Abstract
Many nuclear hormones have physiological effects that are too rapid to be explained by changes in gene expression and are often attributed to unidentified or novel G protein-coupled receptors. Thyroid hormone is essential for normal human brain development, but the molecular mechanisms responsible for its effects remain to be identified. Here, we present direct molecular evidence for potassium channel stimulation in a rat pituitary cell line (GH4C1) by a nuclear receptor for thyroid hormone, TRβ, acting rapidly at the plasma membrane through phosphatidylinositol 3-kinase (PI3K) to slow the deactivation of KCNH2 channels already in the membrane. Signaling was disrupted by heterologous expression of TRβ receptors with mutations in the ligand-binding domain that are associated with neurological disorders in humans, but not by mutations that disrupt DNA binding. More importantly, PI3K-dependent signaling was reconstituted in cell-free patches of membrane from CHO cells by heterologous expression of human KCNH2 channels and TRβ, but not TRα, receptors. TRβ signaling through PI3K provides a molecular explanation for the essential role of thyroid hormone in human brain development and adult lipid metabolism.
Keywords: neuronal development, phosphatidylinositol 3-kinase, potassium channels, Rac, KCNH2
The thyroid hormone, l-3,5,3′triiodothyronine (T3), plays an essential role in the development and metabolism of many tissues and organs (1). In particular, mammalian brain development is disrupted when T3 signaling is reduced by inherited mutations in the T3 receptor gene, THRβ, or when the synthesis or transport of the T3 prohormone thyroxine (tetraiodothyronine) is reduced by dietary iodine deficiency or environmental toxicants (2–4). T3 also regulates its own circulating levels through negative feedback on pituitary thyrotropes, which secrete thyroid stimulating hormone (1). Two genes, THRα and THRβ, encode four alternately spliced ligand-binding zinc-finger receptor proteins from the c-erbA family, which mediate T3 actions on gene expression (5), but specific T3-regulated genes involved directly in neural development remain to be identified. Recently, however, attention has shifted to the rapid, nongenomic signaling by T3 (6) and other thyroxine metabolites (7).
For example, we previously reported a Rac-dependent effect of T3 on voltage-activated potassium channels encoded by the ether-a-go-go-related gene KCNH2 in a rat pituitary cell line (8). KCNH2 proteins are voltage-activated potassium channels that regulate spike frequency in electrically excitable cells, such as the endocrine cells of the pituitary, through their unique kinetics of inactivation and recovery (9). Because they inactivate almost immediately after activation at positive voltages during the peak of each spike, and then reactivate before closing when the membrane repolarizes after the spike, they contribute to determining the interspike interval. Thus, increasing KCNH2 activity decreases excitability and secretion. Rapid effects of thyroid hormone have been reported previously on other ion channels in guinea pig cardiac myocytes, but the receptor and its mechanism of action were not identified (10, 11).
The Rac GTPase, like thyroid hormone, is essential for human neural development (12, 13), but thyroid hormone has not been reported to stimulate Rac activity. However, Rac is a well known effector of phosphatidylinositol 3-kinase (PI3K) (14), and other nuclear receptors have been reported to bind to and stimulate PI3K (15). Therefore, we postulated that the Rac-dependent effect of T3 on KCNH2 channel activity was mediated by activation of PI3K. We demonstrated that KCNH2 stimulation was blocked by wortmannin (8), an active site inhibitor of PI3K (16), but we did not identify the receptor for these effects of T3. Subsequently, PI3K has also been implicated in other rapid effects of thyroid hormone (17, 18), but the limited temporal and spatial resolution of the biochemical assays in all these studies precludes direct evidence for thyroid hormone receptor action in the cytoplasm. Here, we use the unique temporal and spatial resolution of the patch-clamp technique (19) in clonal rat pituitary cells, the original in vitro model system for T3 action (20), to demonstrate directly that the nongenomic effect of T3 on KCNH2 channel activity is mediated by a nuclear T3 receptor acting rapidly at the plasma membrane through PI3K.
Results
Rapid Stimulation of KCNH2 Channels Involves a Conventional Nuclear Receptor.
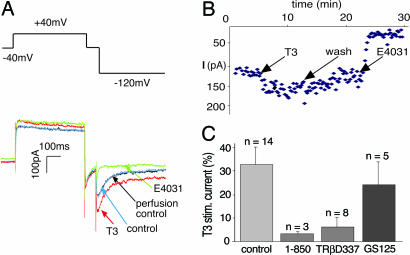
In metabolically intact GH4C1 cells that were voltage-clamped through gramicidin- or nystatin-perforated patches, superfusing the cells with T3 rapidly increased the KCNH2 current amplitude by 52 ± 6% (n = 6) within 3 min (Fig. 1B). In dialyzed cells, the responses were smaller (Fig. 1C), but, in both cases, the potency and pharmacology of T3 action on KCNH2 channels in GH4C1 cells were consistent with its rapid effect being mediated by a nuclear receptor for T3. Smaller and slower increases in KCNH2 currents were produced in some cells by concentrations of T3 as low as 1 pM, but its reverse isomer, 3,3′,5′-triiodothyronine produced no stimulation at concentrations as high as 100 nM (data not shown). Therefore, we routinely used 100 nM T3 to rapidly and maximally activate all T3 receptors. Nevertheless, even at this supramaximal concentration, KCNH2 stimulation by T3 was blocked completely by 3 μM 1-850 (Fig. 1C), an antagonist of the nuclear thyroid hormone receptors in GH4C1 cells (21). We also tested the involvement of nuclear receptors more directly by transfecting GH4C1 cells with mutated receptors, which have been shown to disrupt thyroid hormone signaling (4). Deleting T337 in the C terminus of TRβ2, which also disrupts neural development in mice (22), blocked KCNH2 stimulation by T3 in GH4C1 cells (Fig. 1C). In contrast, the receptor protein, which had been mutated in the P box (E125G-G126S) to prevent binding to thyroid receptor-specific DNA response elements (23), had an insignificant effect on KCNH2 channel stimulation by T3 (Fig. 1C), ruling out nonspecific effects of the transfection protocol. These results implicate nuclear receptors for thyroid hormone in KCNH2 stimulation by T3, but they do not identify the mechanism of action at the plasma membrane.
Fig. 1.
Rapid stimulation of KCNH2 currents by T3 in GH4C1 cells. (A) Voltage protocol and resulting currents elicited from a GH4C1 cell. Four current traces from the same cell are superimposed: control (in blue); perfusion control (in black); 5 min after application of 100 nM T3 (in red); and 8 min after 5 μM E4031 (in green), a selective KCNH2 inhibitor, which completely blocks the T3-stimulated current at −120 mV. (B) Time course of changes in KCNH2 current amplitude measured by voltage protocol repeated at 10-s intervals. (C) Percentage increase in KCNH2 current in the presence of 3 μM 1-850, an antagonist of T3 binding to TRβ, or after overnight transfection with mutant TRβ receptors.
T3 Stimulates Activity of Existing Channels Without Trafficking or Synthesis.
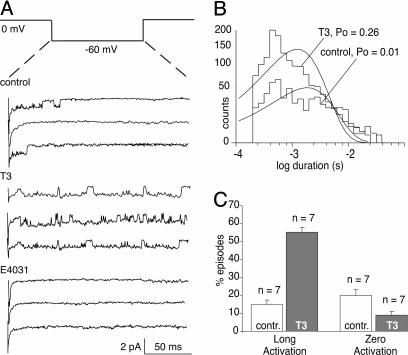
In principle, the increase in KCNH2 current illustrated in Fig. 1 could result from a combination of changes in channel protein synthesis, insertion into the plasma membrane, or posttranslational modification. However, new synthesis seems unlikely because T3 increases maximal whole-cell currents by ≈50% within 3 min (Fig. 1), but similar effects on this time scale have been shown to involve vesicular insertion of nascent channels in other systems (24, 25). Here, we use the patch-clamp technique to rule out insertion or trafficking of KCNH2 channels into the membrane by recording the effect of T3 on individual KCNH2 channels already gating in the plasma membrane (Fig. 2). In control patches held at 0 mV to activate all of the KCNH2 channels, brief steps to −60 mV, repeated at 3-s intervals on the same patch of membrane, elicited channel activity in 80% of the steps. However, in 84% of the steps with activity the channels returned to the closed state within 75 msec. In contrast, application of T3 to the bath outside the patch pipette produced a large increase in open probability (Fig. 2B), which resulted from both a reduction in the percentage of blank traces from 20% to 9% and an increase from 16% to 55% in the percentage of episodes in which channel activity persisted for the entire duration of the step (Fig. 2C, n = 7/7). Thus, T3 stimulates the activity of individual channels that are already present in the plasma membrane.
Fig. 2.
T3 stimulates individual KCNH2 channels already gating in cell-attached patches of plasma membrane on GH4C1 cell voltage-clamped to 0 mV in high potassium. (A) Voltage protocol repeated at 10-s intervals and 9 representative traces (out of 100) from the same patch of a single KCNH2 channel: 3 traces under control conditions, 3 traces 2 min after addition of 100 nM T3, and 3 traces 2 min after addition of 5 μM E4031. (B) Channel open-time histograms at −60 mV for control and thyroid hormone, T3, treated patches, and the open probability (PO) is indicated. (C) Summary of changes in seven such patches plotting percentage of traces, or “episodes” (100 per patch) where no activity was visible (zero activation) or where activity was sustained for more than half the duration of the voltage-step (long activation).
T3 Stimulates KCNH2 Activity Through PI3K.
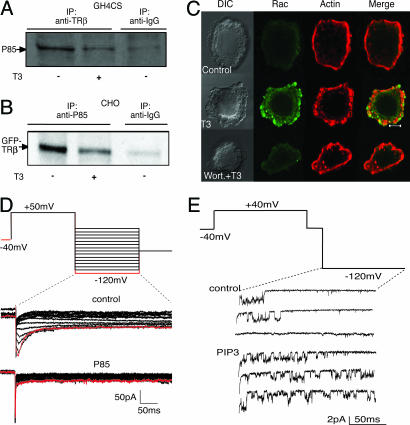
We demonstrated previously that the rapid effect of T3 on KCNH2 channels in GH4C1 cells is mediated by a Rac GTPase (8), and we postulated that T3 stimulated Rac activity through PI3K because the effects of T3, but not Rac, were blocked by 20 nM wortmannin, an active site inhibitor of PI3K (16). Here, we show that TRβ associates with the regulatory p85 subunit of PI3K in GH4C1 cells (Fig. 3). Immunoprecipitation of native TRβ protein in GH4C1 cells pulled down native p85 in the absence of hormone (Fig. 3A). We also transfected CHO cells, which do not express detectable levels of TRβ (see below), with recombinant GFP-TRβ chimeras, which we pulled down by immunoprecipitation of native p85 (Fig. 3B). In both cases, association was reduced by pretreatment of the cells with 100 nM T3 for 5 min. Nevertheless, T3-dependent reduction of the TRβ association with p85 seems to increase signaling through PI3K as reflected by reorganization of the actin cytoskeleton. As illustrated in Fig. 3C, T3 rapidly stimulates recruitment of Rac to the plasma membrane, actin reorganization, and lamellipodia formation. All of these effects are prevented by preincubation with 50 nM wortmannin. Further evidence for an essential role of PI3K in KCNH2 stimulation by T3 was obtained by overexpression of the p85 regulatory subunit of PI3K, which reduced basal KCNH2 currents in GH4C1 cells from −24.2 ± 3.7 pA/pF (n = 4) to −10.9 ± 2.2 pA/pF (n = 6, Fig. 3D). Conversely stimulating KCNH2 channels in cell-free patches from GH4C1 cells with 5 μM phosphatidylinositol 3,4,5-triphosphate (PIP3), the immediate product of PI3K action at the plasma membrane (14), produced comparable changes in KCNH2 activity (Fig. 3E) as those observed for T3 on KCNH2 channels in cell-attached patches (Fig. 2). On average, PIP3 increased activity by 61 ± 8.8% (n = 3) when 1.0 mM ATP and 0.1 mM GTP were present in the solution bathing the former cytoplasmic side of the membrane, but not when they were omitted (n = 3). Thus, PI3K activity is a major determinant of KCNH2 activity.
Fig. 3.
T3 stimulates KCNH2 channels in GH4C1 cells through PI3K. (A) Immunoprecipitation of native TRβ from GH4C1 cell lysates pulls down native p85, but this association is reduced after 5-min treatment with 100 nM T3. (B) Immunoprecipitation of native p85 from CHO cell lysates pulls down heterologously expressed GFP-TRβ1, and this association is also reduced after 5 min in 100 nM T3. (C) Confocal images of GH4C1 cells illustrate the effect of 100 nM T3 for 15 min on lamellipodia [differential interference contrast microscopy (DIC)], Rac distribution (green), and F-actin organization (red). Pretreatment with 50 nM wortmannin prevents the T3-induced changes. (D) Whole-cell currents elicited by the illustrated family of voltage steps from a control cell (Upper) and from a cell transfected overnight with p85 (Lower). (E) Single channel recording of KCNH2 activity in cell-free patch. Control records and response to 5 μM PIP3.
Reconstitution of KCNH2 Stimulation by TRβ.
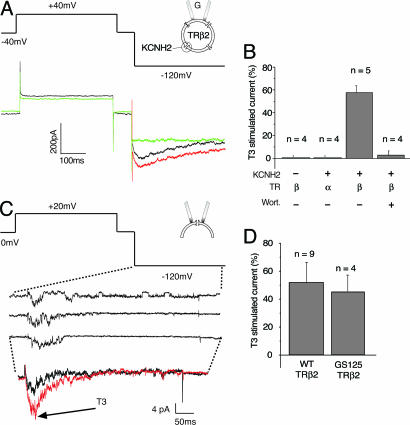
Direct molecular evidence for the involvement of TRβ in T3 action at the plasma membrane was obtained by reconstituting KCNH2 channel stimulation in CHO cells (Fig. 4). When human genes encoding KCNH2 channels and TRβ2 receptors were coexpressed heterologously in CHO cells, robust, E-4031-sensitive KCNH2 currents were measured with characteristic voltage-dependence (Fig. 4A). Superfusion with 100 nM T3 increased the KCNH2 current amplitude in CHO cells by 58% (Fig. 4B). In contrast, coexpression of TRα1 was unable to reconstitute T3-dependent stimulation of KCNH2 (Fig. 4B), demonstrating that the effects of T3 on KCNH2 activity in CHO cells are mediated by TRβ and not TRα or other unidentified receptors (6, 7). Furthermore, transfecting CHO cells with TRβ2 alone did not produce any E-4031-sensitive current (Fig. 4B), ruling out effects of TRβ2 expression on native KCNH2 expression or trafficking in CHO cells. Finally, we used a unique feature of the patch-clamp method to obtain direct evidence for TRβ2 action at the plasma membrane by excising small patches of membrane (2–5 μm2 in area) with its underlying cytoskeletal matrix, and applying T3 directly to the cytoplasmic side of the patch (Fig. 4 C and D). When the bath solution contained ATP, GTP, and phosphatidylinositol 4,5-bisphosphate (PIP2) to support PIP3 synthesis and Rac activation, T3 rapidly increased recombinant channel activity in cell-free patches (Fig. 4C). Similar results were obtained with the mutated TRβ2 E125G-G126S receptor (Fig. 4D), which does not bind to thyroid-specific response elements in the nucleus (23). Thus, neither DNA binding nor the nucleus is required for TRβ2 to stimulate KCNH2 channels in cell-free patches of membrane.
Fig. 4.
KCNH2 stimulation by T3 is reconstituted in CHO cells with recombinant TRβ2. (A) Voltage-protocol and representative currents from a CHO cell expressing human KCNH2 and TRβ2. T3 (100 nM, red) increases control current (black), and 5 μM E4031 blocks all of the current at −120 mV. (B) Histograms comparing current stimulated by T3 in cells expressing TRβ2 or TRα1. (C) Voltage protocol and representative single-channel currents elicited from a cell-free patch taken from a CHO cell expressing KCNH2 and TRβ2. Ensemble averages of 10 traces before and after 100 nM T3 are displayed. (D) Histograms showing the percentage increase in ensemble channel activity produced by 100 nM T3 in cells transfected with TRβ2 WT or GS125.
TRβ stimulates recombinant KCNH2 channels in cell-free patches from CHO cells through the same signaling cascade we identified in GH4C1 cells. KCNH2 stimulation was blocked by inhibition of PI3K with 50 nM wortmannin (Fig. 4B) and also by removing ATP and GTP from the solution on the cytoplasmic side of the cell-free patches (data not shown). In the presence of 1.0 mM ATP and 0.1 mM GTP, addition of 5 μM PIP3 directly to the cytoplasmic side of the patch was as effective as T3 at stimulating KCNH2 activity, rapidly increasing activity by 56 ± 10.8% (n = 5, data not shown). In contrast inositol 1,4,5-trisphosphate (IP3), the product of phosphatidylinositol 4,5-bisphosphate (PIP2) hydrolysis by phospholipase C, had no effect on channel activity (n = 5, data not shown). Therefore, we conclude that the entire signaling cascade from TRβ through PI3K to Rac remains associated with the plasma membrane in mammalian cells (Fig. 5).
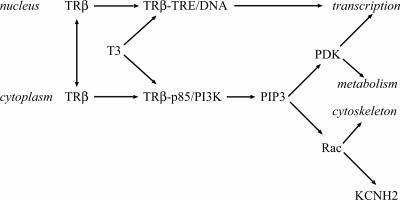
Fig. 5.
TRβ signaling in the nucleus through DNA binding or in the cytoplasm through PI3K can lead to changes in gene expression. Production of PIP3 also stimulates cell metabolism through the PIP3-dependent protein kinase (PDK) and cytoskeleton through guanine exchange factors for the Rac GTPase. The mechanism of KCNH2 stimulation by Rac is addressed in a separate article (37).
Discussion
The central conclusion of this article is the demonstration that a nuclear receptor for thyroid hormone, TRβ2, regulates the activity of KCNH2 potassium channels in the plasma membrane through a rapid signaling mechanism that does not require the nucleus or binding to DNA (Fig. 5). This conclusion stands in contrast to the prevailing view in the field that attributes such rapid nongenomic actions of nuclear hormones to unidentified or G protein-coupled receptors (26). Although there have been previous reports of other nuclear hormone receptor proteins signaling in the cytoplasm (15, 27), the patch-clamp technique has allowed us to demonstrate this phenomenon with unusual temporal and spatial resolution. This effect on KCNH2 channels in GH4C1 cells is specific for TRβ2 (Fig. 4) and TRβ1 receptors (S.G. and D.L.A., unpublished results) but is not reconstituted with TRα receptors. Gene knockout studies have identified TRβ2 as the primary receptor responsible for negative feedback effects of T3 on thyroid-stimulating hormone secretion from thyrotropes (5), and increasing potassium channel activity is consonant with this physiological function. TRβ also regulates potassium channel activity during development in sensory hair cells of the mammalian cochlea (3), but the mechanism has not been addressed. Of more relevance to brain development, however, mutations in the THRβ gene that reduce the receptors’ affinity for hormone also produce learning deficits in humans and mice (28, 29). The most severe mutation in humans, deletion of T337 (4), blocks KCNH2 stimulation by T3 in GH4C1 cells (Fig. 1C) and disrupts neural development in mice (22).
We have identified the target of TRβ action at the plasma membrane as the phosphatidylinositol 3-kinase, PI3K. Low concentrations of wortmannin, an active site inhibitor of PI3K (16), completely prevent T3 action, and the immediate product of PI3K action, PIP3, recapitulates the effects of T3 on KCNH2 activity. This result is consistent with previous reports implicating phosphoinositides and PIP3-stimulated effectors in KCNH2 regulation (30, 31). We have observed that TRβ associates with the p85 regulatory subunit of PI3K in the absence of hormone, which is consistent with the ability of recombinant mutant receptors to block signaling by native wild type receptors (Fig. 1). We postulate that this is a direct interaction because mammalian isoforms of TRβ, but not TRα, have a conserved tyrosine residue in a consensus sequence, pYVXM, for binding to the Src homology 2 (SH2) domains of p85 (32). We also observe that T3 reduces the association of TRβ and p85 (Fig. 3). Disinhibition of unliganded TRβ action by T3 is the predominant mechanism of T3 signaling in the nucleus as reflected by the inability of THRβ gene knockouts to fully phenocopy removal of thyroid hormone (5). If the inhibitory association with PI3K were stimulated by tyrosine phosphorylation of TRβ and relieved by T3 binding, it would provide a novel and powerful mechanism for regulating growth responses, which is the classical function of T3 action in many tissues (1). In this context, it is interesting that selective TRβ agonists, which we would predict to stimulate PIP3 production, are as effective at reducing fat accumulation and its related changes in cholesterol and triglycerides in mice (33) as the elimination of the lipid phosphatase, SHIP2, which dephosphorylates PIP3 (34).
TRβ signaling through PI3K is also consistent with our earlier work demonstrating the requirement for Rac GTPase activity in the stimulation of KCNH2 channels by T3 (8). Many Rac exchange factors are recruited and/or stimulated at the plasma membrane by PIP3 (14), and T3 rapidly stimulates Rac accumulation at the plasma membrane of GH4C1 cells (Fig. 3). In view of the essential role of Rac in human neural development (12, 13), the existence of this signaling cascade in neurons would provide a molecular explanation for some of the most important physiological roles of TRβ receptors in brain development and its susceptibility to disruption by mutation or by environmental toxicants (2, 4).
Finally, TRβ signaling through PI3K raises the possibility that many of the effects of T3 on gene transcription are mediated not by TRβ binding to DNA, but by downstream effectors of PI3K (Fig. 5). For example, deletion of the Tub gene, which is a T3-regulated, phosphoinositide-binding, transcription factor in the brain (35), produces many of the same effects on the developing sensory nervous system as removing thyroid hormone (3). Both the human tubby protein H-tulp-1 (amino acids 296–347) and the human KCNH2 channel protein H-erg-1 (amino acids 12–62) contain homologous phosphoinositide-binding domains, which could target them to a signaling complex surrounding PI3K at the plasma membrane.
Materials and Methods
Cell Culture and Transfection.
Cells were plated onto glass coverslips (Deutsche Spiegelglas, Carolina Biological Supply) and cultured in 5% CO2 in a humidified incubator at 37°C for at least 24 h before transfection or patch-clamp recordings were made. GH4C1 pituitary cells (ATCC CCL-82.2) were grown in DMEM/F-12 with 10% calf serum (HyClone), and CHO fibroblasts were grown in DMEM with 10% FBS (HyClone). Cells were transfected the day before recording by using Lipofectamine 2000 (Life Technologies, Rockville, MD) in OPTI-MEM (Life Technologies). Baseline currents depend on days in culture and serum; therefore, every experiment has same-day controls for comparison.
Plasmids.
Human KCNH2 (NM_000238) was obtained from Barry Ganetzky (University of Wisconsin, Madison); mouse TRα (CAA30575) was obtained from Graham R. Williams (Imperial College, London); human TRβ2 (NP_000452) was obtained from Fred Wondisford (University of Chicago, Chicago); the p85 subunit of PI3K was obtained from James Liao (Harvard University, Cambridge, MA). The plasmid constructs were cotransfected at a ratio of 10:1 with a plasmid encoding GFP (Clontech). Successfully transfected cells were detected with UV fluorescence 12–36 h later. Control currents were measured from cells transfected with GFP alone.
Electrophysiological Recordings and Analysis.
Potassium currents were recorded under voltage-clamp with either an EPC9 patch-clamp amplifier interface (HEKA Electronics, Lambrecht/Pfalz, Germany) or an Axopatch 200 amplifier (Axon Instruments, Foster City, CA). For data acquisition, pulse generation, and analysis, a computer (Dell, Round Rock, TX) running either pulse software (HEKA Electronics) or pclamp 9.0 software (Axon Instruments) was used. Single-channel records were sampled at 20 KHz and filtered at 1.5 or 2 KHz and idealized with tac and tacfit (Bruxton, Seattle). All experiments were conducted at room temperature (20–24°C). Data are presented as mean ± SE of n experiments.
The patch pipettes were made from Corning type 7052 glass (Garner Glass, Claremont, CA) and coated with Sylgard 184 (Dow-Corning) to improve the signal-to-noise ratio and capacitance compensation. To record single-channel currents from cell-attached patches, the pipette contained 140 mM KCl, 1 mM CaCl2, 2 mM MgCl2, and 10 mM Hepes, at pH 7.4. To obtain cell-free recordings, the patch pipette was excised in the inside-out configuration into a bath containing 140 mM KCl, 2 mM MgCl2, 10 mM Hepes, 10 mM glucose, 1 mM dibromoBAPTA, 1 mM ATP, 0.1 mM GTP, and 5 μM phosphatidylinositol 4,5-bisphosphate (PIP2). Whole-cell currents were obtained conventionally from dialyzed cells voltage-clamped through ruptured membrane patches or from metabolically intact cells voltage-clamped through nystatin- or gramicidin-perforated patches. In both cases, the solution bathing the cells contained 140 mM KCl, 0.1 mM CaCl2, 2 mM MgCl2, 10 mM Hepes, and 10 mM glucose, at pH 7.4. For dialyzed cells, the pipette contained 140 mM KCl, 2 mM MgCl2, 10 mM Hepes, 1 mM dibromoBAPTA, 1 mM ATP, and 0.1 mM GTP. For perforated patches, the pipettes contained 110 mM K+-gluconate, 35 mM KCl, 2 mM MgCl2, 1 mM CaCl2, and 10 mM Hepes at pH 7.4 and either 50 μg/ml nystatin, which worked best on GH4C1 cells, or 100 μg/ml gramicidin, which worked best on CHO cells. Both methods gave similar results although, not unexpectedly, the response to thyroid hormone was smaller and more transient in dialyzed cells. However, voltage-clamp is also established more quickly in dialyzed cells, so it was used in experiments where many cells were sampled, but all results were confirmed with cells voltage-clamped through perforated patches.
Current through KCNH2 channels was isolated as a slowly decaying inward tail current at negative voltages with a voltage protocol illustrated in the figures. The pulse protocol was run a minimum of 10 times before the perfusion control and 10 times before addition of T3 to ensure that the currents were stable. Successful isolation was confirmed in every experiment with the class III anti-arrhythmic methane-sulfonanilide E-4031 (10 μM; Biomol, Plymouth Meeting, PA), which selectively blocks KCNH2 current (36).
Coimmunoprecipitation and Immunoblotting.
Cell lysates were prepared from cells that were washed twice with ice-cold PBS and resuspended in solubilization buffer: 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 mM PMSF, and a complete protease inhibitor mixture (mfr). After homogenization by sonication and centrifugation, the pellet was discarded. Protein concentration in the supernatant was measured by the Bradford method (Bio-Rad reagents). For immunoprecipitation, either anti-TRβ antibody (MA1-215; Affinity BioReagents, Golden CO) or anti-p85 antibody (06-497; Upstate, Charlottesville, VA) was added to the supernatant and incubated overnight at 4°C. Agarose beads conjugated with either protein G or protein A were added, and the lysates were incubated further for 1 h at 4°C. Immunoprecipitates were washed four times, and proteins were boiled in SDS/PAGE sample buffer and analyzed by SDS/PAGE. Proteins were transferred to nitrocellulose membranes, blotted with the antibody indicated, and visualized with enhanced chemiluminescence (ECL, Amersham Pharmacia Bioscience).
Confocal Fluorescence Microscopy.
A laser scanning confocal microscope (LSM 510, Carl Zeiss) was used to image cell morphology, Rac distribution, and the actin cytoskeleton through an oil-immersion Plan-Apo 100 objective (NA1.4). Cells were treated as described, then fixed with 4% paraformaldehyde for 10 min, washed twice with PBS, blocked with Superblock (Pierce) for 30 min, and washed twice again. To visualize the actin cytoskeleton, cells were incubated with Alexa Fluor 568 phalloidin (Molecular Probes) diluted 1:500 for 1 h at 25°C. Cells were then rinsed twice and kept in PBS, and the actin and tubulin cytoskeleton was visualized with a 543-nm line of the laser while collecting phalloidin fluorescence with a 560-nm long pass filter. For Rac visualization, cells were incubated overnight with primary anti-Rac antibody 1:250 (Transduction Laboratories, Lexington, KY and BD Biosciences) and then incubated with goat anti-mouse secondary antibody (1:1,000) (Molecular Probes) for 1 h at 25°C. The secondary antibody was excited at 488 nM whereas collection of emission was at 519 nM with a bandpass 500- to 550-nM filter. All cells were imaged across a plane as close as possible to where they were attached to the coverslip. Representative cells are shown in the figures.
Acknowledgments
We thank Barry Ganetzky, James Liao, Graham Williams, and Fred Wondisford for sharing plasmids. This work was supported by the National Institutes of Health intramural program at the National Institute of Environmental Sciences.
Abbreviations
- PI3K
phosphatidylinositol 3-kinase
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- T3
l-3,5,3′-triiodothyronine.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Yen P. M. Physiol. Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 2.Colborn T. Environ. Health Perspect. 2004;112:944–949. doi: 10.1289/ehp.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrest D., Reh T. A., Rusch A. Curr. Opin. Neurobiol. 2002;12:49–56. doi: 10.1016/s0959-4388(02)00289-1. [DOI] [PubMed] [Google Scholar]
- 4.Yen P. M. Trends Endocrinol. Metab. 2003;14:327–333. doi: 10.1016/s1043-2760(03)00114-0. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea P. J., Williams G. R. J. Endocrinol. 2002;175:553–570. doi: 10.1677/joe.0.1750553. [DOI] [PubMed] [Google Scholar]
- 6.Bassett J. H., Harvey C. B., Williams G. R. Mol. Cell. Endocrinol. 2003;213:1–11. doi: 10.1016/j.mce.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 7.Scanlan T. S., Suchland K. L., Hart M. E., Chiellini G., Huang Y., Kruzich P. J., Frascarelli S., Crossley D. A., Bunzow J. R., Ronca-Testoni S., et al. Nat. Med. 2004;10:638–642. doi: 10.1038/nm1051. [DOI] [PubMed] [Google Scholar]
- 8.Storey N. M., O’Bryan J. P., Armstrong D. L. Curr. Biol. 2002;12:27–33. doi: 10.1016/s0960-9822(01)00625-x. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz J. R., Bauer C. K. J. Cell. Mol. Med. 2004;8:22–30. doi: 10.1111/j.1582-4934.2004.tb00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaguchi Y., Cui G., Sen L. Endocrinology. 1996;137:4744–4751. doi: 10.1210/endo.137.11.8895342. [DOI] [PubMed] [Google Scholar]
- 11.Sen L., Sakaguchi Y., Cui G. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H2119–H2129. doi: 10.1152/ajpheart.00326.2002. [DOI] [PubMed] [Google Scholar]
- 12.Ramakers G. J. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 13.Govek E. E., Newey S. E., Van Aelst L. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 14.Welch H. C., Coadwell W. J., Stephens L. R., Hawkins P. T. FEBS Lett. 2003;546:93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- 15.Simoncini T., Hafezi-Moghadam A., Brazil D. P., Ley K., Chin W. W., Liao J. K. Nature. 2000;407:538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker E. H., Pacold M. E., Perisic O., Stephens L., Hawkins P. T., Wymann M. P., Williams R. L. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 17.Lei J., Mariash C. N., Ingbar D. H. J. Biol. Chem. 2004;279:47589–47600. doi: 10.1074/jbc.M405497200. [DOI] [PubMed] [Google Scholar]
- 18.Cao X., Kambe F., Moeller L. C., Refetoff S., Seo H. Mol. Endocrinol. 2005;19:102–112. doi: 10.1210/me.2004-0093. [DOI] [PubMed] [Google Scholar]
- 19.Neher E., Sakmann B. Sci. Am. 1992;266:44–51. doi: 10.1038/scientificamerican0392-44. [DOI] [PubMed] [Google Scholar]
- 20.Samuels H. H., Tsai J. S., Cintron R. Science. 1973;181:1253–1256. doi: 10.1126/science.181.4106.1253. [DOI] [PubMed] [Google Scholar]
- 21.Schapira M., Raaka B. M., Das S., Fan L., Totrov M., Zhou Z., Wilson S. R., Abagyan R., Samuels H. H. Proc. Natl. Acad. Sci. USA. 2003;100:7354–7359. doi: 10.1073/pnas.1131854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto K., Curty F. H., Borges P. P., Lee C. E., Abel E. D., Elmquist J. K., Cohen R. N., Wondisford F. E. Proc. Natl. Acad. Sci. USA. 2001;98:3998–4003. doi: 10.1073/pnas.051454698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibusawa N., Hollenberg A. N., Wondisford F. E. J. Biol. Chem. 2003;278:732–738. doi: 10.1074/jbc.M207264200. [DOI] [PubMed] [Google Scholar]
- 24.Bezzerides V. J., Ramsey I. S., Kotecha S., Greka A., Clapham D. E. Nat. Cell Biol. 2004;6:709–720. doi: 10.1038/ncb1150. [DOI] [PubMed] [Google Scholar]
- 25.Viard P., Butcher A. J., Halet G., Davies A., Nurnberg B., Heblich F., Dolphin A. C. Nat. Neurosci. 2004;7:939–946. doi: 10.1038/nn1300. [DOI] [PubMed] [Google Scholar]
- 26.Losel R., Wehling M. Nat. Rev. Mol. Cell Biol. 2003;4:46–56. doi: 10.1038/nrm1009. [DOI] [PubMed] [Google Scholar]
- 27.Kousteni S., Bellido T., Plotkin L. I., O’Brien C. A., Bodenner D. L., Han L., Han K., DiGregorio G. B., Katzenellenbogen J. A., Katzenellenbogen B. S., et al. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- 28.Hauser P., Zametkin A. J., Martinez P., Vitiello B., Matochik J. A., Mixson A. J., Weintraub B. D. N. Engl. J. Med. 1993;328:997–1001. doi: 10.1056/NEJM199304083281403. [DOI] [PubMed] [Google Scholar]
- 29.McDonald M. P., Wong R., Goldstein G., Weintraub B., Cheng S. Y., Crawley J. N. Learn. Mem. 1998;5:289–301. [PMC free article] [PubMed] [Google Scholar]
- 30.Bian J., Cui J., McDonald T. V. Circ. Res. 2001;89:1168–1176. doi: 10.1161/hh2401.101375. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Wang H., Wang J., Han H., Nattel S., Wang Z. FEBS Lett. 2003;534:125–132. doi: 10.1016/s0014-5793(02)03804-8. [DOI] [PubMed] [Google Scholar]
- 32.Songyang Z., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., Lechleider R. J., et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 33.Baxter J. D., Webb P., Grover G., Scanlan T. S. Trends Endocrinol. Metab. 2004;15:154–157. doi: 10.1016/j.tem.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Sleeman M. W., Wortley K. E., Lai K. M., Gowen L. C., Kintner J., Kline W. O., Garcia K., Stitt T. N., Yancopoulos G. D., Wiegand S. J., Glass D. J. Nat. Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- 35.Carroll K., Gomez C., Shapiro L. Nat. Rev. Mol. Cell Biol. 2004;5:55–63. doi: 10.1038/nrm1278. [DOI] [PubMed] [Google Scholar]
- 36.Herzberg I. M., Trudeau M. C., Robertson G. A. J. Physiol. (London) 1998;511:3–14. doi: 10.1111/j.1469-7793.1998.003bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentile S., Darden T., Erxleben C., Romeo C., Russo A., Martin N., Rossie S., Armstrong D. L. Proc. Natl. Acad. Sci. USA. 2006;103:5202–5206. doi: 10.1073/pnas.0600080103. [DOI] [PMC free article] [PubMed] [Google Scholar]