Abstract
Prostaglandin endoperoxide H synthases (PGHSs) 1 and 2 convert arachidonic acid to prostaglandin H2 in the committed step of prostanoid biosynthesis. These enzymes are pharmacological targets of nonsteroidal antiinflammatory drugs and cyclooxygenase (COX) 2 inhibitors. Although PGHSs function as homodimers and each monomer has its own COX and peroxidase active sites, the question of whether there is cross-talk between monomers has remained unresolved. Here we describe two heterodimers in which a native subunit of human PGHS-2 has been coupled to a subunit having a defect within the COX active site at some distance from the dimer interface. Native/G533A PGHS-2, a heterodimer with a COX-inactive subunit, had the same specific COX activity as the native homodimer. Native/R120Q PGHS-2, a heterodimer in which both subunits can oxygenate arachidonic acid but in which the R120Q subunit cannot bind the COX inhibitor flurbiprofen, was inhibited by flurbiprofen to about the same extent as native PGHS-2. These results imply that native PGHS-2 exhibits half-of-sites reactivity. Isothermal titration calorimetry established that only one monomer of the native PGHS-2 homodimer binds flurbiprofen tightly. In short, binding of ligand to the COX site of one monomer alters its companion monomer so that it is unable to bind substrate or inhibitor. We conclude that PGHS monomers comprising a dimer, although identical in the resting enzyme, differ from one another during catalysis. The nonfunctioning subunit may provide structural support enabling its partner monomer to catalyze the COX reaction. This subunit complementarity may prove to be characteristic of other dimeric enzymes having tightly associated monomers.
Keywords: aspirin, flurbiprofen, heme, ibuprofen, prostaglandin
Prostaglandin endoperoxide H synthases (PGHSs) 1 and 2 catalyze the committed step in prostaglandin biosynthesis (1–3). They are both targets of nonsteroidal antiinflammatory drugs, and PGHS-2 is the target of cyclooxygenase (COX) 2 inhibitors (4, 5). The enzymes have two catalytic activities: (i) a COX that converts arachidonic acid (AA) to a prostaglandin endoperoxide, prostaglandin G2 (PGG2), and (ii) a peroxidase (POX) that reduces PGG2 to PGH2. Oxidation of the heme group at the POX active site by a peroxide is required to activate the COX by generating a tyrosyl radical at Tyr-385 at the COX active site (1–3). The Tyr-385 radical is involved in abstracting the hydrogen atom from the fatty acid substrate in the initial and rate-determining step in COX catalysis.
PGHS-1 and PGHS-2 function only as homodimers although each subunit has both a POX and a COX active site (1–3, 6). It is not clear whether each monomer functions independently or whether cross-talk occurs between monomers comprising a dimer. Titrations of PGHS-1 with time-dependent COX inhibitors such as flurbiprofen (FBP) and indomethacin indicate that complete inhibition requires only one inhibitor molecule to be bound per dimer (7). Other studies with PGHS-1 using low concentrations of PGHS-2-specific inhibitors in the presence of aspirin or indomethacin have suggested that there may be interactions between binding sites of the monomers (8–10).
To investigate interactions between PGHS monomers, we prepared and analyzed native and mutant human PGHS-2 (huPGHS-2) homodimers and heterodimers composed of a native monomer with either a G533A or an R120Q huPGHS-2 monomer. Gly-533 is at the distal end of the COX binding channel and plays a key role in aligning the ω8 proS hydrogen of fatty acid substrates with the Tyr-385 radical involved in COX catalysis (11). G533A mutants of PGHSs oxygenate fatty acids with low efficiencies (11–15). Arg-120 is found near the mouth of the COX active site of PGHSs and is involved in binding the carboxyl group of fatty acid substrates (11, 16, 17). An R120Q huPGHS-2 mutant can oxygenate AA but not eicosapentaenoic acid (EPA) (18); furthermore, FBP is ≈1,000 times less potent as an inhibitor of R120Q huPGHS-2 than native huPGHS-2.
Results
huPGHS-2 Heterodimers.
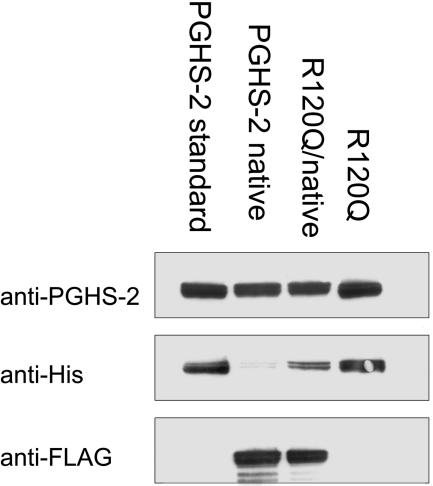
Sf21 cells were infected with baculovirus constructs encoding (i) native huPGHS-2, (ii) mutant huPGHS-2, or (iii) both a native and a mutant subunit. The constructs were designed with a FLAG tag on the N terminus of the native monomers after removal of the signal peptide and a hexahistidine tag near the N terminus of the mutant subunit. After the proteins were expressed, the homo- and heterodimers were purified by using an appropriate combination of Ni-NTA affinity and anti-FLAG antibody chromatography. The samples were tested for purity by using antibodies to the 18-aa cassette of PGHS-2 (19) and to the FLAG and hexahistidine epitope tags. This is illustrated for the native/native, R120Q/R120Q, and native/R120Q huPGHS-2 dimers in Fig. 1. As expected, all of the enzyme forms were reactive with the anti-PGHS-2 antibody. The native/native homodimer and native/R120Q heterodimer were reactive with anti-FLAG antibodies. R120Q/R120Q homodimers were reactive with anti-histidine antibodies.
Fig. 1.
Western transfer blotting of native/native, R120Q/R120Q, and native/R120Q huPGHS-2. Homo- and heterodimeric proteins were expressed and purified as described in Materials and Methods. Purified protein (1–2.5 μg) was subjected to Western transfer blotting by using (i) an anti-peptide antibody to the 18-aa cassette of PGHS-2, (ii) an anti-hexahistidine antibody, or (iii) an anti-FLAG antibody. In this particular experiment, a small amount of anti-histidine immunoreactivity attributable to a small amount of contamination by native/R120Q huPGHS-2 heterodimer was detected in the preparation of native/native huPGHS-2. The numbering of residues in huPGHS-2 follows the numbering system used for oPGHS-1 (25, 38).
For all experiments involving direct comparisons of specific COX and POX activities between native and heterodimeric huPGHS-2, densitometry was performed to estimate the ratio of the native to the mutant subunit in the heterodimer. This involved comparing the relative intensities of the native huPGHS-2 dimer and the heterodimer with anti-PGHS-2 and anti-FLAG antibodies. In all cases, the ratio of the staining intensity of the heterodimer with anti-FLAG antibodies versus anti-PGHS-2 antibodies was 50 ± 5% of the ratio obtained with the FLAG-tagged native enzyme.
Kinetic Properties of huPGHS-2 Heterodimers.
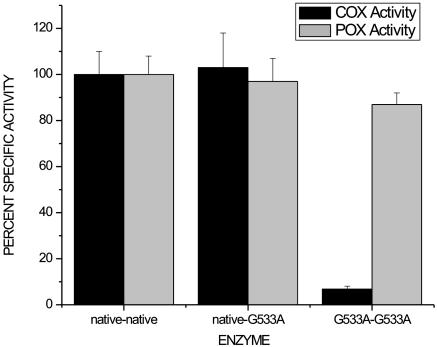
Consistent with previous, related observations (11–15), the specific COX activity of the G533A huPGHS-2 homodimer with AA as the substrate was ≈5% of the specific activity of the native huPGHS-2 homodimer (Fig. 2). Thus, we were surprised to find that the specific COX activity of the native/G533A huPGHS-2 heterodimer averaged 97% of that of the native huPGHS-2 homodimer (Fig. 2). Moreover, the Km values for AA with the native/native huPGHS-2 and native/G533A huPGHS-2 were both ≈5 μM, the same as the value reported previously for native huPGHS-2 (20). Finally, the POX activities of native/native, G533A/G533A, and native/G533A huPGHS-2 were similar to one another.
Fig. 2.
Relative specific COX and POX activities of native huPGHS-2 homodimer, native/G533A huPGHS-2 heterodimer, and G533A/G533A huPGHS-2 homodimer. COX activity was measured by using an oxygen electrode with 100 μM AA as the substrate as described in Materials and Methods. POX activity was measured spectrophotometrically by using 0.1 mM H2O2 and 4 mM guaiacol as substrates. Measurements were performed in triplicate, and the experiment was repeated three times with similar results. Activities were normalized to the milligram of enzyme protein as quantified by densitometry of Western blots.
The pattern of results observed with AA was not unique to this fatty acid. When the native/G533A huPGHS-2 heterodimer was examined with two other, less efficient substrates, linoleic acid (18:2 ω6) and α-linolenic acid (18:3 ω3) (20), the heterodimer had COX activities that averaged 71% of native activity with linoleic acid and 88% of native activity with α-linolenic acid in three experiments (data not shown). The expected percent of specific activities for a heterodimer with one COX-active and one COX-inactive monomer were 50% and 52% for linoleic acid and α-linolenic acid, respectively.
The patterns of products formed from [1-14C]AA by native/native huPGHS-2 and native/G533A huPGHS-2 were indistinguishable when compared by using simple thin layer chromatography (21) with PGG2-derived products accounting for 95% of the oxygenated radioactive products (data not shown).
The results of the studies comparing different combinations of native and G533A huPGHS-2 monomers suggest that the COX active site of only one monomer of a PGHS dimer can function at any given time (i.e., half-of-sites reactivity).
It is known that the COX activity of PGHSs undergoes a suicide inactivation that is directly proportional to the number of turnovers (22). Interestingly, the extent of the COX reaction per mole of enzyme was the same for both the native homodimer and the native/mutant heterodimers. This finding suggests that suicide inactivation of one monomer of the native homodimer also leads to the loss of catalytic activity in the partner monomer.
Inhibition of huPGHS-2 Heterodimers.
Microsomal preparations of the R120Q huPGHS-2 homodimer have >90% of the COX-specific activity of the native huPGHS-2 homodimer when AA is used as the substrate (18). The specific activities of highly purified forms of R120Q/R120Q huPGHS-2 and native/R120Q huPGHS-2 were 87–90% of that of native/native huPGHS-2 with AA and had comparable levels (80–110%) of POX activity (data not shown).
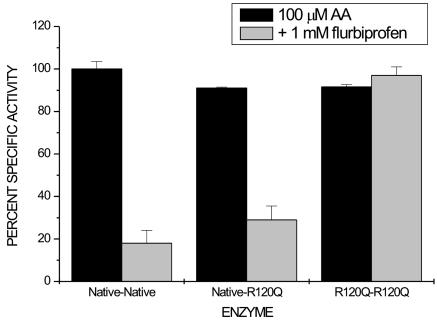
FBP is a nonsteroidal antiinflammatory drug of the 2-phenylpropionic acid class. It is an effective time-dependent inhibitor of huPGHS-2 (23) but has been found not to inhibit R120Q huPGHS-2 even at concentrations of 1 mM (18). Nonetheless, FBP time-dependently inhibited the COX activities of both the native/R120Q huPGHS-2 heterodimer and native/native huPGHS-2 homodimer to similar extents (≈29% and 18%, respectively) without inhibiting the R120Q/R120Q homodimer (Fig. 3). The small amount of COX activity (≈18%) seen after pretreatment of the native/native homodimer with FBP is attributable to the relatively rapid reversibility of this inhibitor (7, 23). Simple additivity of inhibition predicts that FBP would cause 59% inhibition of the heterodimer in the experiment depicted in Fig. 3. These results are in line with the notion that the binding of a single inhibitor molecule to one of the monomers is sufficient for the inhibition of dimer activity.
Fig. 3.
Inhibition of native huPGHS-2 homodimer, native/R120Q huPGHS-2 heterodimer, and R120Q/R120Q huPGHS-2 homodimer by FBP. Purified enzymes were preincubated with 1 mM FBP for 10 min, and an aliquot was removed and assayed immediately for COX activity with 100 μM AA as the substrate. The final concentration of FBP in the COX assay mixture was 1 μM. Each measurement was performed in duplicate, and the experiment was repeated three times with similar results.
Physical Properties of the Native/R120Q huPGHS-2 Heterodimer.
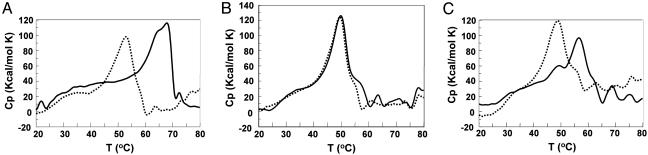
Studies performed with ovine PGHS-1 >20 years ago indicated that time-dependent inhibition of COX activity by inhibitors such as FBP and indomethacin involves the binding of only one inhibitor molecule per dimer to the holoenzyme (7). However, observation of the crystal structures of PGHS-1 and PGHS-2 dimers suggests that both subunits are identical and both bind substrates and inhibitors (14, 15, 17, 24, 25). We performed differential scanning calorimetry (DSC) on the native huPGHS-2 homodimer, the R120Q huPGHS-2 homodimer, and the native/R120Q huPGHS-2 heterodimer in the presence and absence of FBP, a time-dependent COX inhibitor, and S-ibuprofen (S-IBP), a simple competitive inhibitor, to determine whether we could obtain physical evidence for a nonuniformity of subunits (Fig. 4). It should be noted that all experiments were performed in all cases with apoenzyme because adding stoichiometric amounts of heme in dimethyl sulfoxide had inconsistent effects on the melting curves attributable to even small amounts of this solvent. We also point out that the experiments were conducted with 0.1% C10E6 detergent and that lower detergent concentrations led to marked broadening of the melting curves. The melting curve for native/native huPGHS-2 (5 μM) was approximately symmetrical with a melting temperature (Tm) of ≈53°C (Fig. 4A); melting was irreversible. A 10-fold molar excess of S-IBP (500 μM) increased the Tm values by ≈15°C and yielded an asymmetric melting curve (Fig. 4A); similar results were observed with FBP (data not shown). Without inhibitor, the R120Q huPGHS-2 homodimer exhibited a symmetrical melting profile but had a somewhat lower Tm (≈50°C) than the native enzyme (Fig. 4B). Adding a 10- or 100-fold excess of S-IBP did not change the properties of the melting curve; again, similar results were obtained with FBP (data not shown). These results were expected because neither FBP (18) (Fig. 3) nor S-IBP (data not shown) inhibits the R120Q huPGHS-2 homodimer. The native/R120Q huPGHS-2 heterodimer exhibited properties more closely resembling those of the native huPGHS-2 homodimer in the calorimetry experiments (Fig. 4C). In the absence of inhibitor, a symmetrical peak was observed with a Tm of ≈50°C. Addition of inhibitor to the heterodimer led to an asymmetric melting curve with a higher Tm. In brief, the melting curves for the native huPGHS-2 and the native/R120Q heterodimer were qualitatively similar with and without inhibitor and were asymmetric only in the presence of an inhibitor. These observations and the fact that an R120Q monomer would not be expected to bind FBP or S-IBP suggest that only one molecule of inhibitor binds per dimer in the native enzyme.
Fig. 4.
DSC of native/native huPGHS-2 homodimer, native/R120Q huPGHS-2 heterodimer, and R120Q/R120Q huPGHS-2 homodimer with and without S-IBP. DSC was performed as detailed in Materials and Methods by using 5 μM PGHS dimer with (solid line) and without (dotted line) 50 μM S-IBP. Cp, excess heat capacity. (A) Native/native huPGHS-2 homodimer. (B) Native/R120Q huPGHS-2 heterodimer. (C) R120Q/R120Q huPGHS-2 homodimer.
Finally, we examined the melting curves for native and R120Q homo- and heterodimers of huPGHS-2 in the presence of 50 μM AA. In all cases, addition of AA caused a small shift in the melting curves (≈4°C) but no pronounced changes in the shapes of the curves (data not shown).
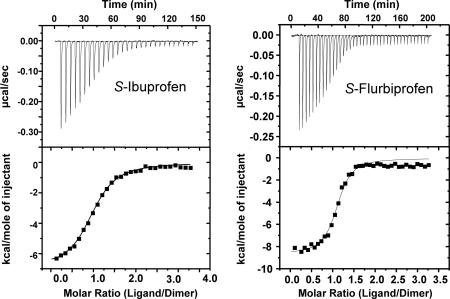
Isothermal titration calorimetry (ITC) was performed to determine the stoichiometry of S-IBP and S-FBP native apo-huPGHS-2 homodimers (Fig. 5). In each case the stoichiometry was ≈0.5 ligand molecule per protein monomer. The Kd values for S-IBP (1.3 μM) and S-FBP (0.3 μM) were similar to Ki values for S-IBP (0.2–0.9 μM; ref. 26) and S-FBP (1 μM; ref. 23) determined by using kinetic analyses or fluorescence quenching. The experiments depicted in Fig. 5 were performed by using apoenzyme, but similar results were obtained when the titrations were performed in the presence of 10 μM hematin; this is consistent with the observation that heme has relatively little effect on the binding of inhibitors to native PGHS-2 (26). ITC was also performed with AA but with inconsistent results that we attributed to the detergent effects of AA on enzyme depleted of detergent in preparation for ITC.
Fig. 5.
ITC of native huPGHS-2 homodimer with S-IBP (Left) and S-FBP (Right). ITC was performed as described in Materials and Methods by titrating 5–7 μM purified histidine-tagged native huPGHS-2 with either 200 μM S-IBP or 200 μM S-FBP. The ligand solution was added to the protein in the cuvette. Different protein preparations were used in the two experiments depicted here, which are representative of three separate titrations with each ligand. (Upper) Kd values and values for moles of ligand/mole protein dimer (n) for S-IBP are Kd = 2.3 μM and n = 0.52 ± 0.02. (Lower) Kd values and values for moles of ligand/mole protein dimer (n) for S-FBP are Kd = 0.30 μM and n = 0.55 ± 0.02. Protocols for determining these values are noted in Materials and Methods.
Discussion
PGHS-1 and PGHS-2 are homodimers with an extensive interface and numerous potential interactions between monomers (24, 25). Disruption of the interface occurs only under denaturing conditions that also perturb the major folding domains of the proteins and eliminate catalytic activity (6). To detect possible interactions between PGHS monomers, we investigated the relationship between the monomers in two different huPGHS-2 heterodimers, native/G533A huPGHS-2 and native/R120Q huPGHS-2. The G533A mutation was chosen because these mutants have little COX activity with AA, linoleic acid, or α-linolenic acid but retain native POX activity (12, 13). The R120Q monomer was used to generate heterodimers because the R120Q huPGHS-2 homodimer has COX kinetic properties similar to those of the native huPGHS-2 homodimer with AA, but, unlike native enzyme R120Q/R120Q, huPGHS-2 is not inhibited effectively by FBP (18).
As anticipated, all of the homodimers and heterodimers exhibited similar POX activities, but, unexpectedly, the COX activities of the heterodimers comprised of native/G533A or native/R120Q subunits had kinetic properties very similar to those of native enzyme and unlike those of the mutant subunits. For example, the native/G533A heterodimer was able to oxygenate AA with an efficiency that approached that of native enzyme, and the native/R120Q heterodimer was effectively inhibited by FBP in a time-dependent manner. The results of our kinetic studies implied that native huPGHS-2 exhibits half-of-sites reactivity (27–35), with only one of the two COX active sites of a dimer catalyzing an oxygenation reaction at a given time. In addition, our results are consistent with earlier data with PGHS-1 suggesting that COX inhibition by standard nonsteroidal antiinflammatory drugs like FBP and indomethacin involves the binding of inhibitor to only one of the two COX sites comprising a PGHS dimer (7). As a further test of this concept, we analyzed the effects of substrate and inhibitor binding on the physical properties of huPGHS-2 forms using calorimetry. Results from DSC suggested that binding of inhibitors by huPGHS-2 leads to two different and more stable folding domains, whereas there is a single, less stable entity in the absence of inhibitors. ITC provided direct evidence that huPGHS-2 exhibits high-affinity binding for only one inhibitor molecule per dimer. It should be noted that these experiments have also yielded the first equilibrium binding measurements of Kd values for inhibitors with PGHSs. The Kd values determined for the S-IBP and S-FBP are similar to previously reported Ki values (5, 7, 23, 26).
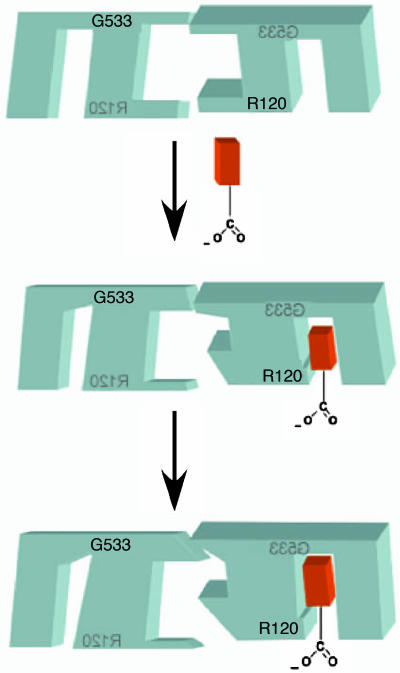
Crystal structures of PGHSs, most of which have been generated with PGHSs incubated in the presence of excess substrate or inhibitor, suggest that the monomers are identical. Our present results establish that the monomer structures do differ during catalysis, with one of the two PGHS subunits binding a substrate molecule and catalyzing its oxygenation. Because a dimer is required for catalysis, we suggest that the subunit that does not bind substrate plays an enabling role. The model shown in Fig. 6 illustrates our concepts about the events occurring during substrate binding. In the absence of substrate, monomers 1 and 2 are the same. Substrate binding causes a change in the monomer 2 active site that, in turn, alters the structure of a portion of monomer 2 where it interacts with monomer 1. This structural change at the interface between monomers 1 and 2 is sensed by monomer 1, leading to constriction of the monomer 1 COX site, thereby preventing substrate binding to monomer 1. As depicted in the final step in Fig. 6, further changes in monomer 1 affect another part of the interface between monomers 1 and 2, causing a change in the monomer 2 active site that molds it around the substrate and enables monomer 2 to oxygenate bound substrate. We imagine that a similar set of structural changes arises when monomer 2 binds an inhibitor. Moreover, we assume that monomers 1 and 2 are equivalent and that a monomer 1a–monomer 2x structure could form in which monomer 1 binds substrate or inhibitor.
Fig. 6.
Depiction of ligand binding to the two monomers of huPGHS-2. Before ligand binding, PGHS monomers are identical. However, as depicted in the first step, fatty acid or inhibitor binding to one monomer causes changes at the interface between monomers, which, in turn, lead to a change in the COX site of the second monomer that prevents ligand binding. In the second step (which occurs at least in the case of substrate binding), there are further rearrangements at the interface between monomers that foster oxygenase catalysis by the occupied monomer.
Our data establish that, in the cases of S-IBP and S-FBP, huPGHS-2 binds only one inhibitor or substrate molecule with high affinity. There is indirect evidence that this is true for ovine PGHS-1 as well (7), and so we presume that this is a general property of both PGHS-1 and PGHS-2. However, there are several observations that raise the possibility that specific PGHS-2 inhibitors can reside on the PGHS-1 inactive monomer without altering the activity of the substrate-bound monomer (8, 36).
As alluded to earlier, there are numerous examples of proteins that function as dimers and some examples of half-of-sites reactivity (27–35), but, in most cases, the nature of the interaction between the monomers is unknown. We suggest that the subunit complementarity or partnering seen with huPGHS-2 may be a common feature of tightly associated monomers of homodimers.
Materials and Methods
Anti-PGHS-2 antibodies directed against the 18-aa insert unique to PGHS-2 were as described (19). Anti-histidine, anti-FLAG antibodies and FLAG peptide, FLAG-affinity resin, hemin, and FBP were from Sigma. Horseradish POX-conjugated secondary antibody (goat anti-rabbit IgG and goat anti-mouse IgG) were from Bio-Rad. All nonradioactive fatty acids and S-FBP were from Cayman Chemical (Ann Arbor, MI). S-IBP was from Calbiochem. [1-14C]AA (40–60 mCi/mmol; 1 Ci = 37 GBq) was purchased from NEN. Sf21 insect cells and Antibiotic–Antimycotic (100×) culture medium were from Invitrogen. Oligonucleotide primers were purchased from Invitrogen or synthesized by the Macromolecular Structure, Sequencing, and Synthesis Facility at Michigan State University (East Lansing, MI); all DNA and N-terminal amino acid sequencing was also performed by this facility. Quantitative amino acid analysis was performed by the University of Michigan (Ann Arbor, MI) Protein Structure Facility. The FBS and HyQ SFX-Insect medium used for cell growth and protein expression were from HyClone. The nonionic detergent C10E6 was from Anatrace (Maumee, OH). Ni-NTA Superflow resin was purchased from Qiagen (Valencia, CA). BCA protein reagent was from Pierce. All other reagents were from common commercial sources.
Preparation of a pFastBac Dual Vector Encoding FLAG-Tagged huPGHS-2.
Starting with hexahistidine-tagged huPGHS-2 in pFastBac Dual vector (Invitrogen), site-directed mutagenesis was performed by using the Stratagene QuikChange mutagenesis kit according to the manufacturer's instructions and using the following oligonucleotide primers: 5′-CTCAGCCATACAGCAAATGACTACAAGGACGACGACAAGCCTTGCTGTTCCCACCCA-3′ and 3′-GAGTCGGTATGTCGTTTACTGATGTTCCTGCTGCTGTTCGGAACGACAAGGGTGGGT-5′.
The mutation was confirmed by sequencing. The FLAG-tagged huPGHS-2 insert in pFastBac Dual vector was removed by digestion with StuI and KpnI. This was subcloned into a pFastBac Dual vector that had been cleaved with SmaI and KpnI, thereby yielding FLAG-tagged huPGHS-2 in pFastBac Dual vector. Amino acid sequencing of the purified protein indicated that after cleavage of the signal peptide the N-terminal sequence was N-D-Y-K-D-D-D-K-P-C-C-S, whereas the N-terminal sequence of the native protein is A-N-P-C-C-S.
Preparation of pFastBac Dual Vectors Encoding Hexahistidine-Tagged huPGHS-2 Mutants.
G533A and R120Q mutations of huPGHS-2 were prepared by site-directed mutagenesis, starting with hexahistidine-tagged huPGHS-2 in pFastBac Dual vector and by using the Stratagene QuikChange mutagenesis kit. The presence of the mutations was confirmed by sequencing. The primers used to prepare the G533A and R120Q mutations were as follows: G533A, 5′-CATTCTCCTTGAAAGCACTTATGG-3′ and 3′-GTAAGAGGAACTTTCGTGAATACCC-5′; R120Q, 5′-GTGTTGACATCCCAATCACATTTGATTG-3′ and 3′-CAATCAATGTGATTGGGATGTCAACAC-5′.
Amino acid sequencing of the purified protein R120Q huPGHS-2 indicated that after cleavage of the signal peptide the N-terminal sequence was N-H-H-H-H-H-H-P-C.
Preparation of pFastBac Dual Vectors Encoding Native/Mutant huPGHS-2 Heterodimers.
Mutated hexahistidine-tagged huPGHS-2 in pFastBac Dual vector was digested with HindIII and EcoRI, and the insert was subcloned into pFastBac Dual vector, which had been cleaved with the same restriction enzymes. A combined construct encoding both FLAG-tagged native huPGHS-2 and a mutant hexahistidine-tagged huPGHS-2 was created by digesting the FLAG-tagged huPGHS-2 in pFastBac Dual vector and the mutated hexahistidine-tagged huPGHS-2 in pFastBac Dual vector with EcoRI and HindIII. The hexahistidine-tagged mutant huPGHS-2 was then subcloned into the pFastBac Dual vector containing the FLAG-tagged native huPGHS-2. The correct orientation of the insert was confirmed by restriction digestion.
Transfection and Expression of Baculovirus in Sf21 Insect Cells.
By following the Life Technologies (Grand Island, NY) Bac-to-Bac expression system protocol, bacmid DNA was generated and used to transfect Sf21 insect cells. Virus was harvested and used to infect the cultures of Sf21 cells. After 72–96 h of infection, the cells were collected and used to purify protein (22).
Purification of huPGHS-2 Heterodimers from Sf21 Insect Cells.
Sf21 cell pellets were resuspended in Ni-solubilization buffer (25 mM sodium phosphate and 100 mM NaCl, pH 7.4) containing Complete EDTA-Free Protease Inhibitor (Roche Applied Science). The enzyme was solubilized with C10E6 (0.8% vol/vol), and insoluble material was removed by centrifugation. The solubilized supernatant was purified by using chromatography on fast-flow Ni-NTA resin (Qiagen). The Ni-NTA flow-through fraction containing FLAG/FLAG-tagged homodimers was further purified by FLAG-affinity chromatography by using FLAG-affinity resin (Sigma) following the manufacturer's protocols. Bound protein was eluted with five column volumes of FLAG peptide (100 μg/ml) in Tris-buffered saline, and the eluant was concentrated to ≈1 ml by using a Millipore centrifugal filter with a cutoff molecular weight of 50,000. The imidazole eluant from the Ni-NTA column was also subjected to FLAG-affinity chromatography to isolate the FLAG/hexahistidine-tagged heterodimers.
The yields of histidine/histidine-tagged mutant homodimers from cells infected with virus encoding both native and mutant enzymes were low. Therefore, when larger amounts of hexahistidine-tagged mutant homodimers were required (e.g., for calorimetry), the mutant proteins were expressed independently in Sf21 cells and purified by Ni-NTA column chromatography followed by chromatography on a HiTrap Q column as detailed (22). Protein concentrations were determined by using the BCA reagent (Pierce) with BSA as a standard.
Western Transfer Blotting.
Western transfer blotting was performed as described (19) by using anti-histidine antibody (1:1,000 dilution), anti-FLAG antibody (1:10,000 dilution), or an antibody against PGHS-2 (1:3,000 dilution) and a 1:2,000 dilution of the appropriate horseradish POX-conjugated secondary antibody. After washing, the nitrocellulose membranes were incubated with Bio-Rad ECL reagents and exposed to film for chemiluminescence.
COX and POX Assays.
COX and POX assays were performed, and profiles of prostanoid products formed from [1-14C]AA were determined as detailed (11, 22).
DSC.
All DSC measurements were performed on a CSC Nano II Differential Scanning Calorimeter (Calorimetry Sciences, Lindon, UT). The sample cell was filled with a protein solution, typically 5 μM protein in the same buffer as that used in reference cell (100 mM sodium phosphate, pH 7.4, containing 0.1% C10E6). All buffers and protein solutions were degassed for at least 15 min before starting an experiment. All samples were subjected to a range of temperatures from 10°C to 100°C at a scanning rate of 0.5°C/min, typically at 3 atm of pressure (1 atm = 101.3 kPa). A buffer-versus-buffer baseline experiment was first recorded, and these values were subtracted from the sample curve as recommended by the manufacturer (Calorimetry Sciences).
ITC.
A VP-ITC system (MicroCal, Amherst, MA) with a computer-controlled stirrer-syringe was used for ITC. Proteins for ITC were dialyzed extensively against 100 mM sodium phosphate, pH 7.6. Ligand solutions of S-IBP (200 μM) and S-FBP (200 μM) were also prepared in 100 mM sodium phosphate, pH 7.6. Typically, aliquots (6 μl) of ligand solution prepared at 10× the protein concentration were injected into a 1.42-ml reaction cell containing ≈10 μM dimer. All experiments were conducted at 20°C. Control experiments, including titrating ligands into buffer alone or proteins into buffer alone, involved separate titrations and, where applicable, were subtracted from the experimental data. Calorimetric data analysis was performed with origin software provided by MicroCal. Binding parameters including the binding constants (KA, M−1), stoichiometry of binding (n), and thermodynamics of binding (ΔH and TΔS, kcal/mol) were determined by fitting the experimental binding isotherms. N-terminal sequence analysis performed by the University of Michigan Protein Structure Facility was used to quantify the amounts of protein used for the ITC experiments.
Acknowledgments
Dr. Raymond Trievel provided guidance in our ITC studies. This work was supported in part by National Institutes of Health Grant R01 GM68848.
Abbreviations
- PGHS
prostaglandin endoperoxide H synthase
- human PGHS
huPGHS
- COX
cyclooxygenase
- AA
arachidonic acid
- POX
peroxidase
- FBP
flurbiprofen
- DSC
differential scanning calorimetry
- IBP
ibuprofen
- ITC
isothermal titration calorimetry.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Smith W. L., DeWitt D. L., Garavito R. M. Annu. Rev. Biochem. 2000;69:149–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 2.Rouzer C., Marnett L. Chem. Rev. (Washington, D.C.) 2003;103:2239–2304. doi: 10.1021/cr000068x. [DOI] [PubMed] [Google Scholar]
- 3.van der Donk W., Tsai A., Kulmacz R. Biochemistry. 2002;41:15451–15458. doi: 10.1021/bi026938h. [DOI] [PubMed] [Google Scholar]
- 4.DeWitt D. L. Mol. Pharmacol. 1999;55:625–631. [PubMed] [Google Scholar]
- 5.Prusakiewicz J., Felts A., Mackenzie B., Marnett L. Biochemistry. 2004;43:15439–15445. doi: 10.1021/bi048534q. [DOI] [PubMed] [Google Scholar]
- 6.Xiao G., Chen W., Kulmacz R. J. J. Biol. Chem. 1998;273:6801–6811. doi: 10.1074/jbc.273.12.6801. [DOI] [PubMed] [Google Scholar]
- 7.Kulmacz R. J., Lands W. E. M. J. Biol. Chem. 1985;260:12572–12578. [PubMed] [Google Scholar]
- 8.Rosenstock M., Danon A., Rimon G. Biochim. Biophys. Acta. 1999;1440:127–137. doi: 10.1016/s1388-1981(99)00105-5. [DOI] [PubMed] [Google Scholar]
- 9.Rosenstock M., Danon A., Rubin M., Rimon G. Eur. J. Pharmacol. 2001;412:101–108. doi: 10.1016/s0014-2999(00)00931-6. [DOI] [PubMed] [Google Scholar]
- 10.Burde T., Rimon G. Eur. J. Pharmacol. 2002;453:167–173. doi: 10.1016/s0014-2999(02)02450-0. [DOI] [PubMed] [Google Scholar]
- 11.Thuresson E. D., Lakkides K. M., Rieke C. J., Sun Y., Wingerd B. A., Micielli R., Mulichak A. M., Malkowski M. G., Garavito R. M., Smith W. L. J. Biol. Chem. 2001;276:10347–10357. doi: 10.1074/jbc.M009377200. [DOI] [PubMed] [Google Scholar]
- 12.Shimokawa T., Smith W. L. J. Biol. Chem. 1992;267:12387–12392. [PubMed] [Google Scholar]
- 13.Rowlinson S. W., Crews B. C., Lanzo C. A., Marnett L. J. J. Biol. Chem. 1999;274:23305–23310. doi: 10.1074/jbc.274.33.23305. [DOI] [PubMed] [Google Scholar]
- 14.Malkowski M. G., Thuresson E. D., Lakkides K. M., Rieke C. J., Micielli R., Smith W. L., Garavito R. M. J. Biol. Chem. 2001;276:37547–37555. doi: 10.1074/jbc.M105982200. [DOI] [PubMed] [Google Scholar]
- 15.Thuresson E. D., Malkowski M. G., Lakkides K. M., Rieke C. J., Mulichak A. M., Ginell S. L., Garavito R. M., Smith W. L. J. Biol. Chem. 2001;276:10358–10365. doi: 10.1074/jbc.M009378200. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharyya D. K., Lecomte M., Rieke C. J., Garavito M., Smith W. L. J. Biol. Chem. 1996;271:2179–2184. doi: 10.1074/jbc.271.4.2179. [DOI] [PubMed] [Google Scholar]
- 17.Malkowski M. G., Ginell S., Smith W. L., Garavito R. M. Science. 2000;289:1933–1937. doi: 10.1126/science.289.5486.1933. [DOI] [PubMed] [Google Scholar]
- 18.Rieke C. J., Mulichak A. M., Garavito R. M., Smith W. L. J. Biol. Chem. 1999;274:17109–17114. doi: 10.1074/jbc.274.24.17109. [DOI] [PubMed] [Google Scholar]
- 19.Spencer A. G., Woods J. W., Arakawa T., Singer I. I., Smith W. L. J. Biol. Chem. 1998;273:9886–9893. doi: 10.1074/jbc.273.16.9886. [DOI] [PubMed] [Google Scholar]
- 20.Laneuville O., Breuer D. K., Xu N., Huang Z. H., Gage D. A., Watson J. T., Lagarde M., DeWitt D. L., Smith W. L. J. Biol. Chem. 1995;270:19330–19336. doi: 10.1074/jbc.270.33.19330. [DOI] [PubMed] [Google Scholar]
- 21.Thuresson E. D., Lakkides K. M., Smith W. L. J. Biol. Chem. 2000;275:8501–8507. doi: 10.1074/jbc.275.12.8501. [DOI] [PubMed] [Google Scholar]
- 22.Song I., Ball T. M., Smith W. L. Biochem. Biophys. Res. Commun. 2001;289:869–875. doi: 10.1006/bbrc.2001.6071. [DOI] [PubMed] [Google Scholar]
- 23.Laneuville O., Breuer D. K., Dewitt D. L., Hla T., Funk C. D., Smith W. L. J. Pharmacol. Exp. Ther. 1994;271:927–934. [PubMed] [Google Scholar]
- 24.Picot D., Loll P. J., Garavito M. Nature. 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 25.Kurumbail R. G., Stevens A. M., Gierse J. K., McDonald J. J., Stegeman R. A., Pak J. Y., Gildehaus D., Miyashiro J. M., Penning T. D., Seibert K., et al. Nature. 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 26.Houtzager V., Ouellet M., Falgueyret J., Passmore L., Bayly C., Percival M. Biochemistry. 1996;35:10974–10984. doi: 10.1021/bi960053m. [DOI] [PubMed] [Google Scholar]
- 27.Izard T., Geerlof A. EMBO J. 1999;18:2021–2030. doi: 10.1093/emboj/18.8.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoylaerts M. F., Manes T., Millan J. L. J. Biol. Chem. 1997;272:22781–22787. doi: 10.1074/jbc.272.36.22781. [DOI] [PubMed] [Google Scholar]
- 29.Klotz I. M., Hunston D. L. Proc. Natl. Acad. Sci. USA. 1977;74:4959–4963. doi: 10.1073/pnas.74.11.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinha S. C., Wetterer M., Sprang S. R., Schultz J. E., Linder J. U. EMBO J. 2005;24:663–673. doi: 10.1038/sj.emboj.7600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W., Olson J. S., Phillips G. N., Jr Biophys. J. 2005;88:2801–2814. doi: 10.1529/biophysj.104.047936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts C., Chlebowski J. J. Biol. Chem. 1984;259:3625–3632. [PubMed] [Google Scholar]
- 33.Bild G., Janson C., Boyer P. J. Biol. Chem. 1980;255:8109–8115. [PubMed] [Google Scholar]
- 34.Izard T. J. Bacteriol. 2003;185:4074–4080. doi: 10.1128/JB.185.14.4074-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill T. L. Proc. Natl. Acad. Sci. USA. 1978;75:1101–1105. doi: 10.1073/pnas.75.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalgutkar A., Crews B., Marnett L. Biochemistry. 1996;35:9076–9082. doi: 10.1021/bi9605752. [DOI] [PubMed] [Google Scholar]
- 37.Spencer A. G., Thuresson E. A., Otto J. C., Song I., Smith T., DeWitt D. L., Garavito R. M., Smith W. L. J. Biol. Chem. 1999;274:32936–32942. doi: 10.1074/jbc.274.46.32936. [DOI] [PubMed] [Google Scholar]
- 38.Kiefer J., Pawlitz J., Moreland K., Stegeman R., Hood W., Gierse J., Stevens A., Goodwin D., Rowlinson S., Marnett L., et al. Nature. 2000;405:97–101. doi: 10.1038/35011103. [DOI] [PubMed] [Google Scholar]