mitochondrial fractions by using Western blotting, and Elk-1 increased its association with mitochondria following proapoptotic stimuli. Consistent with a role for Elk-1 in neuron viability, overexpression of Elk-1 in primary neurons decreased cell viability, whereas Elk-1 siRNA-mediated knockdown increased cell viability. This decrease in viability induced by Elk-1 overexpression was blocked with application of a PTP inhibitor. These results show an association of the nuclear transcription factor Elk-1 with the mitochondrial PTP and suggest an additional extranuclear function for Elk-1 in neurons.
Keywords: dendrite, mitochondria, viability
E-26-like protein 1 (Elk-1) belongs to the ETS domain transcription factor family and the ternary complex factor subfamily (1). In the nucleus, Elk-1 forms a ternary complex with the serum response factor (SRF) protein and the serum response element promoter region. In addition to an N-terminal DNA-binding domain, Elk-1 contains a “B box” mediating its interaction with SRF, a “C domain” acting as a transcriptional activation domain, two repression domains, and two domains that act as docking sites for multiple mitogen-activated protein kinases, including extracellular signal-regulated kinase and c-Jun N-terminal kinase (1). In addition to roles in neuronal differentiation (2), cell proliferation (1), tumorigenesis (3), and apoptosis (4), Elk-1 phosphorylation has been shown to be up-regulated in response to the induction of both long-term depression and long-term potentiation in the hippocampus in vivo, suggesting a potential role for Elk-1 in synaptic plasticity (5,6).
Sgambato et al. (7) first reported Elk-1 protein localization in dendrites of adult rat brain sections, raising the possibility that Elk-1 may have additional functions outside the nucleus. Previous research on transcription factor p53 has shown that, in addition to a role in transcription, p53 can also interact with mitochondrial proteins to regulate cell viability (8–10). Similarly, research on NF-κB and IκB has shown that these transcription factors can localize at the permeability transition pore (PTP) and impact cell viability (11–13). The PTP is a protein complex forming a functional pore that, once open, allows molecules up to 1.5 kDa to move across the mitochondrial inner membrane nonselectively (14). PTP opening can induce apoptosis or necrosis through initiation of mitochondrial swelling, mitochondrial membrane potential depolarization, ATP hydrolysis, and release of stored soluble proteins such as calcium and cytochrome C (15). Here, we show that Elk-1 associates with mitochondria at the level of the PTP and demonstrate a role for Elk-1 in neuron viability, a finding with significant implications for the functioning of Elk-1.
Results
Elk-1 Interacts with Proteins That Form the Mitochondrial PTP Complex.
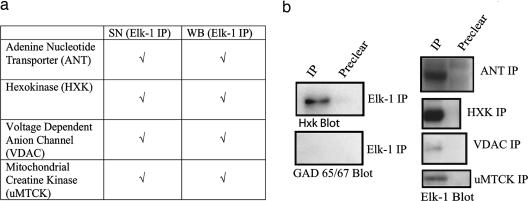
To investigate the potential function(s) of Elk-1 in the cytoplasm, we looked for novel protein-binding partners using coimmunoprecipitation (co-IP) from adult mouse whole-brain (WB) tissue and synaptoneurosome (SN) fractions. SNs, although not pure preparations, represent fractions enriched in pre- and postsynaptic aspects of neurons, and we used these fractions to detect potential interactions with low-abundance synaptic proteins. This allowed us to look for protein interactions from a primarily synaptic area in addition to WB lysate. Two Elk-1 antibodies raised against different epitopes (Santa Cruz Biotechnology) were used for co-IP. Immunoprecipitates were separated by SDS/PAGE, and Coomassie-stained bands (Fig. 5, which is published as supporting information on the PNAS web site) were submitted for peptide sequencing at the University of Pennsylvania Proteomics Core Facility. Using co-IP from both WB and SN fractions, we found an association between Elk-1 and a group of mitochondrial PTP proteins (Fig. 1), including the adenine nucleotide transporter (ANT) and voltage-dependent anion channel (VDAC), both integral membrane proteins, and the ubiquitous mitochondrial creatine kinase (uMTCK) and hexokinase (HXK), both enzymes important for energy metabolism. All four proteins have been implicated in the functioning of the PTP (16). Immunoprecipitation with an Elk-1 antibody followed by Western blotting with a HXK antibody was used to confirm the mass spectrometry data, and immunoprecipitation with an Elk-1 antibody followed by Western blotting with a glutamic acid decarboxylase (GAD) 65/67 antibody was used as a negative control (Fig. 1b). Interaction of Elk-1 with these proteins was confirmed through reciprocal co-IP with antibodies against ANT (n = 4), VDAC (n = 2), HXK (n = 3), and uMTCK (n = 2), followed by Western blotting with an Elk-1 antibody (Fig. 1b).
Fig. 1.
Elk-1 associates with mitochondrial PTP proteins. (a) Co-IPs from mouse WB and SN fractions were performed by using Elk-1 antibodies (Santa Cruz Biotechnology), and immunoprecipitates were submitted to the University of Pennsylvania Proteomics Core Facility for peptide sequencing. Listed are mitochondrial proteins identified through peptide sequencing, with check marks representing the antibody and tissue conditions that led to their identification. (b) Shown are co-IPs carried out with an Elk-1 antibody (Santa Cruz Biotechnology) and immunoprecipitates blotted with antibodies to HXK as a positive control, and GAD 65/67 as a negative control. To confirm the co-IP data, reciprocal co-IP was carried out by using antibodies against adenine nucleotide transporter, VDAC, and HXK, and ubiquitous mitochondrial creatine kinase and immunoprecipitates were then analyzed by Western blot using an Elk-1 antibody (Cell Signaling Technology). Shown are the reciprocal co-IPs from SN fractions.
Elk-1 Is Localized Around Mitochondria in Cell Somas and Dendrites.
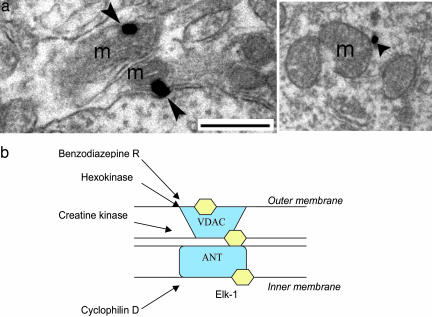
In light of the interaction between Elk-1 and mitochondrial proteins, electron microscopy was used to confirm the mitochondrial localization of Elk-1. Using immunogold–silver detection in adult rat hippocampal sections, Elk-1 signal was visualized around mitochondrial membranes in dendrites (Fig. 2a) and cell somas, showing a definitive association of Elk-1 with mitochondria in both subcellular compartments. The images shown in Fig. 2a are representative of results seen across tissue sections from multiple animals. Although these data do not allow us to distinguish whether Elk-1 interacts directly with proteins on the inner vs. the outer mitochondrial membrane, it does confirm our previous biochemical finding of Elk-1 association with mitochondrial proteins. Potential sites of interaction between Elk-1 and the mitochondrial PTP are shown in Fig. 2b. Taken together, these data demonstrate an association between Elk-1 and mitochondria, a finding that significantly broadens the potential functioning of Elk-1 in neurons.
Fig. 2.
Elk-1 associates with mitochondria in adult rat brain section. (a) Immunogold–silver labeling for Elk-1 (arrowhead) is associated with mitochondria (m) from adult rat brain (n = three rats). Images show labeling in a neuronal dendrite. (Scale bar, 500 nm.) (b) Potential interactions of Elk-1 (hexagon) with proteins that form the mitochondrial PTP.
Elk-1 Increases Its Association with Mitochondria After Application of DNA Damage Agents.
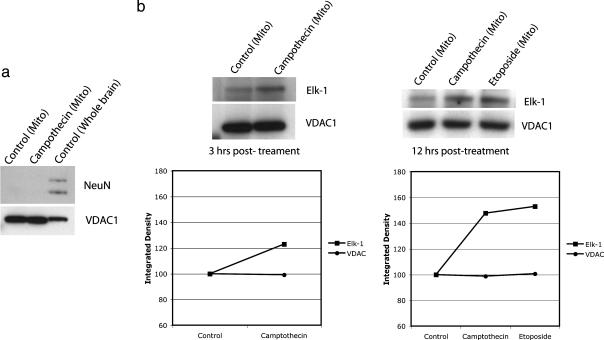
Given the association of Elk-1 with mitochondria in neurons, we next examined whether specific cellular insults could increase or decrease this association. Primary rat cortical neurons were exposed to a variety of compounds known to induce cell death in neurons, and mitochondrial fractions were then isolated (Pierce) and used for Western blotting (Fig. 3a). Using this paradigm, we found that Elk-1 increased its association with mitochondria after application of camptothecin (n = 4; Fig. 3b), an inhibitor of topoisomerase I, or etoposide (n = 3; Fig. 3b), an inhibitor of topoisomerase II, both known to induce DNA damage and apoptosis in neurons (17–19). This increase in association could be detected as soon as 3 hours following drug application (Fig. 3b Left), consistent with the time course in which DNA damage and subsequent cell death are initiated and continued to be detected 12 hours following drug application (Fig. 3b Right).
Fig. 3.
Elk-1 increases its association with mitochondria following application of camptothecin and etoposide. (a) Mitochondrial fractions were obtained from primary rat cortical neurons 14–21 days in vitro according to the manufacturer's instructions (Pierce). Shown is a Western blot for mitochondrial lysate with an absence of signal for the nuclear protein NeuN (Abcam) in the mitochondrial fractions, compared with WB fractions. Also shown is the presence of signal for the mitochondrial protein VDAC1 (Santa Cruz Biotechnology) in the mitochondrial fractions. (b) Three hours after treatment with camptothecin or etoposide (15 μM), an increase in Elk-1 signal was detected in the mitochondrial fractions by Western blot. VDAC1 was used as a loading control. Integrated densities measured using image j (National Institutes of Health) are shown below Western blots. Values have been normalized to control to show relative change in density after drug treatment for bands corresponding to Elk-1 and VDAC.
Elk-1 Overexpression Leads to Decreased Cell Viability, Whereas Elk-1 siRNA-Mediated Knockdown Leads to Increased Cell Viability.
Given that mitochondria are critically involved in cell survival and death, and that Elk-1 associates with mitochondria, we hypothesized that Elk-1 may be able to influence neuron viability. We used several constructs to determine the effects of Elk-1 overexpression or knockdown throughout whole cells in culture. Primary rat hippocampal neurons were electroporated in suspension immediately after harvesting with either a mammalian expression vector containing Elk-1 cDNA under cytomegalovirus promoter control or the empty mammalian expression vector without Elk-1 (pcDNA 3.1; control). Additionally, neurons were electroporated with one of two siRNAs recognizing different regions of Elk-1 (siRNA A or siRNA B; Ambion, Austin, TX), or a control siRNA with no significant sequence similarity to the rat genome (Ambion pSilencer Negative Control siRNA). The use of two distinct siRNAs is a common method to decrease the chance that the observed biological effects result from off-target siRNA effects. Using electroporation, we obtained average transfection efficiencies of ≈50% (based on GFP transfections; data not shown). All subsequent analysis was performed on mixed cultures that contained both transfected and untransfected neurons, and as a result, our findings underestimate the effects of Elk-1 on neuron viability.
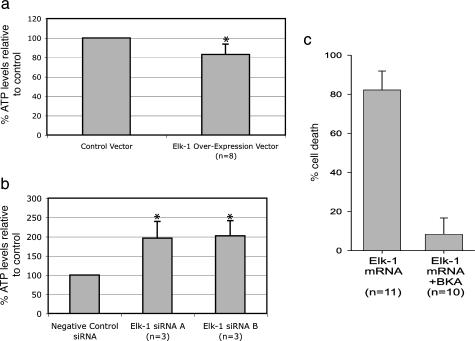
Using a luminescent ATP cell viability assay (Promega), we measured whole-cell ATP levels to estimate the relative change in number of viable cells between different treatment conditions in culture. Specifically, we found a decrease in the number of viable neurons 48 h after Elk-1 overexpression compared with control (n = 8; Fig. 4a). Conversely, we found an increase in the number of viable neurons 48 h following Elk-1 siRNA-mediated knockdown compared with control (n = 3; Fig. 4b). These findings are consistent with an overabundance of Elk-1 enhancing cell death and a reduction of Elk-1 enhancing cell survival. As shown, similar results were obtained for both Elk-1 siRNA constructs. Immunocytochemistry (n = 4; Fig. 5B) and Western blots (n = 3; Fig. 5C) were used to confirm that Elk-1 siRNA (Ambion) electroporated into primary rat hippocampal neurons decreased Elk-1 protein expression. That Elk-1 siRNA did not produce detrimental cellular effects is consistent with previous data on Elk-1 knockout mice, which showed reduced immediate early gene activation but no other obvious phenotype in a limited number of tests (20).
Fig. 4.
Elk-1 overexpression leads to decreased cell viability, whereas Elk-1 siRNA-mediated knockdown leads to increased cell viability. (a) Graph shows whole-cell ATP levels from primary rat hippocampal neurons 48 h following electroporation with either Elk-1 in a pcDNA3.1 vector or pcDNA3.1 vector alone (control). (b) Graph shows whole-cell ATP levels from primary rat hippocampal neurons 48 h following electroporation with either one of two Elk-1 siRNAs (Ambion) or pSilencer Negative Control siRNA (Ambion). ATP levels are normalized to control. Error bars represent standard deviations, and ∗ indicates P < 0.05 when comparing siRNA-mediated knockdown to control or Elk-1 overexpression to control. (c) Graph shows percent of cell death in primary rat hippocampal neurons after transfection with Elk-1 mRNA under control conditions or after preincubation with 25 μM BKA. Note the decrease in cell death after preincubation with BKA.
Elk-1-Induced Cell Death Can Be Blocked with a PTP Inhibitor.
To assess the dependence of Elk-1-induced cell death on mitochondrial PTP function, we preincubated primary rat hippocampal neurons 14–21 days in vitro with the mitochondrial PTP inhibitor, bongkrekic acid (BKA), and then transfected neurons with Elk-1 mRNA to achieve Elk-1 overexpression. BKA has been applied to primary neuronal culture in concentrations ranging from 2 to 50 μM to inhibit PTP opening and to demonstrate the mitochondrial dependence or independence of cell death pathways (21–24). We were unable to use a second common PTP inhibitor, cyclosporin A (CSA), as CSA also inhibits calcineurin, which was identified as a primary Elk-1 phosphatase (25) and is therefore not specific enough for PTP inhibition in this case. By preincubating neurons with BKA, we were able to inhibit the cell death induced by Elk-1 (Fig. 4c), indicating that the Elk-1 cell death pathway depends upon mitochondrial PTP function.
Discussion
The association of Elk-1 with the mitochondrial PTP suggests a transcriptionally independent mechanism of action for Elk-1 in neurons. Numerous ligands for PTP proteins have been shown to facilitate or regulate PTP opening (15), and it is reasonable to hypothesize that Elk-1 can facilitate or regulate PTP opening as well and thus impact cell viability. Our finding that camptothecin and etoposide increase Elk-1's association with mitochondria suggests that specific types of cellular stressors can dictate Elk-1 localization. It may be that Elk-1 associates with mitochondria to aid in the initiation of cell death in response to specific signals, such as DNA damage, while Elk-1 moves into the nucleus to regulate transcription in response to other types of cellular inputs. Future studies will help to tease apart the cellular consequences that can be attributed to Elk-1's nuclear vs. mitochondrial association as well as the relationship between these two associations. How Elk-1 is directed to a particular subcellular region is currently unknown. Elk-1 can be phosphorylated at multiple S/T residues (26) in response to a diverse array of cellular input, and it is possible that differences in posttranslational modifications such as phosphorylation may regulate Elk-1 protein–protein interactions, protein–DNA interactions, and/or direct movement of Elk-1 to different subcellular regions.
Here, we showed that Elk-1 overexpression decreased viability in primary neurons, which is consistent with previous data implicating Elk-1 in cell death in rat fibroblast and human breast cancer cell lines (4). By using the PTP inhibitor BKA in this study, we were also able to show that cell death induced by Elk-1 overexpression in neurons depended on PTP functioning. One possible explanation for this result is that Elk-1 was modulating mitochondria directly to induce cell death. A second possible explanation is that Elk-1 indirectly affected mitochondrial function through transcription of genes that themselves act on the level of the mitochondria. It has been shown that p53 can transcribe proapoptotic genes as well as induce mitochondrial permeabilization independent of transcription (9), although the relationship between these two diverse functions has not been well characterized. Elk-1 may similarly be able to effect cell viability through its mitochondrial as well as its nuclear association. Although it is clear that there is a mitochondrial component to Elk-1-mediated cell death, it does not preclude the involvement of transcriptional events in this process as well. At the level of the nucleus, Elk-1 can act as both a transcriptional activator and a transcriptional repressor (1) and to date, not all of the gene targets of Elk-1 have been identified. Because of this, teasing apart the specific downstream transcriptional consequences of Elk-1 will require a more detailed analysis of the cadre of genes regulated by Elk-1. However, it is clear from these data that functions previously ascribed solely to Elk-1 transcriptional activity may need to be reevaluated in light of Elk-1's association with the mitochondrial PTP.
In addition to Elk-1, numerous other transcription factor proteins have been found localized in neuronal dendrites or synapses, including CREB (27), p53 (28), NF-κB (29), and ATF4 (30). Of these, p53, NF-κB and now Elk-1 have been found to associate with the mitochondrial PTP, and it is possible that other transcription factors found in dendrites may also share in this unique association. Based upon the data presented here, we are now exploring how these proteins are regulated to effect neuron viability, and what the functional significance might be of Elk-1's association with mitochondria in distal dendrites vs. mitochondria in the cell body. The potential parallels and divergences between these proteins is an area of great interest, and in general, this field represents an exciting area for future ventures into the complex multifunctional nature of proteins.
Methods
Cell Culture.
Neuron-enriched primary rat hippocampal cultures from embryonic day 18.5 embryos were plated at 100,000 per milliliter in neurobasal medium (Invitrogen) with B-27 supplement (Sigma) on 12-mm round German Spiegelglas coverslips (Bellco Glass) or gridded coverslips (Eppendorf), as previously described.
co-IP.
Mouse WB and SN fractions were lysed in the presence of protease inhibitors under nondenaturing conditions, precleared with Protein A Agarose (Invitrogen) and immunoprecipitated with one of two polyclonal Elk-1 antibodies (Santa Cruz Biotechnology). Lysis buffer contained: 50 mM Tris·HCl, 150 mM NaCl, 0.5% Triton X-100, 0.5% octyl-β-d1-thioglucopyranoside, 10 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM sodium orthovanidate, 1 mM PMSF, and 100 μg/ml N-tosyl-l-phenylalanyl chloromethyl ketone. Immunoprecipitates were fractionated by SDS/PAGE, stained with Coomassie blue, and bands were submitted to the University of Pennsylvania Proteomics Core Facility for peptide sequencing using an LCQ Deca XP Plus mass spectrometer. Reciprocal co-IP from WB and SN fractions using antibodies against HXK (Chemicon), VDAC (Santa Cruz Biotechnology), adenine nucleotide transporter (MitoSciences, Eugene, OR), and ubiquitous mitochondrial creatine kinase (Santa Cruz Biotechnology) followed by Western blotting with an Elk-1 antibody (Cell Signaling Technology, Beverly, MA) was carried out on binding partners detected through peptide sequencing. co-IP was also carried out with an Elk-1 antibody (Santa Cruz Biotechnology) and immunoprecipitates were blotted with an HXK antibody (Santa Cruz Biotechnology) or a GAD65/67 antibody (Santa Cruz Biotechnology).
SN Fractionation for co-IP.
SN isolation was based on the established technique of Booth and Clark (31). Briefly, 6- to 8-week-old male C57BL/6 mice were killed by CO2 followed by cervical dislocation. After removal of the cerebellum, the brain was homogenized with a large dounce homogenizer (Wheaton Scientific) in cold isolation medium containing 320 mM sucrose, 10 mM Tris·HCl, and 1 mM EDTA. The homogenate was spun at 3,500 rpm in a Sorvall RC2B contrifuge for 3 min and the supernatant collated and respun at 10,000 rpm for 10 min. After resuspension of the pellet in cold isolation medium, the solution was further homogenized and mixed with 12% Ficoll (Sigma). Seven percent Ficoll and isolation medium were layered on the homogenate, and solutions were spun at 27,000 rpm for 35 min (Beckman L8–55M). SN fractions collected between the 12% and 7% Ficoll layers were used immediately for co-IP or Western blotting experiments.
Western Blotting.
Protein lysates were run on NuPAGE 10% Bis-Tris precut gels (Invitrogen), transferred to poly(vinylidene difluoride) membrane (BioRad), rinsed with PBS, blocked with 3% BSA and 0.1% Tween in PBS, and incubated overnight with primary antibody. Membranes were then washed in 0.1% Tween in PBS, incubated for 1 hour in horseradish-peroxidase-conjugated secondary antibodies (PerkinElmer), and visualized using chemiluminescence (PerkinElmer).
Electron Microscopy.
Male rats (250 g; n = 3) were anesthetized with sodium pentobarbital (60 mg/kg) and perfused transcardially with 50 ml of 3.8% acrolein (Electron Microscopy Sciences, Fort Washington, PA) and 200 ml of 2% paraformaldehyde in 0.1 M phosphate buffer (PB; pH 7.4). The brains were removed, cut into 1- to 3-mm coronal slices, and postfixed for an additional 30 min. Sections (40 μm) were cut through the rostrocaudal extent of the hippocampal formation by using a Vibratome and collected into 0.1 M PB. These were placed in 1% sodium borohydride to remove reactive aldehydes and rinsed in 0.1 M PB before the primary antibody incubation. The sections were incubated in an Elk-1 antibody (Cell Signaling Technology) for 15–18 h at room temperature. Control sections were run in parallel in which the primary antisera was omitted, but the rest of the processing procedure was identical. Sections were rinsed in 0.1 M PBS and incubated in the secondary antiserum at room temperature. For peroxidase detection of Elk-1, sections were incubated in biotinylated donkey anti-rabbit IgG (1:400; Jackson ImmunoResearch) for 30 min followed by incubation in avidin–biotin complex (Vector Laboratories). This was visualized by reaction with 3–3′ diaminobenzidine (Aldrich) and H2O2 in 0.1 M PBS. For immunogold–silver detection of Elk-1, sections were incubated in 0.01 M PBS containing 0.1% gelatin and 0.8% BSA for 30 min. Sections were then incubated in donkey anti-rabbit IgG conjugated to 1-nm gold particles (Amersham Pharmacia) for 2 h at room temperature. These were rinsed and incubated in 1.25% glutaraldehyde (Electron Microscopy Sciences) followed by a wash in 0.01 M PBS and then in 0.2 M sodium citrate buffer (pH 7.4). Silver intensification of the gold particles was achieved by using a silver enhancement kit (Amersham Pharmacia). Sections were rinsed and incubated in 2% osmium tetroxide (Electron Microscopy Sciences) for 1 h, rinsed, dehydrated, and flat-embedded in Epon 812 (Electron Microscopy Sciences). Thin sections (80–100 nm) were cut from the outer surface of the tissue, collected on grids, and examined with an FEI transmission electron microscope.
Mitochondrial Fractionation for Western Blots.
A mitochondrial isolation kit (Pierce) was used according to the manufacturer's instructions to isolate mitochondria from primary cortical neurons 14–21 days in vitro. Once isolated, mitochondria were lysed in buffer containing: 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris, and Pierce protease inhibitor cocktail. Lysate was then used for Western blotting with the following antibodies: Elk-1 (Santa Cruz Biotechnology), VDAC1 (Santa Cruz Biotechnology), and NeuN (Abcam Ltd., Cambridge, U.K.).
Camptothecin/Etoposide Treatment.
Primary rat cortical neurons 14–21 days in vitro were incubated at 37°C in extracellular solution (pH 7.4) containing: 100 mM NaCl, 3 mM KCl, 2 mM CaCl2, 10 mM glucose, 100 μM glycine, and 10 mM Hepes (32). Following application of camptothecin or etoposide (15 μM; Calbiochem), mitochondrial fractions were isolated according to the manufacturer's instructions (Pierce), and lysate was then used for Western blotting.
ATP Cell Viability Measurements.
Elk-1 in a pcDNA3.1A vector (Invitrogen) or pcDNA3.1A alone, or Elk-1 siRNA or Silencer Negative Control siRNA (Ambion) was introduced into primary rat hippocampal neurons in suspension using electroporation according to the manufacturer's instructions (Amaxa, Gaithersburg, MD). Two different Elk-1 siRNAs were introduced, corresponding to different regions of Elk-1 (siRNA A target sequence: GGTGAGCGGCCAGAAGTTT, siRNA B target sequence: AGTTGGTGGATGCAGAGGA). Neurons were then plated on glass coverslips, and a luminescent cell viability assay (Promega) was used to measure total cellular ATP. Neurons were lysed with CellTiter-Glo reagent (Promega), equilibrated at room temperature for 30 min, and luminosity was measured with a spectrofluorometer (Tecan, Durham, NC).
BKA Inhibition.
Primary rat hippocampal neurons 14–21 days in vitro were preincubated for 30 min with 25 μM BKA (Calbiochem), and individual neurons were transfected with full-length Elk-1 mRNA for overexpression (produced by using Ambion mMessage mMachine high-yield capped RNA transcription kit). Four hours later, cell death was measured in neurons by using a live-dead cell viability-cytotoxicity assay according to the manufacturer's instructions (Molecular Probes).
Supplementary Material
Acknowledgments
We thank M. Maronski for the preparation of primary neuronal cultures, R. Magtoto and M. Schmid for help with tissue processing, and Drs. R. Pittman and C. Lai and past and present members of the Eberwine lab for helpful discussions. This work was funded by National Institutes of Health Grants MH74169 (to L.E.B.), DA015395 (to E.J.V.B.), DA09082 (to E.J.V.B.), MH071705 (to P.G.H.), AG9900 (to J.H.E.), and MH58561 (to J.H.E.).
Abbreviations
- Elk-1
E-26-like protein 1
- PTP
permeability transition pore
- co-IP
coimmunoprecipitation
- VDAC
voltage-dependent anion channel
- HXK
hexokinase
- BKA
bongkrekic acid
- WB
whole brain
- SN
synaptoneurosome.
Footnotes
Conflict of interest statement: L.E.B., J.Y.S., P.G.H., and J.H.E. are inventors on a patent that has been submitted on aspects of this work.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sharrocks A. D. Biochem. Soc. Trans. 2002;30:1–9. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 2.Sharrocks A. D. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 3.Chai Y. L., Chipitsyna G., Cui J., Liao B., Liu S., Aysola K., Yezdani M., Reddy E. S. P., Rao V. N. Oncogene. 2001;20:1357–1367. doi: 10.1038/sj.onc.1204256. [DOI] [PubMed] [Google Scholar]
- 4.Shao N., Chai Y., Cui J., Wang N., Aysola K., Reddy E. S. P., Rao V. N. Oncogene. 1998;17:527–532. doi: 10.1038/sj.onc.1201931. [DOI] [PubMed] [Google Scholar]
- 5.Thiels E., Kanterewicz B. I., Norman E. D., Trzaskos J. M., Klann E. J. Neurosci. 2002;22:2054–2062. doi: 10.1523/JNEUROSCI.22-06-02054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis S., Vanhoutte P., Pages C., Caboche J., Laroche S. J. Neurosci. 2000;20:4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sgambato V., Vanhoutte P., Pages C., Rogard M., Hipskind R., Besson M. J., Caboche J. J. Neurosci. 1998;18:214–226. doi: 10.1523/JNEUROSCI.18-01-00214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchenko N. D., Zaika A., Moll U. M. J. Biol. Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 9.Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 10.Schuler M., Green D. R. Trends Genet. 2005;21:182–187. doi: 10.1016/j.tig.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Bottero V., Rossi F., Samson M., Mari M., Hofman P., Peyrom J. F. J. Biol. Chem. 2001;276:21317–21324. doi: 10.1074/jbc.M005850200. [DOI] [PubMed] [Google Scholar]
- 12.Cogswell P. C., Kashatus D. F., Keifer J. A., Guttridge D. C., Reuther J. Y., Bristow C., Roy S., Nicholson D. W., Baldwin A. S., Jr J. Biol. Chem. 2003;278:2963–2968. doi: 10.1074/jbc.M209995200. [DOI] [PubMed] [Google Scholar]
- 13.Zamora M., Merono C., Vinas O., Mampel T. J. Biol. Chem. 2004;279:38415–38423. doi: 10.1074/jbc.M404928200. [DOI] [PubMed] [Google Scholar]
- 14.Kim J. S., He L., Lemasters J. J. Biochem. Biophys. Res. Commun. 2003;304:463–470. doi: 10.1016/s0006-291x(03)00618-1. [DOI] [PubMed] [Google Scholar]
- 15.Halestrap A. P., McStay G. P., Clarke S. J. Biochemie. 2002;84:153–166. doi: 10.1016/s0300-9084(02)01375-5. [DOI] [PubMed] [Google Scholar]
- 16.Beutner G., Ruck A., Riede B., Brdiczka D. Biochim. Biophys. Acta. 1998;1368:7–18. doi: 10.1016/s0005-2736(97)00175-2. [DOI] [PubMed] [Google Scholar]
- 17.Morris E. J., Geller H. M. J. Cell Biol. 1996;134:757–770. doi: 10.1083/jcb.134.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck W. T., Mo Y. Y., Bhat U. G. Biochem. Soc. Trans. 2001;29:702–703. doi: 10.1042/0300-5127:0290702. [DOI] [PubMed] [Google Scholar]
- 19.Besirli C. G., Johnson E. M., Jr J. Biol. Chem. 2003;278:22357–22366. doi: 10.1074/jbc.M300742200. [DOI] [PubMed] [Google Scholar]
- 20.Cesari F., Brecht S., Vintersten K., Vuong L. G., Hofmann M., Klingel K., Schnorr J. J., Arsenian S., Schild H., Herdegen T., et al. Mol. Cell. Biol. 2004;24:294–305. doi: 10.1128/MCB.24.1.294-305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hans G., Malgrange B., Lallemend F., Crommen J., Wislet-Gendebien S., Belachew S., Robe P., Rogister B., Moonen G., Rigo J. M. Neuropharmacology. 2005;48:105–117. doi: 10.1016/j.neuropharm.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Jiang D., Sullivan P. G., Sensi S. L., Steward O., Weiss J. H. J. Biol. Chem. 2001;276:47524–47529. doi: 10.1074/jbc.M108834200. [DOI] [PubMed] [Google Scholar]
- 23.Tang T. S., Slow E., Lupu V., Stavrovskaya I. G., Sugimori M., Llinas R., Kristal B. S., Hayden M. R., Bezprozvanny I. Proc. Natl. Acad. Sci. USA. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan J., Galindo M. F., Gonzalez-Garcia C., Cena V. Neuroscience. 2003;122:707–715. doi: 10.1016/j.neuroscience.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 25.Sugimoto T., Stewart S., Guan K. L. J. Biol. Chem. 1997;272:29415–29418. doi: 10.1074/jbc.272.47.29415. [DOI] [PubMed] [Google Scholar]
- 26.Cruzalegui F. H., Cano E., Treisman R. Oncogene. 1999;18:7948–7957. doi: 10.1038/sj.onc.1203362. [DOI] [PubMed] [Google Scholar]
- 27.Crino P., Khodakhah K., Becker K., Ginsberg S., Hemby S., Eberwine J. Proc. Natl. Acad. Sci. USA. 1998;95:2313–2318. doi: 10.1073/pnas.95.5.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilman C. P., Chan S. L., Guo Z., Zhu X., Greig N., Mattson M. P. Neuromol. Med. 2003;3:159–172. doi: 10.1385/NMM:3:3:159. [DOI] [PubMed] [Google Scholar]
- 29.Lerner-Natoli M., Montpied P., Rousset M. C., Bockaert J., Rondouin G. Epilepsy Res. 2000;41:141–154. doi: 10.1016/s0920-1211(00)00132-7. [DOI] [PubMed] [Google Scholar]
- 30.Vernon E., Meyer G., Pickard L., Dev K., Molnar E., Collingridge G. L., Henley J. M. Mol. Cell. Neurosci. 2001;17:637–645. doi: 10.1006/mcne.2000.0960. [DOI] [PubMed] [Google Scholar]
- 31.Booth R. F., Clark J. B. Biochem. J. 1978;176:365–370. doi: 10.1042/bj1760365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng H. C., Dichter M. A. Neurobiol. Dis. 2005;19:77–83. doi: 10.1016/j.nbd.2004.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.