Abstract
We have investigated the Rac-dependent mechanism of KCNH2 channel stimulation by thyroid hormone in a rat pituitary cell line, GH4C1, with the patch-clamp technique. Here we present physiological evidence for the protein serine/threonine phosphatase, PP5, as an effector of Rac GTPase signaling. We also propose and test a specific molecular mechanism for PP5 stimulation by Rac-GTP. Inhibition of PP5 with the microbial toxin, okadaic acid, blocked channel stimulation by thyroid hormone and by Rac, but signaling was restored by expression of a toxin-insensitive mutant of PP5, Y451A, which we engineered. PP5 is unique among protein phosphatases in that it contains an N-terminal regulatory domain with three tetratricopeptide repeats (TPR) that inhibit its activity. Expression of the TPR domain coupled to GFP blocked channel stimulation by the thyroid hormone. We also show that the published structures of the PP5 TPR domain and the TPR domain of p67, the Rac-binding subunit of NADPH oxidase, superimpose over 92 α carbons. Mutation of the PP5 TPR domain at two predicted contact points with Rac-GTP prevents the TPR domain from functioning as a dominant negative and blocks the ability of Y451A to rescue signaling in the presence of okadaic acid. PP5 stimulation by Rac provides a unique molecular mechanism for the antagonism of Rho-dependent signaling through protein kinases in many cellular processes, including metastasis, immune cell chemotaxis, and neuronal development.
Keywords: KCNH2, tetratricopeptide repeat, neuronal development, potassium channel, thyroid hormone
Rac and Rho are members of the same family of Ras-related monomeric GTPases that are best known for their regulation of the actin cytoskeleton (1), but they also participate in many other important cellular processes (2), such as the innate immune response that stimulates superoxide production in neutrophils through Rac-dependent activation of the NADPH oxidase (3). In many of these other processes, Rac and Rho generally produce antagonistic effects, particularly in the developing nervous system, where they have opposite effects on neuronal polarity, neurite outgrowth, and synaptic plasticity (4). Although Rho signals primarily through several protein serine/threonine (S/T) kinases (5), none of the Rac effectors identified thus far provide a simple explanation for the functional antagonism between Rac and Rho.
We reported previously that Rac and Rho mediate the opposing effects of the thyroid hormone, 3,5,3′-triiodothyronine (T3), and thyrotropin-releasing hormone (TRH) on KCNH2 channel activity in pituitary cells (6), but the mechanism of channel regulation by Rac or Rho was not determined. The conclusions from that study are summarized in Fig. 1A. KCNH2 proteins are voltage-activated potassium channels that regulate spike frequency in electrically excitable cells, such as cardiac myocytes and the endocrine cells of the pituitary, through their unique kinetics of inactivation and recovery (7). KCNH2 channels are known to be phosphorylated and inhibited by the cAMP-dependent protein kinase (8), and recovery from inhibition by TRH requires protein phosphatase activity (9). Consequently, we postulated that Rho inhibits the channels via a protein S/T kinase, whereas Rac stimulates the channels via a protein S/T phosphatase. Here we demonstrate that the Rac-dependent effects of thyroid hormone on KCNH2 potassium channel activity in a rat pituitary cell line (6) are mediated by the protein S/T phosphatase, PP5 (10), which is expressed in all tissues, including the brain (11, 12). We also propose and test a structural model for direct PP5 activation by Rac-GTP, which involves residues conserved in all three Rac isoforms, but not in the Rho or Cdc42 family members. PP5 is unique among protein phosphatases in that it contains three tetratricopeptide repeats (TPRs) (13), which inhibit its activity (14, 15). We show that the published structures of the PP5 TPR domain (13) and the first three TPRs of p67, the Rac-binding subunit of NADPH oxidase (16), can be superimposed over 92 α carbons. Mutation of PP5 at two predicted contact points with Rac-GTP blocks channel stimulation by thyroid hormone. Rac-dependent stimulation of a protein S/T phosphatase provides a general mechanism for the antagonism between Rac and Rho.
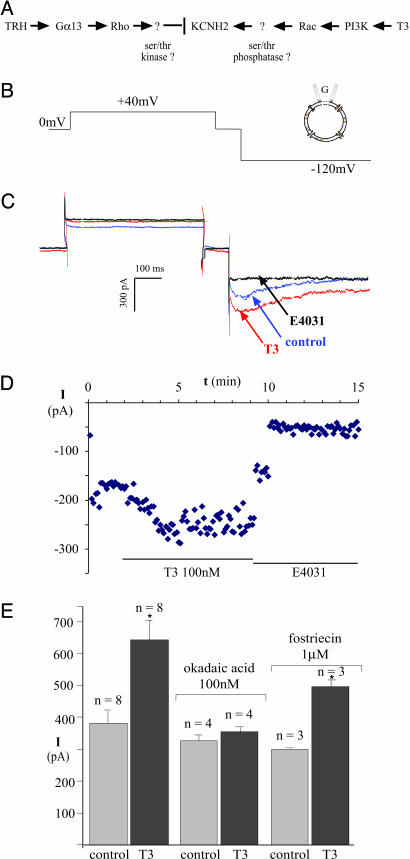
Fig. 1.
KCNH2 channel stimulation by thyroid hormone is blocked by okadaic acid. (A) Diagrammatic summary of ref. 6. KCNH2 stimulation by thyroid hormone (T3) was shown to be Rac-dependent and opposed by Rho. (B) Voltage protocol for isolating current through KCNH2 channels. (C) KCNH2 current at −120 mV is increased by 100 nM T3 and blocked completely by 10 μM E-4031, a selective KCNH2 antagonist. (D) Time course of changes in KCNH2 current amplitude obtained from metabolically intact cells voltage-clamped through gramicidin-perforated patches. (E) Peak amplitude histograms of the E-4031-sensitive current before and 3 min after bath application of 100 nM of the T3 hormone in control cells and in cells exposed to 100 nM okadaic acid or 1 μM fostriecin for 5 min before T3 application.
Results
KCNH2 current amplitude was measured under voltage-clamp through gramicidin-perforated patches on GH4C1 cells, as described in the Materials and Methods. The unique kinetics of KCNH2 channels (7), which inactivate rapidly after activation at positive voltages and then transiently reactivate during repolarization, allow one to separate the potassium current through KCNH2 channels from other potassium currents with the voltage protocol shown in Fig. 1B. Representative currents are shown in Fig. 1C. Note that the class III antiarrhythmic drug E-4031, which selectively inhibits KCNH2 channels (17), completely blocks the KCNH2 currents recorded during the negative step to −120 mV. T3 rapidly increases the amplitude of the E-4031-sensitive KCNH2 current by ≈50%, and this response is sustained in metabolically intact cells voltage-clamped through perforated patches (Fig. 1D).
To test the involvement of protein S/T phosphatases in Rac-dependent stimulation of KCNH2 channels, we exposed the cells to okadaic acid, a dinoflagellate toxin that is concentrated by shellfish during algal blooms. At a concentration of 100 nM, okadaic acid completely inhibits several protein S/T phosphatases, including PP2A, PP4, and PP5, but not other protein S/T phosphatases such as PP1 or PP2B (18). Bath application of 100 nM okadaic acid for 5 min had no significant effect on the amplitude of the basal, E-4031-sensitive current. However, okadaic acid completely prevented the increase in KCNH2 current in response to a subsequent application of T3 (Fig. 1E). In contrast, 1 μM fostriecin, which inhibits PP2A and PP4 but not PP5 (18), had no effect on channel stimulation by T3. In both control conditions and the presence of fostriecin, peak current was increased 1.7-fold by T3 (Fig. 1E). Because PP5 is the only S/T phosphatase that is inhibited completely by 100 nM okadaic acid but not at all by 1 μM fostriecin, we investigated whether PP5 is required for Rac-dependent stimulation of KCNH2 channels by T3.
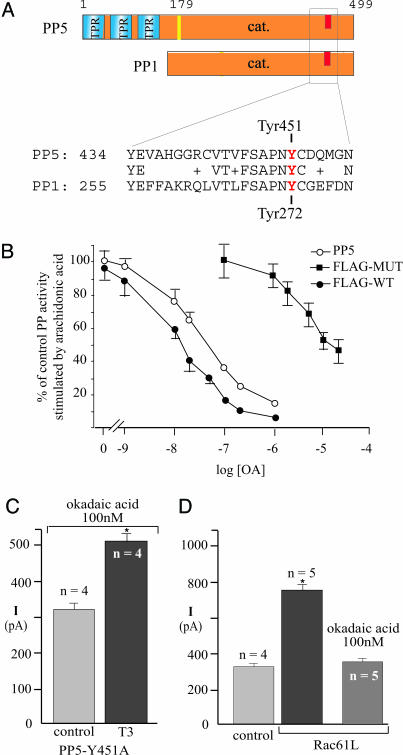
To provide a more direct test for PP5 involvement in channel stimulation by T3, we engineered a mutant form of PP5 with decreased sensitivity to inhibition by okadaic acid. Structural studies have identified the toxin-binding site in the C terminus of PP1 (19), and this sequence is conserved in all of the protein S/T phosphatases inhibited by okadaic acid, including PP5 (Fig. 2A). Y272 in PP1 was reported to be particularly important for inhibition by okadaic acid (20). This residue corresponds to Y451 in rat PP5, so we mutated Y451 to A and measured the sensitivity of its lipid-stimulated enzymatic activity to inhibition by okadaic acid. The concentration of okadaic acid required to produce 50% inhibition of the flag-tagged PP5 is <30 nM, but for the Y451A mutant, it is >10 μM (Fig. 2B). In cells transfected with the Y451A mutant of PP5, T3 increased the KCNH2 current amplitude by >50% in the presence of 100 nM okadaic acid (Fig. 2C). This result demonstrates that PP5 alone is sufficient to restore KCNH2 stimulation in the presence of okadaic acid, strongly supporting the involvement of PP5 in T3 signaling. Furthermore, PP5 is required downstream of Rac in KCNH2 stimulation by T3, because okadaic acid also prevented the stimulation of KCNH2 currents produced by dialyzing the GH4C1 cells with the constitutively active Rac1 Q61L protein (Fig. 2D).
Fig. 2.
PP5 Y451A rescues T3 signaling in the presence of okadaic acid. (A) Schematic diagram showing the domain structure of the protein Ser/Thr phosphatases PP5 and PP1. Blue and orange boxes indicate the TPR and the catalytic domains, respectively. The C-terminal phosphatase sequences implicated in toxin binding are shown. (B) Enzymatic activity of recombinant PP5 constructs stimulated by arachidonic acid in the presence of okadaic acid. Open circles, wild-type PP5; closed squares, Flag-tagged PP5 mutated at Y451A; closed circles, Flag-tagged wild-type PP5. (C) In cells transfected with PP5 Y451A, T3 stimulates KCNH2 channels in the presence of 100 nM okadaic acid. (D) In cells dialyzed with Rac1 Q61L, the basal currents are larger but pretreatment with 100 nM okadaic acid prevented the increase.
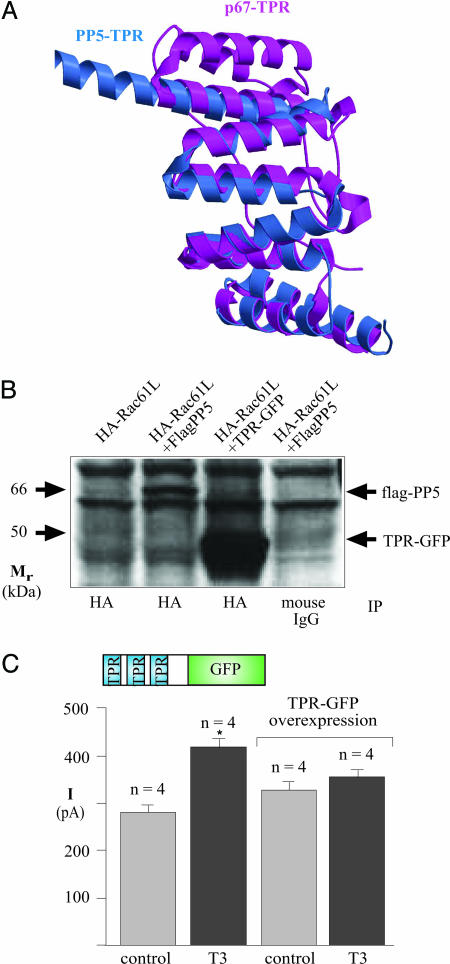
PP5 is unique among protein phosphatases in that it contains three TPRs in the amino terminus (13), which inhibit its phosphatase activity (14, 15). The TPR is a degenerate 34-aa motif that forms two antiparallel helices (21). TPRs are expressed in a wide variety of proteins, usually as arrays of 3–10 tandem repeats, and typically mediate binding to protein partners (21). A well documented effector of Rac, the p67 subunit of the NADPH oxidase, binds Rac-GTP through its own TPR domain (16). Using the o program (22), we superimposed the published structures of the three TPRs of PP5 (13) onto the first three TPRs of p67 (16). The two structures superimpose very closely, with a rms error equal to 0.1 nm over 92 α carbons (Fig. 3A). We confirmed that PP5 associates with Rac by coexpressing hemagglutinin (HA)-tagged Rac1 Q61L and Flag-PP5 in GH4C1 cells. Flag-PP5 was immunoprecipitated together with HA-Rac by the anti-HA antibody, but not by control IgG (Fig. 3B).
Fig. 3.
TPR of PP5 mediate Rac-dependent signaling. (A) Ribbon diagram showing the superposition of the three TPR motifs of PP5 (blue) with the first three TPR motifs of p67 (magenta). The two structures superimpose with a rms error of 0.1 nm over 92 α carbons. (B) Immunoprecipitation of HA-tagged constitutively active Rac Q61L in lysates from GH4C1 cells that had been transfected with Rac1 Q61L alone (lane 1), Rac1 Q61L and Flag-PP5 (lane 2), or Rac1 Q61L and TPR-GFP (lane 3). In contrast, control immunopecipitation with mouse IgG from cells transfected with Rac1 Q61L and Flag-PP5 (lane 4) failed to coprecipitate PP5. (C) In TPR-GFP expressing cells, basal KCNH2 currents are normal, but stimulation by T3 is blocked.
To test the role of the TPR domain in Rac-dependent stimulation of PP5, we constructed a chimera containing residues 1–179 of PP5, which encompasses the three TPRs but lacks the catalytic domain, which we replaced with GFP (Fig. 3C). We reasoned that this construct might function as a dominant negative by competing with native PP5 for protein partners, as others have demonstrated (23). In support of our hypothesis, immunoprecipitation of Rac1 Q61L coprecipitated TPR-GFP from GH4C1 cells transiently expressing both constructs (Fig. 3B). More importantly, transient expression of the PP5 TPR-GFP construct in GH4C1 cells completely blocked KCNH2 stimulation by T3 (Fig. 3C). In contrast, the TPR-GFP construct had no effect on the lipid-stimulated phosphatase activity of wild-type PP5 in vitro (not shown), and KCNH2 stimulation by T3 in cells transfected with GFP alone was normal (not shown). Because the TPR-GFP construct blocked T3 signaling in GH4C1 cells but not PP5 activity in vitro, we conclude that a protein that binds to TPR domains is required for T3 signaling. The Rac GTPase fits both these criteria. It is required for T3 signaling (6) and is known to bind to a similar TPR domain in p67 (16).
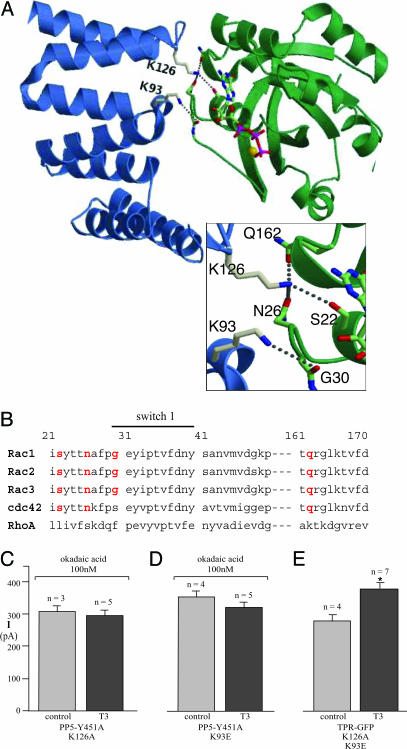
Although PP5 is the only protein S/T phosphatase with an identified TPR domain, many other proteins in mammalian cells contain TPR domains (21), so we investigated the specificity of this effect. Based on the similarity between the secondary structures of the TPR domains in PP5 and p67 (Fig. 3A), we constructed a model substituting the TPR domain of PP5 for that of p67 in the complex with Rac1-GTP (Fig. 4A). In this model, the exposed switch 1 loop of activated Rac-GTP binds to the TPR domain of PP5 on the side opposite the TPR interaction with the PP5 active site (24). If the interaction with Rac pulled the TPR domain out of the active site, it would provide a plausible mechanism for PP5 stimulation by activated Rac. The model also allowed us to identify K126 in rat PP5, corresponding to R102 in p67, as the key contact point with Rac-GTP, hydrogen bonding simultaneously with the side chain oxygens of three different Rac residues, S22, N26, and Q162 (Fig. 4A). We also identified an important contact between D92/K93 in PP5, which correspond to D67/K68 in p67, with the backbone amine and carbonyl of Rac G30 in switch 1. Rac G30 is the only residue contacting p67 that is not conserved in any other Rho family members (Fig. 4B). When we substituted both K93E and K126A in the PP5 TPR-GFP construct, it no longer inhibited KCNH2 channel stimulation (Fig. 4E). Furthermore, when either K93E or K126A was substituted in PP5 Y451A, the construct lost its ability to rescue T3 signaling in the presence of okadaic acid (Fig. 4 C and D), despite being expressed normally (not shown). Thus, interaction with the TPR domain appears to be critical for Rac signaling through PP5.
Fig. 4.
A structural model for PP5 stimulation by Rac-GTP. (A) Ribbon diagram of a model for the PP5-TPR (blue) complex with Rac1-Q61L-GTP (green). GTP is shown as sticks with carbon atoms in green, oxygen atoms in red, and phosphates in magenta. (Inset) The hydrogen bonds between PP5 K126 and the sidechain oxygens of Rac S22, N26, and Q162, and between PP5 K93 and the backbone carbonyl of Rac G30. (B) Sequence alignment of the switch 1 domain of Rho family GTPases. All three Rac isoforms, but neither Rho nor Cdc42, have all four of the critical residues (highlighted in red) that are proposed to contact PP5. (C and D) Peak amplitude histograms of the KCNH2 current in 100 nM okadaic acid from control cells or from cells expressing PP5-Y451A/K126A or PP5-Y451A/K93E after bath application of 100 nM T3. (E) Peak amplitude histograms of the KCNH2 current before and 3 min after bath application of 100 nM of the T3 hormone in cells expressing the TPR domain of PP5 with the mutations K93E and K126A.
Discussion
Together, these results provide structural and physiological evidence for PP5 as a direct molecular effector for activated Rac-GTP. Rho-GTP has been shown to inhibit myosin phosphatase activity in smooth muscle, but this appears to be an indirect mechanism mediated by Rho kinase phosphorylation of a PP1 inhibitor protein (25). The heterotrimeric G proteins, Gα12 and Gα13, have been reported to stimulate PP5 in vitro (26), but neither the structural mechanism nor the physiological significance of this effect has been established. Gα13 is unlikely to stimulate PP5 in GH4C1 cells because, unlike T3 and Rac, which require PP5 to stimulate KCNH2 channels (Fig. 2), constitutively active G13 reduces KCNH2 current amplitude (6). The mechanism we propose is not unique. It is based on the well documented stimulation of NADPH oxidase by Rac through binding to the TPR domain of p67 (3, 16). Despite the very limited identity between the amino acid sequences of the p67 and PP5 TPR domains, the secondary structures of the TPR domains are essentially identical (Fig. 3A). Thus, other proteins with TPR domains might now be investigated for regulation by Rac.
The evidence presented here also identifies Rac as a hormonally regulated activator for PP5. We initially identified PP5 as a lipid-stimulated protein phosphatase in brain extracts (11), having postulated that pertussis toxin-sensitive heterotrimeric G proteins might increase phosphatase activity by stimulating phospholipase A2 to produce arachidonic acid (27). However, receptor activation of Gαi and Gαo also stimulates the activity of the γ isoform of phosphatidylinositol 3 kinase through Gβγ subunits (28), and phosphatidylinositol 3,4,5 tris phosphate (PIP3) is known to activate guanine exchange factors for Rac (29). In addition, we have obtained separate evidence that channel stimulation by thyroid hormone is mediated by the nuclear receptor for thyroid hormone, TRβ, stimulating phosphatidylinositol 3 kinase (30). Phosphatidylinositol 3 kinase has been implicated in several examples of rapid physiological effects of other nuclear hormones (31). Thus, Rac-dependent stimulation of PP5 could account for many other examples of ion channel regulation by nuclear and G protein-coupled receptors.
Finally, although Rho and Rac signaling pathways interact at many levels (2), Rac-dependent stimulation of PP5 provides a direct molecular mechanism for the antagonism of Rho-dependent signaling through protein kinases in many fundamental cellular processes linked to human disease, including cancer (32), inflammation (3), and mental retardation (33). Rac-dependent stimulation of PP5 also provides a molecular mechanism for antagonism of apoptosis (34) in degenerative diseases of the aging (35–37) nervous system.
Materials and Methods
GH4C1 cells were grown in DMEM/F12 with 10% calf serum/50 mg/liter streptomycin/31 mg/liter penicillin/640 mg/liter NaHCO3. Cells were plated on glass coverslips (Deutsche Spiegelglas, Carolina Biological Supply) and transfected by using Lipofectamine 2000 (Invitrogen) with the PP5 and Rac plasmids together with a separate plasmid encoding GFP at a ratio of 10:1. Fluorescent cells were used for recordings between 12 and 48 h after transfection.
Recombinant PP5 Constructs.
The reference sequence for PP5 cloning was the rat “ppt” (RNPPT) sequence (GenBank accession no. X77237). Rat PP5 was cloned by PCR as a MluI-NotI fragment and inserted into the pCI vector engineered to contain a Flag epitope at the N terminus. The TPR-GFP chimera was created by cutting the Flag-PP5 construct at amino acid 179 with HindIII and NotI and substituting GFP (Clontech) by using the following two oligos to PCR a GFP that could be fused to the TPR domain at the HindIII site: cgc ggg aag ctt gcc ATG GTG AGC AAG GGC GAG GAA CTG TTC (the HindIII site is underlined, uppercase letters are a GFP sequence from CLONTECH); CAT GGC ATG GAC CTG TAC AAG TAA agc ggc cgc ccc gcg (the NotI site underlined). Single mutations in PP5 were introduced by PCR with QuickChange XL site-directed mutagenesis kit (Stratagene). Mutant constructs were sequenced to confirm the presence of the desired mutations and the absence of additional mutations.
Biochemical Assay of PP5 Activity.
Recombinant PP5, Flag-PP5, and Flag-PP5(Y451A) were expressed as GST-fusion proteins, purified, and assayed by using 32P-casein as substrate as described (11). Flag-PP5 and Flag-PP5(Y451A) exhibited specific activities in the range of 10–40 nmol Pi released per min per mg of phosphatase protein, which is similar to the activity of wild-type recombinant PP5.
Electrophysiological Recordings and Analysis.
Potassium currents were recorded at room temperature (20–24°C) under voltage clamp with an EPC9 patch-clamp amplifier interface (HEKA Electronics, Lambrecht/Pfalz, Germany) by using pulse software (HEKA Electronics) for data acquisition, pulse generation, and analysis. Data are presented as mean ± SE of n experiments. The patch pipettes were made from Corning type 7052 glass (Garner Glass, Claremont, CA). Whole-cell currents were obtained conventionally from dialyzed cells voltage-clamped through ruptured membrane patches or from metabolically intact cells voltage-clamped through gramicidin-perforated patches. No leak subtraction was applied. In both cases, the solution bathing the cells contained 140 mM KCl, 0.1 mM CaCl2, 2 mM MgCl2, 10 mM Hepes, and 10 mM glucose, pH 7.4. For dialyzed cells, the pipette contained 140 mM KCl, 2 mM MgCl2, 10 mM Hepes, 1 mM dibromoBAPTA [BAPTA, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate], 1 mM ATP, and 0.1 mM GTP. For perforated patches, pipettes contained 110 mM K+-gluconate, 35 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 10 mM Hepes, pH 7.4, and 100 μg/ml gramicidin. The pulse protocol was run a minimum of 10 times before the perfusion control and 10 times before the addition of T3 to ensure that the currents were stable. Successful isolation was confirmed in every experiment with the class III antiarrhythmic methane-sulfonanilide, E-4031 (10 μM, Biomol, Plymouth Meeting, PA). Averaged data are presented as mean ± SE. Differences were evaluated with Student's t test. P ≤ 0.05 is indicated with an asterisk.
Immunoprecipitation.
GH4C1 cells were transfected with HA-Rac-Q61L alone or in combination with either Flag-PP5 or TPR-GFP. After 24 h, cells were washed twice with ice-cold PBS and sonicated briefly in lysis buffer (40 mM Tris, pH 7.6/5 mM Hepes/50 mM NaCl/1 mM EDTA/1 mM EGTA/5 mM NaF/1% glycerol/0.1% Triton X-100), complete protease inhibitor mixture (Roche Diagnostics) at 4°C. The lysate was centrifuged (4°C, 15 min, 20,000 × g) and protein in the supernatant quantified by using the Bradford method (Bio-Rad). Equal amounts of supernatant protein were mixed with HA monoclonal antibody (Covance, Berkeley, CA) or mouse IgG overnight at 4°C, followed by Protein-G-agarose for 1 h more. Immunoprecipitates were washed four times with lysis buffer. Samples were subjected to SDS/PAGE (10% acrylamide), transferred to nitrocellulose, and PP5 detected by using a PP5 antiserum (12) and enhanced chemiluminescence (Amersham Pharmacia Bioscience).
Structure Modeling.
The models were created by using the o program (22). The proposed complex between the TPR domain of PP5 and Rac-GTP was built by a simple docking process, by using the experimentally determined structures of the TPR domain of PP5 alone (ref. 13; Protein Data Bank ID code 1A17) and of the TPR domain of P67 in complex with Rac-GTP (ref. 16; Protein Data Bank ID code 1E96). The initial model was adjusted by small rigid rotations of PP5 together with rotamer searches for K126, K93, and a few other PP5 residues to optimize contacts involving these sidechains while attempting to preserve contacts analogous to those found in the experimentally determined p67-Rac-GTP structure. Figs. 1–3 were prepared by using molscript (38).
Acknowledgments
We thank Angela Everhart and Erica Scappini for technical assistance and Lutz Birnbaumer, John O'Bryan, and Fernando Ribeiro-Neto for helpful suggestions. This work was supported by the National Institutes of Health intramural program at the National Institute for Environmental Health Sciences and by National Institutes of Health Grant NS031221 (to S.R.).
Abbreviations
- S/T
serine/threonine
- T3
3,5,3′-triiodothyronine
- TPR
tetratricopeptide repeat
- HA
hemagglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ridley A. J. Trends Cell Biol. 2001;11:471–477. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- 2.Burridge K., Wennerberg K. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 3.Bokoch G. M. Trends Cell Biol. 2005;15:163–171. doi: 10.1016/j.tcb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Govek E. E., Newey S. E., Van Aelst L. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Z. S., Manser E. Biochem. J. 2005;386:201–214. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storey N. M., O'Bryan J. P., Armstrong D. L. Curr. Biol. 2002;12:27–33. doi: 10.1016/s0960-9822(01)00625-x. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz J. R., Bauer C. K. J. Cell Mol. Med. 2004;8:22–30. doi: 10.1111/j.1582-4934.2004.tb00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas D., Zhang W., Karle C. A., Kathofer S., Schols W., Kubler W., Kiehn J. J. Biol. Chem. 1999;274:27457–27462. doi: 10.1074/jbc.274.39.27457. [DOI] [PubMed] [Google Scholar]
- 9.Barros F., Mieskes G., del Camino D., de la Pena P. FEBS Lett. 1993;336:433–439. doi: 10.1016/0014-5793(93)80851-k. [DOI] [PubMed] [Google Scholar]
- 10.Chinkers M. Trends Endocrinol. Metab. 2001;12:28–32. doi: 10.1016/s1043-2760(00)00335-0. [DOI] [PubMed] [Google Scholar]
- 11.Skinner J., Sinclair C., Romeo C., Armstrong D., Charbonneau H., Rossie S. J. Biol. Chem. 1997;272:22464–22471. doi: 10.1074/jbc.272.36.22464. [DOI] [PubMed] [Google Scholar]
- 12.Bahl R., Bradley K. C., Thompson K. J., Swain R. A., Rossie S., Meisel R. L. Brain Res. Mol. Brain Res. 2001;90:101–109. doi: 10.1016/s0169-328x(01)00089-4. [DOI] [PubMed] [Google Scholar]
- 13.Das A. K., Cohen P. W., Barford D. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M. X., Cohen P. T. FEBS Lett. 1997;400:136–140. doi: 10.1016/s0014-5793(96)01427-5. [DOI] [PubMed] [Google Scholar]
- 15.Sinclair C., Borchers C., Parker C., Tomer K., Charbonneau H., Rossie S. J. Biol. Chem. 1999;274:23666–23672. doi: 10.1074/jbc.274.33.23666. [DOI] [PubMed] [Google Scholar]
- 16.Lapouge K., Smith S. J., Walker P. A., Gamblin S. J., Smerdon S. J., Rittinger K. Mol. Cell. 2000;6:899–907. doi: 10.1016/s1097-2765(05)00091-2. [DOI] [PubMed] [Google Scholar]
- 17.Herzberg I. M., Trudeau M. C., Robertson G. A. J. Physiol. 1998;511:3–14. doi: 10.1111/j.1469-7793.1998.003bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honkanen R. E., Golden T. Curr. Med. Chem. 2002;9:2055–2075. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- 19.Maynes J. T., Bateman K. S., Cherney M. M., Das A. K., Luu H. A., Holmes C. F., James M. N. J. Biol. Chem. 2001;276:44078–44082. doi: 10.1074/jbc.M107656200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L., Zhang Z., Long F., Lee E. Y. Biochemistry. 1996;35:1606–1611. doi: 10.1021/bi9521396. [DOI] [PubMed] [Google Scholar]
- 21.D'Andrea L. D., Regan L. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Chen M. S., Silverstein A. M., Pratt W. B., Chinkers M. J. Biol. Chem. 1996;271:32315–32320. doi: 10.1074/jbc.271.50.32315. [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Roe S. M., Cliff M. J., Williams M. A., Ladbury J. E., Cohen P. T., Barford D. EMBO J. 2005;24:1–10. doi: 10.1038/sj.emboj.7600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somlyo A. P., Somlyo A. V. Physiol. Rev. 2003;83:1325–1358. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi Y., Katoh H., Mori K., Negishi M. Curr. Biol. 2002;12:1353–1358. doi: 10.1016/s0960-9822(02)01034-5. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong D. L., White R. E. Trends Neurosci. 1992;15:403–408. doi: 10.1016/0166-2236(92)90192-b. [DOI] [PubMed] [Google Scholar]
- 28.Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L. C. Trends Biochem. Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Welch H. C., Coadwell W. J., Stephens L. R., Hawkins P. T. FEBS Lett. 2003;546:93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- 30.Storey N. M., Gentile S., Ullah H., Russo A., Muessel M., Erxleben C., Armstrong D. L. Proc. Natl. Acad. Sci. USA. 2006;103:5197–5201. doi: 10.1073/pnas.0600089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losel R. M., Falkenstein E., Feuring M., Schultz A., Tillmann H. C., Rossol-Haseroth K., Wehling M. Physiol. Rev. 2003;83:965–1016. doi: 10.1152/physrev.00003.2003. [DOI] [PubMed] [Google Scholar]
- 32.Malliri A., Collard J. G. Curr. Opin. Cell Biol. 2003;15:583–589. doi: 10.1016/s0955-0674(03)00098-x. [DOI] [PubMed] [Google Scholar]
- 33.Ramakers G. J. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 34.Morita K., Saitoh M., Tobiume K., Matsuura H., Enomoto S., Nishitoh H., Ichijo H. EMBO J. 2001;20:6028–6036. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanekura K., Hashimoto Y., Kita Y., Sasabe J., Aiso S., Nishimoto I., Matsuoka M. J. Biol. Chem. 2005;280:4532–4543. doi: 10.1074/jbc.M410508200. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y., Su Y., Li B., Liu F., Ryder J. W., Wu X., Gonzalez-DeWhitt P. A., Gelfanova V., Hale J. E., May P. C., et al. Science. 2003;302:1215–1217. doi: 10.1126/science.1090154. [DOI] [PubMed] [Google Scholar]
- 37.Liu F., Iqbal K., Grundke-Iqbal I., Rossie S., Gong C. X. J. Biol. Chem. 2005;280:1790–1796. doi: 10.1074/jbc.M410775200. [DOI] [PubMed] [Google Scholar]
- 38.Kraulis P. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]