Abstract
A class of diuretic/aquaretic agents based on mirror-image oligonucleotides (so-called Spiegelmers) has been identified. These molecules directly bind and inhibit the neuropeptide vasopressin (AVP). AVP is the major regulatory component of body fluid homeostasis mediated through binding to the renal V2 receptor. Elevated plasma levels of AVP are implicated in several pathological conditions, mainly cardiovascular diseases. In congestive heart failure, AVP is part of a neuroendocrine imbalance that is responsible for progressive worsening of the disease. Employing in vitro selection techniques, RNA aptamers that bind to the unnatural d-configuration of AVP were isolated. The best aptamer displayed an affinity to d-AVP of ≈560 pM at 37°C. The corresponding Spiegelmer, a 38-mer mirror-image oligonucleotide (l-RNA) termed NOX-F37, inhibits vasopressin-dependent activation of V1a as well as V2 receptors with IC50 values of 6.1 nM and 1 nM, respectively. NOX-F37 administered to healthy rats effectively neutralized AVP and increased diuresis dose-dependently for 24 h. The mode of action was strictly aquaretic, i.e., the increase in urine volume was not accompanied by an increase in electrolytes. These results clearly prove the in vivo efficacy of NOX-F37 and points out its potential as a drug in the treatment of diseases that are associated with body fluid overload.
Keywords: aptamer, congestive heart failure, diureses, water homeostasis, Spiegelmer
The cyclic nonapeptide vasopressin (AVP) is a neurohormone that is synthesized in the hypothalamus and stored in the hypophysis from where it is released into the circulation (1). AVP is also known as antidiuretic hormone because its main peripheral function is the maintenance of volume and osmolality of body fluids by regulating the free water reabsorption in the kidneys. Under physiological concentrations, AVP acts vasoconstrictive. AVP’s actions are mediated through specific G protein-coupled receptors (2). The V2 receptor, mainly expressed on cells of the basolateral membrane of the collecting duct in the kidneys, mediates the antidiuretic effect via enhanced cAMP levels (3). As a result, aquaporin channels are inserted into the cytoplasma membranes, and the transcription of aquaporin mRNA is induced. The V1a receptor, mainly expressed on vascular smooth muscle cells and on cells of the myocardium, exerts vasoconstrictive effects through the second messengers inositoltrisphosphate and diacylglycerol. Subsequently, intracellular Ca2+ is mobilized from the endoplasmic reticulum, and Ca2+ channels as well as Na+/H+ exchangers are activated (4). In addition to vasoactive effects, binding of AVP to the V1a receptor also stimulates cell growth and proliferation in vascular smooth muscle cells through activation of the mitogen-activated protein kinase pathway (5).
AVP release into the circulation is tightly regulated by osmoreceptors in the hypothalamus and baroreceptors in the heart and large arteries. Whereas osmoreceptors respond to small changes (1%) in osmolality, baroreceptors respond only to significant variations (10%) in blood volume and pressure (6, 7). In congestive heart failure (CHF) patients, AVP plasma levels are approximately two to three times higher than in healthy subjects (8). It is not well understood why the AVP release is inadequately coupled to the decreased plasma osmolality (9), but, as a consequence, patients show symptoms of chronic volume overload, hyponatremia, congestion, and increased systemic vascular resistance (10). A central point in CHF is the progression of left ventricular dysfunction and chamber remodeling (11). AVP may be involved in these aspects of CHF because AVP can induce structural changes in the myocardium through activation of the V1a receptor (12, 13). Elevated AVP levels are also found in the syndrome of inappropriate antidiuretic hormone secretion (14) and late-stage liver cirrhosis (15), which leads in both diseases to dilutional hyponatremia and hypoosmolality. Through its mitogenic properties, AVP can also affect the genesis of tumors. AVP and its receptors are expressed in all small cell lung carcinomas and breast cancer (16). In conclusion, a variety of severe human diseases exist where AVP seems to be dysregulated and for which effective treatments are necessary.
Here we describe the development of a Spiegelmer that binds to AVP with high affinity and neutralizes its action. Spiegelmers are l-enantiomeric aptamers (17) that are identified through a SELEX-based (18) in vitro selection process that involves chiral principles (19). First, the mirror-image configuration of AVP (all d-amino acids) is synthesized. A combinatorial nucleic acid library is screened to identify aptamers that bind to the all-d-AVP. Lastly, an individual aptamer sequence is synthesized in its corresponding mirror-image configuration to give a Spiegelmer (l-oligonucleotide) that binds to the natural AVP. Spiegelmers have been identified already to bind to a variety of different peptide and protein targets (20–22). The mirror-image configuration confers excellent biostability to Spiegelmers without the need for further modifications.
We report the identification of a 38-mer RNA aptamer that binds to AVP with an extraordinarily low dissociation constant of 560 pM at 37°C. The corresponding Spiegelmer NOX-F37 inhibits AVP signaling in cell culture at the V2 and the V1a receptor with IC50 values of 1 and 6.1 nM, respectively. After systemic administration NOX-F37 efficiently induces diuresis in a rat model, demonstrating its potential in neutralizing AVP’s action in vivo.
Results
Identification of d-AVP-Binding RNA Sequences.
The in vitro selection process to identify high-affinity RNA sequences binding to d-AVP was divided into two parts: an automation-based standard selection (23) followed by a manually performed high-stringency protocol that included mutagenic steps within the amplification step. The selection was started with a library of ≈2.4 × 1015 different RNA molecules and individual aptamer sequences were identified after 16 selection rounds. Alignment of the sequences revealed one family of relatives that differ by point mutations (data not shown).
Truncation of the primer binding sites resulted in aptamers consisting of 47–49 nucleotides. The best binding sequence 134-A9 (Fig. 1A) was synthesized in its l-configuration. It inhibits l-AVP-induced cAMP formation in cell culture with an IC50 of ≈50 nM (data not shown). To identify sequences with even higher binding affinity, the enriched pool of round 16 was subjected to additional 13 selection rounds with increased stringency, and a mutagenic amplification protocol was applied in six selection rounds. Sequencing revealed further variants of the originally identified family. An alignment unveiled a 46- to 49-nt consensus motif (Fig. 1B; clustal x; ref. 24) that contains distinct secondary structure elements as predicted by a software algorithm (ref. 25; Fig. 2). The 5′ and 3′ end of the sequences are self-complementary (Helix 1 and Helix 2) and seem to form a helix. Box 1, directly attached to the helix, is joined to Box 2 by a 6- to 9-nt-long region mainly composed of adenosine and uridine residues. This AU-rich region shows the highest degree of diversity. Box 2, defined by four conserved stretches of two (one time), three (two times), or four (one time) guanosine nucleotides, is joined to the helix region at the 3′ end. The guanosine stretches of Box 2 can potentially form a G quartet. Although the sequences that resulted from the high stringency selection were not much different from the originally identified molecules, comparative ranking showed a significant improvement in binding affinity. In a competitive binding assay with an excess of nonlabeled RNA, the best aptamer from the high stringency selection, 157-B4, was determined to bind ≈12-fold better to biotinylated d-AVP than 134-A9 (data not shown).
Fig. 1.
AVP binding sequences. (A) 134-A9 is the best sequence that resulted from automated selection whereas 157-B4 was obtained as the best sequence after applying a mutagenic high-stringency (mhs) protocol; other variants of the mhs protocol are listed in Supporting Appendix, which is published as supporting information on the PNAS web site. (B) Consensus motif of AVP-binding sequences composed of terminal helical regions, two G rich boxes and an AU-rich (6–9 nt) region. The four G stretches in box2 are shown in bold.
Fig. 2.
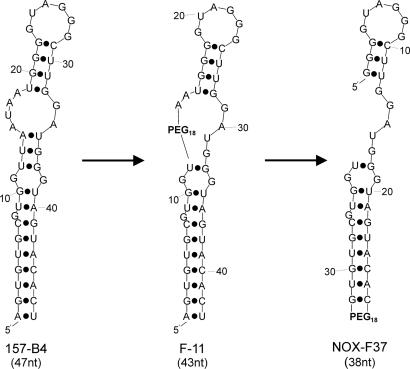
Secondary structure predictions of AVP-binding sequences. Site-directed modification of 157-B4 with a hexaethylene glycol linker (PEG18) led to a series of intermediate sequences with a PEG18 linker located in the AU-rich region (e.g., F-11). In the final sequence NOX-F37, the AU-rich region was completely removed, the secondary structure was opened at this site and closed with the PEG18 linker at the former 5′ and 3′ end.
Optimization of Aptamer 157-B4 by Site-Directed Modification with Hexaethyleneglycol.
Because shorter oligonucleotides are easier and more cost effectively produced, we truncated the best binding aptamer 157-B4 (47-mer) further by replacing nucleotides with a hexaethyleneglycol linker (26). The sequence variability of the AU-rich region was the most promising target to introduce the linker (Fig. 2). Indeed, all different variants in which up to seven nucleotides were replaced are still active (e.g., F-11, a 43-mer). However, replacement of more than three nucleotides showed a slight decrease in affinity toward AVP. Therefore, the structure was “opened” at this site. Seven nucleotides were deleted and new 5′ and 3′ ends were introduced, whereas the structure was “closed” at the former helical 5′ and 3′ ends with the hexaethylene glycol linker. This operation preserved the high affinity to AVP. Moreover, the former 7-bp helix could be truncated additionally to a 6-bp helix so that the resulting RNA oligonucleotide consists of 38 nucleotides plus the linker. The final sequence was synthesized in its mirror-image configuration as Spiegelmer NOX-F37 (Fig. 2) and was further characterized in vitro and in vivo.
Affinity of NOX-F37 Toward AVP.
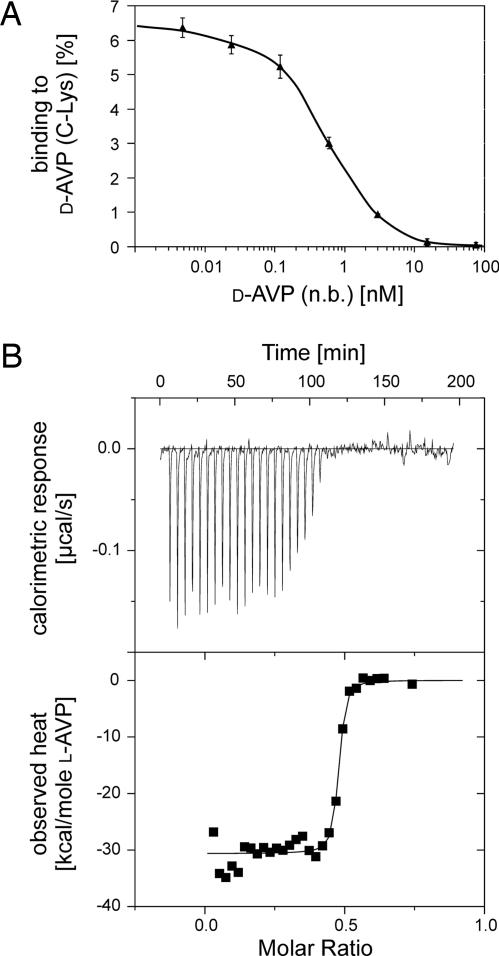
The affinity of aptamer NOX-F37 was measured in a competitive pulldown assay that allowed the determination of affinity constants in solution (27). The concentrations of radioactively labeled aptamer and biotinylated d-AVP were kept constant, whereas the concentration of the nonbiotinylated d-AVP competitor was varied. Assuming a 1:1 stoichiometry, the dissociation constant (Kd) of aptamer NOX-F37 to d-AVP at 37°C was determined to be 560 ± 52 pM (Fig. 3A). Reducing AVP’s disulfide bridge (100 mM DTT) and, thus, destroying its 3D structure resulted in a total loss of aptamer binding, which underlines the oligonucleotides’ ability to specifically recognize a structural element rather than a short sequence stretch of amino acids (data not shown).
Fig. 3.
Affinity of the aptamer and Spiegelmer NOX-F37 sequences to AVP. (A) Competitive pulldown assay of aptamer NOX-F37. The binding of radioactively labeled aptamer to biotinylated d-AVP(C-Lys) was measured in the presence of the indicated concentrations of nonbiotinylated peptide [d-AVP(n.b.)]. The dissociation constant (560 ± 52 pM) was calculated (grafit) by using a 1:1 stoichiometry. (B) Calorimetric profile of Spiegelmer NOX-F37 binding to l-AVP. Results of a typical ITC measurement are shown. The dissociation constant was determined to be 1.7 ± 0.8 nM assuming a 1:1 stoichiometry.
The affinity of the Spiegelmer NOX-F37 was determined by isothermal titration calorimetry (ITC), and the apparent Kd was calculated to be 1.7 ± 0.8 nM at 37°C (Fig. 3B). The 3′ PEGylated [40 kDa polyethylene glycol (PEG)] variant of NOX-F37, which was used for in vivo experiments (22, 28), displayed a Kd of 1.3 ± 0.5 nM, which is in good agreement with the value determined for the unmodified molecule.
Inhibition of l-AVP by NOX-F37 in Cell Culture.
The potential of NOX-F37 to block receptor-mediated cell signaling of l-AVP was tested with two different cell lines. Whereas LLC-PK1 cells (expressing V2 receptors) respond with an increase of intracellular cAMP release, A7r5 smooth muscle cells (expressing V1a receptors) mobilize intracellular Ca2+ after l-AVP stimulation.
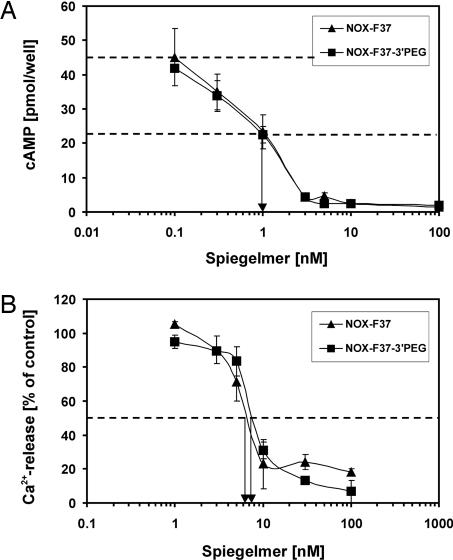
The inhibitory potency of NOX-F37 was determined by using a constant l-AVP stimulus in the EC50 range and increasing concentrations of NOX-F37. The NOX-F37 concentration at 50% inhibition (IC50) was deduced from the respective dose–response curve and calculated to be 1 nM for the V2 receptor and 6.1 nM for the V1a receptor, respectively (Fig. 4). The inhibition was specific because a nonfunctional Spiegelmer that does not bind to AVP did not show any inhibitory effect in the test systems (data not shown). The 3′ PEGylated NOX-F37 showed essentially the same inhibition constant (Fig. 4), confirming the results of the ITC measurements. The different IC50 values of NOX-F37 in the two cell culture assays may be due to the divergent EC50 values.
Fig. 4.
Inhibition of l-AVP mediated receptor activation by NOX-F37 (▴) and its 40-kDa PEGylated variant (■). (A) V2 receptor was stimulated with 1 nM l-AVP in the presence of the indicated concentrations of Spiegelmer, the resulting cAMP formation was measured, and the IC50 (1 nM) was determined at the Spiegelmer concentration of half-maximal cAMP formation. (B) V1a receptor was stimulated with 5 nM l-AVP in the presence of the indicated concentrations of Spiegelmer, the resulting Ca2+ release was measured, and the IC50 (6.1 nM) was determined at the Spiegelmer concentration of half-maximal Ca2+ release.
Effect of NOX-F37 on Diuresis.
In vivo activity of the l-AVP binding and inhibiting Spiegelmer NOX-F37 was tested in a rat diuresis model. Three doses of the 3′ PEGylated NOX-F37 (80, 400, and 2,000 nmol/kg), a nonfunctional control Spiegelmer (3′-PEGylated), and vehicle were administered to conscious, healthy rats i.v. At different time points, urine volume, osmolality, sodium concentration, and water intake was measured.
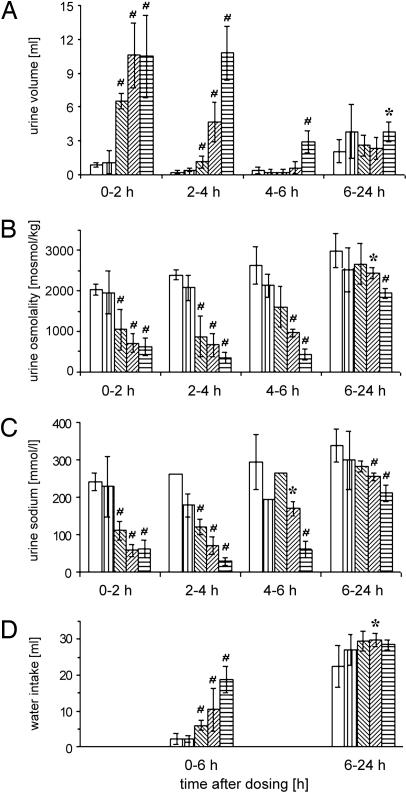
Two hours after NOX-F37 administration, a dose-dependent increase in urine volume with a peak of 6.5 ml for the low dose and peaks of 10.5 ml for the middle and high dose was observed (Fig. 5A). These effects are specific to NOX-F37, because rats treated with vehicle or with control Spiegelmer showed a urine volume of only ≈1 ml in this time interval. For the low and middle dose, the effect of NOX-F37 decreased in the following time interval (2–4 h), and the urine volume reached the level of the control animals in the third interval (4–6 h). However, the highest dose of NOX-F37 induced a very strong response not only in the first (0–2 h) and second (2–4 h), but also in the third time interval (4–6 h). In the final time interval (6–24 h), no major differences between the groups were detectable.
Fig. 5.
Time course and dose–response of NOX-F37 on diuresis and water intake in conscious rats. The different bars represent the following groups: open bars, vehicle (PBS); vertical hatched bars, 2,000 nmol/kg control Spiegelmer; left hatched bars, 80 nmol/kg NOX-F37; right hatched bars, 400 nmol/kg NOX-F37; horizontal hatched bars, 2,000 nmol/kg NOX-F37. Determined parameters are as follows: urine volume (A), urine osmolality (B), urine sodium concentration (C), and water intake (D). Data are the mean ± SEM (n = 5) with the following exceptions: For the urine sodium concentration, only one measured value could be generated in the vehicle group in the interval 2–4 h, and in the control Spiegelmer group and the NOX-F37 group (80 nmol/kg) for the interval 4–6 h. ∗, P < 0.05; #, P < 0.01 indicate statistical significance of effects observed in the treated groups compared with the vehicle group.
Concomitant to the increase in urine volume, the osmolality and the sodium concentration of the urine decreased dose dependently after NOX-F37 administration (Fig. 5 B and C). These effects seem to persist longer than the increase in urine volume. After 6 h, all three doses showed a clear trend to a lower urine osmolality and sodium concentration, and this effect was still significant in the final time interval 6–24 h for the highest dose. The decrease of the electrolyte concentration is clear proof of the Spiegelmers’ ability to interfere with the animal’s AVP system. The urine volume was increased and diluted compared to the control groups, indicating that the inhibition of AVP results in a reduction of reabsorption of water in the collecting duct of the kidneys.
During the study, the animals had free access to water. In parallel to the drastic increase in urine volume, the water intake increased dose dependently in the first measured time interval (0–6 h) (Fig. 5D). In the time interval 6–24 h, the differences between the groups in water intake leveled off.
Discussion
Spiegelmers and aptamers are 3D nucleic acid structures that are able to bind to target molecules conceptually similar to antibodies. Unlike aptamers and other oligonucleotide-based structures, Spiegelmers are nuclease-resistant because of their unnatural l-enantiomeric configuration and, therefore, are biostable without any further modification. This property makes them very well suited for in vivo applications and, thus, for therapeutic use.
After a combination of high-stringency selection protocols and mutagenization steps, we have identified very affine vasopressin-specific RNA oligonucleotides that display picomolar dissociation constants at 37°C. Truncation and site-directed modifications, including the introduction of an internal (18 atoms) PEG linker spanning the former 5′ and 3′ end yielded the final candidate sequence Spiegelmer NOX-F37. NOX-F37 and its terminally PEGylated derivative (40 kDa) display dissociation constants of 1.7 nM and 1.3 nM (measured at 37°C), respectively, which is three orders of magnitude better than the dissociation constant of a previously published 55-mer mirror-image DNA molecule that binds to AVP with a Kd of 1.2 μM (measured at room temperature) (29). In both cases, the binding to nonmodified AVP was determined in solution but with different assay formats (ITC vs. equilibrium dialysis).
Elevated AVP levels have been correlated with severe diseases. The most important and widespread is CHF, a major cardiac disorder that is increasing in prevalence (30–32). The standard therapy comprises the medication with angiotensin-converting enzyme inhibitors, β-adrenoreceptor antagonists, and diuretics (33). Despite the effectiveness of standard medications, the morbidity and mortality of patients suffering from CHF remains dissatisfactorily high (34).
Evidence that points to a significant role of AVP in the progressive worsening of CHF has stimulated the development of AVP receptor antagonists. Various nonpeptidic AVP receptor antagonists have been discovered (35). Most advanced are tolvaptan, described as a specific V2 receptor antagonist, and conivaptan, a dual V1a and V2 receptor antagonist. Both antagonists are benzazepine derivatives that show inhibition constants to the human V2 and V1a receptors of 0.4 and 12.3 nM for tolvaptan and 1.1 and 6.3 nM for conivaptan, respectively (36, 37). In vivo efficacy was demonstrated for both antagonists in preclinical and clinical studies (7, 38–40).
We have demonstrated in vivo efficacy of NOX-F37, a compound that directly binds and inhibits AVP. A single i.v. administration of NOX-F37 increased the urine volume of normally hydrated conscious rats in a 2-h interval >10-fold and decreased urine osmolality and sodium concentration >5-fold at the same time. These aquaretic effects are comparable with reports on the activity of tolvaptan (36) and conivaptan (41) in rats. The advantage of aquaretic drugs in the treatment of CHF is the removal of excessive water from the body without causing serum electrolyte imbalances. A loss of electrolytes, which usually is excited by conventional diuretics (31, 42) can induce or even worsen hyponatremia, one of the most important indicators of cardiovascular mortality (43). Quite the contrary, the normalization of plasma sodium concentrations will result with aquaretically acting compounds. Therefore, NOX-F37 may have the potential for a more optimal diuretic therapy in diseases with increased water load.
In addition to its function in water homeostasis, a solid body of evidence exists that the AVP system(s) may be involved in cardiac muscle hypertrophy and remodeling of the left ventricle, a key aspect in the progression of CHF (10). In this context, the importance of AVP signaling at the V1a receptor still remains to be elucidated. With respect to the low dissociation constant, NOX-F37 might be even more effective in inhibiting AVP signaling via the V1a receptor than the receptor antagonist conivaptan. For conivaptan, a Ki of 6.3 nM has been reported (44). Moreover, the IC50 value in a cell culture assay is higher for conivaptan (44) than for NOX-F37 (14.3 nM vs. 6.1 nM), which is further strengthened by the fact that the IC50 of NOX-F37 was determined in an ≈4-fold less-sensitive cell assay (EC50 ≈ 5 nM) compared with conivaptan (EC50 ≈ 1.3 nM; ref. 37). It can be concluded that NOX-F37 might block AVP-induced V1a receptor activation very efficiently and could reduce AVP-mediated remodeling in the heart of CHF patients accordingly.
NOX-F37 is a nucleic acid-based compound with potentially high therapeutic value for the treatment of severe AVP-conveyed diseases of which CHF remains the most important. To examine whether NOX-F37 can, in addition to its strong aquaretic effects, reduce cardiac hypertrophy, further studies in appropriate animal models will be necessary.
Materials and Methods
Peptides and Nucleic Acids.
All-l-AVP (CYFQNCPRG-NH2) and all-d-AVP were obtained from Bachem (Bubendorf, Switzerland); the latter was custom synthesized nonbiotinylated as well as C-terminally biotinylated. The biotin group was linked to the d-peptide with a PEG linker d-AVP(C-Lys) (linker: amino-ethyloxy-ethyloxy-acetylamino-ethyloxy-ethyloxy-acetyl) and a double-long chain linker d-AVP(LC-LC) (LC-LC: ε-aminohexyl-ε-aminohexyl), respectively. Oligonucleotides were synthesized at NOXXON by using standard phosphoramidite chemistry. l-Phosporamidites and hexaethylene glycol phosporamidites (PEG-linker) were from ChemGenes Corp. (Wilmington, MA). The DNA library with 34 internal random positions had the sequence 5′-GTGGAACCGACTCACCTGAGCGTGC-N34-GCACGCTGCTGTTGTCTAAGCTCC-3′ and was amplified with the forward primer 5′-TCTAATACGACTCACTATAGGAGCTTAGACAACAGCAG-3′ and the reverse primer 5′-GTGGAACCGACTCACCTGAG-3′. For in vivo application, the AVP-binding Spiegelmer NOX-F37 (5′-GGGGUAGGGCUUGGAUGGGUAGUACAC[PEG]18GUGUGCGUGGU-3′) and a nonfunctional control Spiegelmer (5′-UAAGGAAACUCGGUCUGAUGCGGUAGCGCUGUGCAGAGCU-3′) were modified with 40-kDa PEG at the 3′ and 5′ terminus, respectively (22).
In Vitro Selection.
In vitro selection to identify aptamers to d-AVP was started with an automation-based standard protocol (23) followed by 13 rounds of a mutagenic selection protocol. After annealing the forward primer, the ssDNA library (≈2.4 × 1015 molecules) was made double-stranded by using TaqDNA polymerase (Invitrogen). The RNA library was transcribed and labeled with T7 RNA polymerase (Stratagene) from the dsDNA by using [α-32P]-ATP and -GTP (Hartmann Analytic, Braunschweig, Germany). RNA with higher specific radioactivity was obtained by transcription in the presence of 2.5 mM guanosine and 1 mM NTPs and subsequent labeling with [γ-32P]-ATP (Hartmann Analytic) and T4 polynucleotide kinase (Invitrogen). Transcribed and labeled RNA was purified on denaturing (8 M urea) polyacrylamide gels. Before each selection step, RNA was denatured in 20 mM Tris, pH 7.4/150 mM NaCl/5 mM KCl at 94°C for 2 min, and was immediately placed on ice before it was supplemented with 1 mM MgCl2/1 mM CaCl2/0.1% Tween 20 for renaturing at 37°C.
Selection rounds 1–3 were carried out manually with two complexities of the RNA library at 25 μM RNA/25 μM peptide (rounds 1–2) and 10 μM RNA/10 μM peptide concentration (round 3). The library was incubated with biotinylated d-AVP at 37°C, and complexes were immobilized on NeutrAvidin agarose (NAag) or Streptavidin ultralink plus (SAul+) (Pierce). Nonbound RNA was separated by washing the matrix with selection buffer. Bound RNA was eluted, reverse transcribed (SuperScript II, Invitrogen), amplified by TaqDNA polymerase, and in vitro transcribed for the next selection round. A preincubation step with pure matrix was performed starting with round 3. In every round, a different combination of matrix (NAag/SAul+) and peptide [d-AVP(C-Lys)/d-AVP(LC-LC)] was used. The stringency was increased by reducing the peptide concentration to 20 nM. DNA from round 16 was cloned and sequenced (Agowa, Berlin). The same DNA was used as the starting library for the mutagenic protocol.
In the mutagenic protocol, the stringency was increased further by decreasing the peptide concentration from 5 nM (round 17) to 0.4 pM (round 29). In each round, binding reactions with different peptide concentrations and a control reaction without target peptide were conducted. Only the eluate from the most stringent binding reaction that still showed a higher signal than the control reaction without peptide was processed further. In parallel, the RNA concentration was decreased from 10 nM (round 17) to 330 pM (round 29). To increase the complexity of the binding pool, the DNA templates for rounds 17, 18, 20, 22, 24, and 27 were amplified by mutagenic PCR in the presence of MnCl2 and an unbalanced mix of dNTPs according to Fromant et al. (45). The mutation rate of an amplified reference aptamer was determined to be 6%. DNA obtained from round 29 was cloned and sequenced.
Site-Directed Modification and Truncation.
The most affine oligonucleotide was modified to identify shorter variants. Potential dispensable positions were bridged with a hexaethylene glycol linker. The resultant variants were analyzed with a ranking assay using competitive binding of a radioactively labeled reference aptamer and an excess of nonlabeled variant aptamer to d-AVP.
Determination of Binding Affinities.
The affinity of aptamers to d-AVP was measured by competition binding to biotinylated d-AVP vs. nonbiotinylated d-AVP in a pulldown assay at 37°C. After de- and renaturing, samples of 0.2 nM radioactively labeled d-RNA were incubated in selection buffer at 37°C with a constant amount of 0.8 nM biotinylated d-AVP and varying amounts of nonbiotinylated d-AVP for 2–3 h to reach equilibrium at low concentrations. Selection buffer was supplemented with 1 μg/ml human serum albumin (Sigma-Aldrich) and 1 μg/ml yeast RNA (Roche, Mannheim, Germany). After immobilization and washing without any competition, ≈10–15% binding to the biotinylated peptide was observed. To immobilize peptide and peptide–aptamer complexes, a constant amount of 25% SAul+ matrix, preequilibrated in selection buffer (corresponds to 1 μl of 100% SAul+), was added and kept suspended by mixing for 30 min. After detaching the supernatant and washing, the matrix-associated radioactivity was determined in a scintillation counter (LS6500; Beckman Coulter). The percentage of binding was plotted against the concentration of nonbiotinylated d-AVP and dissociation constants were calculated assuming a 1:1 stoichiometry (grafit, Erithacus Software, Surrey, U.K.).
The affinity of NOX-F37 to l-AVP was measured in solution by isothermal titration calorimetry on a VP-ITC instrument (MicroCal, Northampton, MA) at 37°C (23). After de- and renaturing of NOX-F37, Spiegelmer as well as l-AVP solutions were degassed and temperature adjusted in selection buffer (without detergent). 3–5 μM NOX-F37 (1.4 ml) solution was loaded to the instrument’s cell, and 0.25 ml of 25 μM l-AVP was injected in 7.5-μl portions at 6-sec duration for each injection and 5-min intervals between the injections for equilibration. After each injection, the generated heat of the binding reaction was recorded. ITC binding curves were analyzed by using the single-site binding equation in the MicroCal origin software package.
Inhibition of AVP Signaling in Cell Culture.
The inhibitory activity of l-AVP binding NOX-F37 was determined by using cell lines expressing the renal V2 receptor (LLC-PK1 from porcine kidney epithelium; ATCC-CL101) and the vascular V1a receptor (A7r5 from rat aortic smooth muscle; ATCC-CRL-144), respectively.
LLC-PK1 cells were stimulated with 1 nM l-AVP, and cAMP formation was determined in the presence of different NOX-F37 concentrations. Cells were grown overnight at a density of 6 × 104 per well in a 96-well plate in Medium 199 (Invitrogen) supplemented with 10% heat inactivated FCS/4 mM l-alanyl-l-glutamine/50 units/ml penicillin/50 μg/ml streptomycin at 37°C and 5% CO2 atmosphere. Twenty minutes before stimulation, the cells were pretreated with 1 mM isobutyl-1-methylxanthin (IBMX). The IBMX-containing medium was replaced by the stimulation mix (l-AVP with varying concentrations of Spiegelmer in Hank’s balanced salt solution (HBSS)/0.1% BSA/1 mM IBMX), which had been preincubated at 37°C for 30–60 min. After 30 min, the stimulation solution was replaced by lysis buffer from the cAMP-Screen System (Applied Biosystems). The cAMP content of the cells was measured by luminescence detection in a POLARstar Galaxy multidetection microplate reader (BMG Labtech, Offenburg, Germany). IC50 values were determined graphically by plotting the cAMP amount against the concentration of NOX-F37. The EC50 value (1 nM) was determined previously by stimulation with varying concentrations of l-AVP without NOX-F37 (data not shown).
A7r5 cells were stimulated with 5 nM l-AVP, and the mobilization of calcium was determined in the presence of different NOX-F37 concentrations. Approximately 4 × 104 cells per well were grown overnight in a dark 96-well plate with clear bottom (Greiner, Frickenhausen, Germany) in DMEM supplemented with 10% heat-inactivated FCS/4 mM l-alanyl-l-glutamine/50 units/ml penicillin/50 μg/ml streptomycin at 37°C and 5% CO2 atmosphere. Cells were washed once with 200 μl HBSS+ (HBSS supplemented with 0.1% BSA/5 mM probenecid/20 mM Hepes) and were preloaded for 60 min at 37°C with 50 μl of 10 μM fluo-4 indicator dye solution/0.08% pluronic 127 (both from Molecular Probes) in HBSS+. Cells were then washed three times with 180 μl HBSS+, 90 μl HBSS+ was added, and the background fluorescence was measured at 485 nm excitation and 520 nm emission in a POLARstar Galaxy multidetection microplate reader. Ten microliters of stimulation mix (l-AVP with varying concentrations of NOX-F37 in HBSS+), which had been preincubated at 37°C for 15–30 min, was added and the maximum fluorescence signal was measured, corrected for background fluorescence, and plotted against the concentration of NOX-F37. IC50 values were drawn from the graphs. The EC50 value (5 nM) had been determined previously with varying peptide concentrations as described (data not shown).
Diuresis Study in a Rat Model.
The effect of NOX-F37 on diuresis was tested in a rat model (Aurigon Life Science, Tutzing, Germany). Male Sprague–Dawley rats (239–290 g; Elevage Janvier, Le Genest St. Isle, France) were acclimatized to the lab conditions for 9 days. Two days before the treatment, they were placed in individual metabolic cages. Throughout the whole experiment, animals had free access to water and food. The day before treatment, water intake and urine volume was measured. Groups of five animals were treated with NOX-F37 (80, 400, or 2,000 nmol/kg), a nonfunctional control Spiegelmer (2,000 nmol/kg), or vehicle (PBS) by i.v. bolus injection (2 ml/kg) into the tail vein. After administration, the urine was collected for time intervals of 0–2 h, 2–4 h, 4–6 h, and 6–24 h. Water intake was determined for time intervals 0–6 h and 6–24 h. Urine osmolality and sodium concentration was measured at Synlab.vet (Augsburg, Germany) by using a freezing point depression osmometer and a flame photometer, respectively. The experimental design was approved by the local government.
Statistical Analysis.
All data are expressed as mean ± SEM. The values of the diuresis study were analyzed by one-way ANOVA between treated groups and the vehicle group. P < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
We thank the NOXXON chemistry group for providing the oligonucleotides, the biology group for carrying out the cell culture assays, Christine Kaduk and Britta Wlotzka for helpful discussions, and Jenny Fischer for critical reading of the manuscript.
Abbreviations
- AVP
vasopressin
- CHF
congestive heart failure
- ITC
isothermal titration calorimetry
- PEG
polyethylene glycol.
Footnotes
Conflict of interest statement: All authors are employed by the biotechnology company NOXXON Pharma; none of the authors owns significant stock of the company.
This paper was submitted directly (Track II) to the PNAS office. J.K. is a guest editor invited by the Editorial Board.
References
- 1.Zimmerman E. A. Adv. Biochem. Psychopharmacol. 1981;28:63–75. [PubMed] [Google Scholar]
- 2.Thibonnier M., Berti-Mattera L. N., Dulin N., Conarty D. M., Mattera R. Prog. Brain. Res. 1998;119:147–161. doi: 10.1016/s0079-6123(08)61568-x. [DOI] [PubMed] [Google Scholar]
- 3.Lolait S. J., O’Carroll A. M., McBride O. W., Konig M., Morel A., Brownstein M. J. Nature. 1992;357:336–339. doi: 10.1038/357336a0. [DOI] [PubMed] [Google Scholar]
- 4.Thibonnier M., Auzan C., Madhun Z., Wilkins P., Berti-Mattera L., Clauser E. J. Biol. Chem. 1994;269:3304–3310. [PubMed] [Google Scholar]
- 5.Thibonnier M., Conarty D. M., Plesnicher C. L. Am. J. Physiol. 2000;279:H2529–H2539. doi: 10.1152/ajpheart.2000.279.5.H2529. [DOI] [PubMed] [Google Scholar]
- 6.Morris M., Alexander N. Hypertension. 1989;13:110–114. doi: 10.1161/01.hyp.13.2.110. [DOI] [PubMed] [Google Scholar]
- 7.Paranjape S. B., Thibonnier M. Expert Opin. Investig. Drugs. 2001;10:825–834. doi: 10.1517/13543784.10.5.825. [DOI] [PubMed] [Google Scholar]
- 8.Goldsmith S. R., Francis G. S., Cowley A. W., Jr, Levine T. B., Cohn J. N. J. Am. Coll. Cardiol. 1983;1:1385–1390. doi: 10.1016/s0735-1097(83)80040-0. [DOI] [PubMed] [Google Scholar]
- 9.Goldsmith S. R., Francis G. S., Cowley A. W., Jr Am. J. Cardiol. 1986;58:295–299. doi: 10.1016/0002-9149(86)90065-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee C. R., Watkins M. L., Patterson J. H., Gattis W., O’Connor C. M., Gheorghiade M., Adams K. F., Jr Am. Heart J. 2003;146:9–18. doi: 10.1016/S0002-8703(02)94708-3. [DOI] [PubMed] [Google Scholar]
- 11.Sabbah H. N. Heart Fail Rev. 2004;9:91–97. doi: 10.1023/B:HREV.0000046363.59374.23. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y., Haneda T., Osaki J., Miyata S., Kikuchi K. Eur. J. Pharmacol. 2000;391:39–48. doi: 10.1016/s0014-2999(99)00775-x. [DOI] [PubMed] [Google Scholar]
- 13.Fukuzawa J., Haneda T., Kikuchi K. Mol. Cell. Biochem. 1999;195:93–98. doi: 10.1023/a:1006980517557. [DOI] [PubMed] [Google Scholar]
- 14.Baylis P. H. Int. J. Biochem. Cell Biol. 2003;35:1495–1499. doi: 10.1016/s1357-2725(03)00139-0. [DOI] [PubMed] [Google Scholar]
- 15.Martin P. Y., Schrier R. W. Kidney Int. Suppl. 1997;59:S43–S49. [PubMed] [Google Scholar]
- 16.North W. G. Exp. Physiol. 2000;85:27S–40S. doi: 10.1111/j.1469-445x.2000.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 17.Ellington A. D., Szostak J. W. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C., Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Klussmann S., Nolte A., Bald R., Erdmann V. A., Furste J. P. Nat. Biotechnol. 1996;14:1112–1115. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- 20.Purschke W. G., Radtke F., Kleinjung F., Klussmann S. Nucleic Acids Res. 2003;31:3027–3032. doi: 10.1093/nar/gkg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vater A., Jarosch F., Buchner K., Klussmann S. Nucleic Acids Res. 2003;31:e130. doi: 10.1093/nar/gng130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wlotzka B., Leva S., Eschgfaller B., Burmeister J., Kleinjung F., Kaduk C., Muhn P., Hess-Stumpp H., Klussmann S. Proc. Natl. Acad. Sci. USA. 2002;99:8898–8902. doi: 10.1073/pnas.132067399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eulberg D., Buchner K., Maasch C., Klussmann S. Nucleic Acids Res. 2005;33:e45. doi: 10.1093/nar/gni044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicke B. J., Marion C., Chang Y. F., Gould T., Lynott C. K., Parma D., Schmidt P. G., Warren S. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 27.Friguet B., Chaffotte A. F., Djavadi-Ohaniance L., Goldberg M. E. J. Immunol. Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 28.Ostendorf T., Kunter U., Eitner F., Loos A., Regele H., Kerjaschki D., Henninger D. D., Janjic N., Floege J. J. Clin. Invest. 1999;104:913–923. doi: 10.1172/JCI6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams K. P., Liu X.-H., Schumacher T. N. M., Lin H. Y., Ausiello D. A., Kim P. S., Bartel D. P. Proc. Natl. Acad. Sci. USA. 1997;94:11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamani M., Massie B. M. Mayo Clin. Proc.; 1993. pp. 1214–1218. [DOI] [PubMed] [Google Scholar]
- 31.Schrier R. W., Abraham W. T. N. Engl. J. Med. 1999;341:577–585. doi: 10.1056/NEJM199908193410806. [DOI] [PubMed] [Google Scholar]
- 32.Doggrell S. A., Brown L. Expert Opin. Pharmacother. 2002;3:915–930. doi: 10.1517/14656566.3.7.915. [DOI] [PubMed] [Google Scholar]
- 33.Heart Failure Society of America Congest. Heart Fail. 2000;6:11–39. [Google Scholar]
- 34.Russell S. D., DeWald T. Am. J. Cardiovasc. Drugs. 2003;3:13–20. doi: 10.2165/00129784-200303010-00002. [DOI] [PubMed] [Google Scholar]
- 35.Thibonnier M., Coles P., Thibonnier A., Shoham M. Annu. Rev. Pharmacol. Toxicol. 2001;41:175–202. doi: 10.1146/annurev.pharmtox.41.1.175. [DOI] [PubMed] [Google Scholar]
- 36.Yamamura Y., Nakamura S., Itoh S., Hirano T., Onogawa T., Yamashita T., Yamada Y., Tsujimae K., Aoyama M., Kotosai K., et al. J. Pharmacol. Exp. Ther. 1998;287:860–867. [PubMed] [Google Scholar]
- 37.Tahara A., Saito M., Sugimoto T., Tomura Y., Wada K., Kusayama T., Tsukada J., Ishii N., Yatsu T., Uchida W., Tanaka A. Br. J. Pharmacol. 1998;125:1463–1470. doi: 10.1038/sj.bjp.0702220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adis Data Information BV Drugs R D. 2004;5:94–97. [Google Scholar]
- 39.Doggrell S. A. Curr. Opin. Investig. Drugs. 2004;5:977–983. [PubMed] [Google Scholar]
- 40.Miyazaki T., Yamamura Y., Onogawa T., Nakamura S., Kinoshita S., Nakayama S., Fujiki H., Mori T. Endocrinology. 2005;146:3037–3043. doi: 10.1210/en.2004-1590. [DOI] [PubMed] [Google Scholar]
- 41.Wada K., Tahara A., Arai Y., Aoki M., Tomura Y., Tsukada J., Yatsu T. Eur. J. Pharmacol. 2002;450:169–177. doi: 10.1016/s0014-2999(02)02101-5. [DOI] [PubMed] [Google Scholar]
- 42.Golestaneh L., Talreja A., Le Jemtel T. H. Curr. Heart Fail. Rep. 2004;1:190–196. doi: 10.1007/s11897-004-0008-5. [DOI] [PubMed] [Google Scholar]
- 43.Anderson R. J., Chung H. M., Kluge R., Schrier R. W. Ann. Intern. Med. 1985;102:164–168. doi: 10.7326/0003-4819-102-2-164. [DOI] [PubMed] [Google Scholar]
- 44.Tahara A., Saito M., Sugimoto T., Tomura Y., Wada K., Kusayama T., Tsukada J., Ishii N., Yatsu T., Uchida W., Tanaka A. Naunyn Schmiedebergs Arch. Pharmacol. 1998;357:63–69. doi: 10.1007/pl00005139. [DOI] [PubMed] [Google Scholar]
- 45.Fromant M., Blanquet S., Plateau P. Anal. Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.