Abstract
Hsp70s are highly conserved ATPase molecular chaperones mediating the correct folding of de novo synthesized proteins, the translocation of proteins across membranes, the disassembly of some native protein oligomers, and the active unfolding and disassembly of stress-induced protein aggregates. Here, we bring thermodynamic arguments and biochemical evidences for a unifying mechanism named entropic pulling, based on entropy loss due to excluded-volume effects, by which Hsp70 molecules can convert the energy of ATP hydrolysis into a force capable of accelerating the local unfolding of various protein substrates and, thus, perform disparate cellular functions. By means of entropic pulling, individual Hsp70 molecules can accelerate unfolding and pulling of translocating polypeptides into mitochondria in the absence of a molecular fulcrum, thus settling former contradictions between the power-stroke and the Brownian ratchet models for Hsp70-mediated protein translocation across membranes. Moreover, in a very different context devoid of membrane and components of the import pore, the same physical principles apply to the forceful unfolding, solubilization, and assisted native refolding of stable protein aggregates by individual Hsp70 molecules, thus providing a mechanism for Hsp70-mediated protein disaggregation.
Keywords: Brownian ratchet, disaggregation, Hsp90, DnaK, Tim44
Hsp70 is a central component of the chaperone network in the cell with disparate cellular functions. Associated with the ribosome, Hsp70s foster proper de novo protein folding. In the cytoplasm, Hsp70s mediate the deoligomerization and recycling of native protein complexes (1, 2) and control key functions in evolution, cell morphogenesis (2), and apoptosis (3), often in association with Hsp90 (4). Hsp70 also serves as the central translocation motor in the posttranslational import of cytoplasmic proteins into mitochondria (5), chloroplasts (6, 7), and the endoplasmic reticulum (8). Moreover, Hsp70s can actively unfold, solubilize, and reactivate already formed, stable protein aggregates (9,10) and may participate in targeting proteins to the degradation pathway (11, 12).
Existing Models for Hsp70-Mediated Protein Translocation into Mitochondria
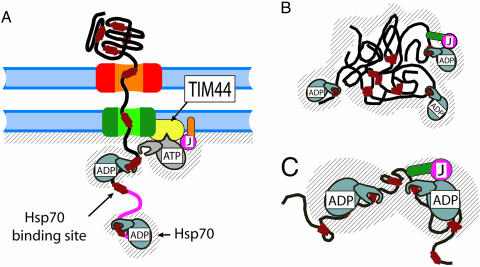
The translocation of proteins across the mitochondrial membrane, through the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) translocation pores, is mediated by the presequence translocase-associated motor (PAM) complex consisting of matrix-localized Hsp70 (mtHsp70), membrane-associated J domain-containing proteins (three identified so far, PAM16/Tim16, PAM18/Tim14, and Mdj2) (13–19) and the nucleotide exchange factor Mge1. In the ATP-bound state, mtHsp70 is in the open (unlocked) state, which is as yet unbound to the translocating protein substrate, whereas mtHsp70 is found anchored to the mitochondrial import channel by way of its transient association with the mitochondrial peripheral inner-membrane protein Tim44. In the ADP-bound state, mtHsp70 is tightly bound (locked) onto the incoming polypeptide and is not associated to the membrane (5, 18, 20, 21) (Fig. 1A). Two mechanisms have been proposed to explain the ability of mtHsp70 to import tightly folded precursor proteins into mitochondria.
Fig. 1.
Schematic view of Hsp70’s role in protein translocation and in protein disaggregation and unfolding. (A) MtHsp70·ATP anchors to the pore-associated protein Tim44 and is, thus, ready to lock to a typical hydrophobic binding segment on an entering polypeptide (red rectangles). The nearby J domain of membrane-anchored PAM16/Tim16 (or PAM18/Tim14) (pink) triggers ATP hydrolysis as soon as a typical hydrophobic binding site exits from the pore. PAM16/18 contains a membrane-anchoring domain (orange). While locked onto the polypeptide, mtHsp70·ADP dissociates from Tim44. The shaded region is forbidden to a binding site on the polypeptide, upon association with mtHsp70. (B) Hsp70 interactions with a stable protein aggregate, as in A, with regions (hatched) of forbidden access for an exposed, chaperone-bound polypeptide loop in a misfolded polypeptide. The protein-binding domain (green) of a soluble J domain cochaperone (such as DnaJ, Hsp40) binds the aggregate and entraps freely diffusing Hsp70s within the entropic pulling region of the aggregated substrate by inducing with its J domain (pink) ATP hydrolysis and the locking of Hsp70 onto the polypeptide. (C) Close-up of the regions of mutually excluding volumes in the case of two Hsp70 molecules bound on either side of a misfolded region in the same misfolded polypeptide monomer.
The Brownian ratchet model assumes that the transient binding of mtHsp70 to an incoming translocating chain forbids backward movements of the latter, because of the large volume of the chaperone that cannot enter the translocation pore, which, alone, would favor forward movements (5, 22, 23). The energy from ATP hydrolysis is used here to fuel a conformational change in the chaperone from the unlocked to the substrate-locked state and does not actively contribute to the movement.
The power-stroke model assumes that the energy from ATP hydrolysis is directly converted into a conformational change within the chaperone, resulting in both a high-affinity state for the substrate and, by using Tim44 as a fulcrum, in a lever-arm movement, generating a mechanical force capable of pulling the polypeptide into the matrix while unfolding it on the cytoplasmic side (21, 24–26).
Existing Models for Hsp70-Mediated Protein Disaggregation
Bacterial Hsp70 (DnaK), regulated by J domain (DnaJ) and nucleotide exchange (GrpE) cochaperones can disaggregate stable protein aggregates without the assistance of other chaperones, such as GroELS or ClpB (9). Moreover, when in at least a 2-fold molar excess over the heat-preaggregated model substrate, DnaK can mediate the reactivation of otherwise stable small enzyme aggregates (9, 27). Based on biochemical evidence and theoretical considerations, individual Hsp70 molecules have been proposed to act as unfolding chaperones using the energy of ATP hydrolysis to disentangle misfolded structures at the surface of stable protein aggregates and gradually unfold misfolded polypeptides into natively refoldable ones (27, 28).
A Single Mechanism Explains the Different Hsp70 Functions
The very high degree of conservation within the Hsp70 family members strongly favors a simple unique molecular mechanism for all Hsp70s, whereas functional differences may depend on modulating cochaperones, such as J domain proteins (29), nucleotide exchange factors, docking proteins (such as Tim44), or other members of the chaperone network such as Hsp100, GroEL, CHIP, and Hsp90 (30).
Here, we argue that, by fueling changes in the conformational freedom of the various components of the system, ATP hydrolysis in the Hsp70 molecule can indirectly drive forceful protein unfolding and translocation by a modified Brownian ratchet mechanism named entropic pulling. We examined by statistical physics the basic thermodynamic principles and molecular geometries that can fit a common mechanism for both Hsp70-mediated protein import across membranes and active unfolding of stable protein aggregates by Hsp70. The entropic pulling mechanism was found to be fully compatible with the disparate cellular activities of Hsp70. The entropic pulling mechanism settled apparent controversial data and arguments favoring either the Brownian ratchet or the power-stroke mechanism and, moreover, provided a mechanism by which Hsp70s can mediate the active unfolding and solubilization of stable protein aggregates in vitro and in the cell.
Entropic Pulling in Protein Translocation
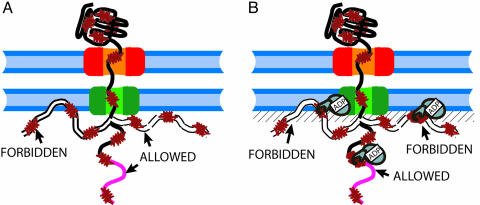
An incoming polypeptide that has emerged into the mitochondrial matrix and not yet bound any mtHsp70 chaperones can access all available polymer conformations, with the exception of those that violate the excluded volume between the polypeptide and the membrane and the other proteins of the pore (Fig. 2A). Because of the large volume of mtHsp70, chaperone binding, thus, greatly increases the excluded-volume constraint, and the emerging preprotein segment is prevented from accessing those conformations for which a chaperone-binding site tightly bound to a mtHsp70 molecule would be closer to the membrane than one chaperone radius, i.e., ≈20–25 Å (31), else the excluded-volume constraint between mtHsp70 and the membrane would be violated (Fig. 2B and shaded region in Fig. 1A).
Fig. 2.
Schematic view of the excluded volume effects without (A) and with (B) mtHsp70 bound to a translocating polypeptide. (A) Polypeptide conformations that occupy only the mitochondrial matrix space are allowed, whereas polypeptide conformations that would partially occupy the volume already taken by the membrane are forbidden. (B) Binding of mtHsp70 further forbids the polypeptide conformations, such that the chaperone would partially occupy the volume taken by the membrane.
The number N(n) of conformations accessible to a preprotein segment composed of n amino acids between the pore exit and the chaperone-binding site is related to its entropy, S(n), by the formula S(n) = kBlnN(n) (32). Because of excluded-volume effects, the binding of mtHsp70 onto the entering preprotein reduces to N70(n) the number of conformations accessible to the preprotein and, thus, its entropy to S70(n) = kBlnN70(n). Consequently, upon chaperone binding, the conformational contribution of the free energy for the entering preprotein changes by an amount ΔF(n) = −T[S70(n) − S(n)] = −kBT ln[N70(n)/N(n)]. ΔF(n) is the intrinsic contribution to the import process of mtHsp70 locking on the peptide.
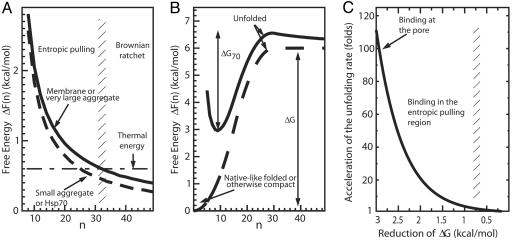
We computed N(n) and N70(n) using realistic values of the persistence length of unfolded proteins (33) and of the size of Hsp70 (31) (see Materials and Methods). Our calculations revealed that, as the number n of residues between the mtHsp70-binding site and the pore exit increased, ΔF(n) decreased as 1/n (Fig. 3A, solid line), because the effect of the excluded volume between the chaperone and the membrane decreases for larger values of n (an explicit derivation using a less realistic but analytically tractable preprotein model provides a rigorous foundation for the inverse proportionality between ΔF(n) and n, see supporting information, which is published on the PNAS web site).
Fig. 3.
Free-energy profiles without and with a bound Hsp70 molecule. (A) Free-energy profiles of polypeptide translocation and aggregate unfolding by action of Hsp70 were calculated as a function of a parameter n corresponding to the number of imported residues in the mitochondrial matrix (as in Fig. 1A, solid line) or of exposed flexible segments flanked by the aggregate on one side and the bound chaperone on the other (as in Fig. 1B, solid line for large aggregates and dashed line for small ones) or of flexible amino acids between two independent chaperones bound to the same misfolded polypeptide (as in Fig. 1C, dashed line). The horizontal dot-dashed line is the energy associated with thermal fluctuations. The vertical shaded region at residues 31–33 separates a region (to the left) where entropic pulling prevails from a region (to the right) where entropic pulling is least effective and where Brownian ratchet may still prevent protein backsliding. (B) Free-energy profile of unfolding for a native-like, misfolded or otherwise compact translocating protein in the absence (dashed line) or presence (solid line) of a polypeptide-bound (locked) Hsp70 chaperone on the matrix side of the membrane. The reaction coordinate n is the number of free residues at the preprotein N terminus available for translocation through the pore. (C) Acceleration of thermally driven unfolding as a function of the reduction of the free-energy barrier of unfolding, ΔG−ΔG70. The binding of Hsp70 at increasing distances from the excluded volume region (larger values of n in A) gives rise to a decreasing rate of pulling-biased spontaneous unfolding. However, as long as binding occurs in the entropic binding region (n < 30), some acceleration still occurs. All values of energies and rate accelerations are computed at T = 25°C.
A thermodynamic constraint for the binding process, and thus for the entropic pulling mechanism to be feasible, is that the overall free-energy change on Hsp70 locking, ΔH + ΔF(n), be negative, where ΔH is the affinity of the chaperone for its substrate. We found that ΔF(n) is, at most, 3 kcal/mol when n is small (Fig. 3A), whereas the affinity of Hsp70 in the ADP form (locked state) for its substrates has been experimentally measured to be ΔH ≈ −9 kcal/mol (20, 26, 34, 35). Therefore, the constraint is verified, and the locking of Hsp70 onto its substrate is a thermodynamically favorable process.
Because, according to thermodynamics, all systems spontaneously tend toward the minimum of their free energy and, in this case, to the minimum of ΔF(n), the number n of residues imported within the mitochondrial matrix must increase because of the locking of mtHsp70 on the entering polypeptide. Therefore, we found that the tight binding of mtHsp70 onto an entering polypeptide, with the concomitant decrease of its affinity for Tim44, can produce an effective pulling force of entropic origin on the polypeptide. The pulling force, which is proportional to the free-energy gradient, was found to be the largest, ranging between 10 and 20 pN, when the bound chaperone is the nearest to the membrane (n ranging from 8 to 15 residues). Thus, recruiting mtHsp70 (in the ATP state) at the pore exit by means of Tim44 binding exploits the full potential of the entropic pulling mechanism. Interestingly, a force of 10–20 pN is of the same order as the force of 20 pN predicted by the power-stroke mechanism of action of GroEL over misfolded proteins, showing that intrinsically different mechanisms can lead to similar effects (36).
Whereas the typical energy of thermal fluctuations ranges ≈0.6 kcal/mol, the free-energy differences induced in the entropic pulling region by mtHsp70 binding on the polypeptide were found to be significantly larger, up to 3 kcal/mol (see Fig. 3A), corresponding to a significant net pulling force applied on the entering polypeptide by the bound mtHsp70. Once the distance between the pore exit and the bound chaperone has exceeded 30 residues, our calculations showed that the free-energy difference ΔF has become smaller than the energy associated with thermal fluctuations (0.6 kcal/mol), and the polypeptide has entered a new region, where no significant force is produced by the bound mtHsp70 molecule. In such a region, a true Brownian ratchet, capable of preventing retrotranslocation may be at work, because backward movements would bring the bound chaperone back into the entropic pulling region, whence it had just been forcefully expelled. This newly defined Brownian ratchet is different from the one originally proposed for protein translocation, because it relies on the inability of mtHsp70 to reenter the entropic pulling region rather than on the size incompatibility between the chaperone and the pore (5).
As in the case of the Brownian ratchet, the release of the chaperone from Tim44 is fundamental to the entropic pulling mechanism, to allow the excluded-volume constraint between mtHsp70 and the membrane to be effective. In contrast, an unfolding and translocation power stroke would have to occur in Hsp70 prior to chaperone dissociation from Tim44. However entropic pulling, similar to the power stroke, can generate a strong pulling force inside the entropic pulling region, up to a length of ≈30 imported residues, whereas the Brownian ratchet cannot actively pull the entering polypeptide (Fig. 3A). Notably, it has been found that, on average, incoming proteins may present a new typical Hsp70-binding site every 25–35 residues (37). Our finding that entropic pulling is effective up to 30 residues warrants that a second mtHsp70 molecule can bind the incoming polypeptide at the pore entrance, often before, or at least as soon as, the first Hsp70 molecule has exhausted its entropic pulling potential. Entropic pulling may, thus, carry a continuous, active pulling force until the whole polypeptide has been unfolded on the cytoplasmic side and has been fully translocated into the matrix.
Entropic Pulling Can Accelerate Protein Unfolding and Translocation
We also found that binding of mtHsp70 on the emerging polypeptide close to the pore exit changes the free-energy profile of unfolding for the preprotein and reduces the unfolding free-energy barrier by as much as 3 kcal/mol.
When a folded (or otherwise compact) protein, blocked against the translocation pore, is pulled by the N terminus, its free energy increases with the C-to-N distance (38), which, in the present context, is related to the number of residues unraveled from the protein structure and inserted into the pore. The shape of the free-energy curve shows an upward curvature, a signature of the cooperative nature of protein unfolding (39), as recently highlighted in single-molecule experiments (40). The dashed line in Fig. 3B represents such a stylized free-energy profile.
The locking of an Hsp70 molecule, at the pore exit, to a preprotein that is in a folded (or otherwise compact) state cannot promote import until the preprotein unfolds. As it is known from single molecule experiments, mechanical pulling of proteins at constant force accelerates protein unfolding by lowering the free-energy barrier of unfolding according to the Bell formula (41), which is an example of the more general Kramers’ escape-rate formula. Entropic pulling applies a force, which is the gradient of ΔF(n) (Fig. 3A, solid line), that is not constant but decreases with the number n of unraveled residues. The resulting free-energy profile is, therefore, obtained by adding the free-energy ΔF(n) associated with the pulling force to the free-energy profile of unfolding, resulting in a new free energy of unfolding (Fig. 3B, solid line) with an equilibrium state that is partially unfolded (the minimum at n = 8).
More importantly, we found that chaperone binding close to the pore exit reduces the free-energy barrier of unfolding of about 3 kcal/mol, from ΔG to ΔG70 (Fig. 3B). Because the rates ku of protein unfolding driven by thermal fluctuations decrease exponentially with the free-energy barrier of unfolding ΔG, according to Kramers’ formula ku(ΔG) = k0 exp(−ΔG/kBT) (39), the reduction of ΔG of 3 kcal/mol by entropic pulling accelerates unfolding events up to 100-fold.
According to these calculations, the acceleration of unfolding should apply equally to loosely and to tightly folded domains, the slower unfolding rates of tightly folded domains (5) being ascribed to intrinsically lower rates of thermally driven unfolding fluctuations. Noticeably, even when chaperone binding may not occur closest to the pore exit, as in the case of Tim44-depleted mitochondria (42, 43) but still in the entropic pulling region, some acceleration of thermally driven unfolding and translocation is possible (Fig. 3C). Hence, even if mtHsp70 was forced to bind the incoming polypeptide (assisted by the soluble Hsp40) halfway from the membrane into the entopic pulling region, an acceleration of translocation would yet be in the order of 20-fold higher than if only the Brownian ratchet was in effect.
Because protein folding/unfolding is an all-or-none cooperative process (39, 44), domains on the cytoplasmic side are either fully folded or unfolded and, thus, available for translocation. The same force accelerating unfolding will, thus, immediately pull into the mitochondrial matrix any preprotein that would spontaneously visit the unfolded state and, so, hinder subsequent refolding events on the cytoplasmic side. A single mechanism can thus explain both the mtHsp70-mediated ATP-consuming unfolding ability, previously seen as a power-stroke signature, and the prevention of refolding of the protein on the cytoplasmic side, previously attributed to the Brownian ratchet in its targeted version (45).
Entropic Pulling Applies to Hsp70-Mediated Disaggregation and Unfolding of Misfolded Monomers
Contrary to protein import across membranes, Hsp70-mediated protein disaggregation takes place without a membrane pore and membrane-anchoring components. Yet, we found that the same entropic pulling mechanism described above can apply to Hsp70-mediated unfolding/disaggregation and reactivation reactions. Here, ends and loops exposed at the surface of the aggregate cannot access the volume of the aggregate itself. DnaK can specifically bind to extended hydrophobic protein segments at the surface of the stable protein aggregates (9): In this case, upon Hsp70 binding, more space becomes forbidden (Fig. 1C, shaded region).
An aggregate can be modeled in a simplified way as a forbidden sphere with exposed polypeptide loops, accessible to Hsp70, some of which with chaperone-binding sites. A sphere of infinite radius would correspond to a very large aggregate or to a membrane (Fig. 3A, solid line). Our thermodynamic calculations showed that, as the size of the aggregate (the sphere in the mathematical model) decreased, different free-energy profiles were obtained. For the sake of clarity, we show in Fig. 3A only the curve for the smallest aggregate (dashed line), corresponding to the stably misfolded 56-kDa glucose-6-phosphate dehydrogenase (G6PDH) monomer (9, 27). For all aggregate sizes, the free-energy change caused by Hsp70 binding was found to be significantly reduced by increasing the number n of free residues between the aggregate (the inaccessible sphere) and the chaperone-binding site on a given exposed peptide segment (Fig. 3A). Increasing n, i.e., unraveling residues from the aggregate, is, thus, thermodynamically favorable and leads to progressive local unfolding and disaggregation.
Experimentally, Ben-Zvi et al. (27) showed that aggregates are effectively reduced by substoichiometric amounts of DnaK but that the complete Hsp70-mediated reactivation of misfolded enzyme monomers is obtained only in the presence of at least a 2-fold molar excess of chaperones per misfolded polypeptide substrate (27). We modeled here two Hsp70 molecules concomitantly bound to the opposite ends of a misfolded region on the same polypeptide (Fig. 1C) and found a similar free-energy landscape as in the case of a small misfolded monomer (of the same size as Hsp70) bound to a single chaperone molecule (Fig. 3A, dashed line). Our computations explained the biochemical evidence of Ben-Zvi et al. (27) and confirmed that a pair of Hsp70 molecules can, indeed, cooperate at applying a stretching force by entropic pulling, allowing, upon chaperone release, the spontaneous native refolding of the newly unfolded region.
Thus, because several Hsp70 molecules can bind to the same misfolded polypeptide substrate, the general mechanism of entropic pulling that we uncovered in the case of protein translocation does not require a specific anchor protein to exert a pulling force: Such role can be played by the aggregate itself or, in the case of misfolded monomers, by other chaperone molecules bound elsewhere on the same polypeptide. Here, entropic pulling does not need a pore to act, as would be in the case of a Brownian ratchet, because thermal fluctuations are rectified by the free-energy gradient (Fig. 3A).
Experimental Confirmation That MtHsp70 Can Disaggregate Misfolded Proteins
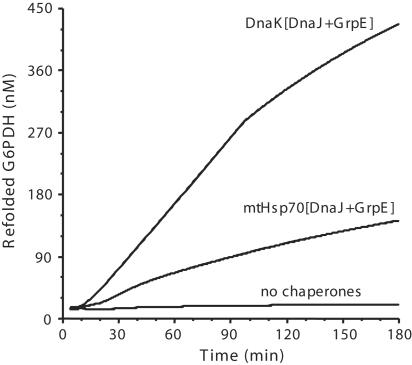
We presented here thermodynamic arguments for a basic molecular mechanism that must be common to different Hsp70s, despite apparent disparities in their cellular functions. Here, we addressed the ability of mtHsp70, without the ClpB homologue, to disaggregate a stable model protein substrate, G6PDH, under conditions where DnaK, DnaJ, and GrpE efficiently unfold and reactivate this enzyme (27). When mtHsp70 was substituted in the refolding assay to DnaK, the mitochondrial chaperone was found to effectively mediate in vitro protein disaggregation and native G6PDH refolding up to 55% yields (Fig. 4). Although less efficient than the control reaction with bacterial DnaK (and DnaJ and GrpE), we conclude that mtHsp70 is clearly able to function as a disaggregating and unfolding Hsp70 chaperone, even without ClpB (46) and Tim44 and when membrane-anchored PAM18/Tim14 is replaced by soluble DnaJ. Because mtHsp70 is unquestionably the motor driving the translocation of nuclear-encoded proteins into mitochondria, this strongly suggests that the conserved Hsp70 molecules in the various compartments of the cells operate by means of a similar simple mechanism, which is likely to be the forceful pulling and local unfolding of misfolded, alternatively folded, or translocating protein substrates (28). Moreover, this result supports a role for Tim44 that is less general and essential as compared with that of GrpE and of the J domain protein, as suggested by the delayed but effective protein import in Tim44-depleted mitochondria (42, 43).
Fig. 4.
Time-dependent reactivation of stable G6PDH aggregates. Stable G6PDH were formed at 52°C and reactivated at 30°C in the presence of bacterial DnaK or yeast mtHsp70, the bacterial cochaperones DnaJ, GrpE, and ATP.
The Role of Hsp70 Cochaperones
Effective entropic pulling requires a delicate balance of the time the chaperone spends in the locked state, during which local unfolding may occur, and the time the chaperone spends in the unlocked state, during which local native refolding of the substrate may occur. Two classes of cochaperones regulate this delicate balance.
J domain proteins catalyze ATP hydrolysis in Hsp70s, essential for the tight locking of the chaperone on its substrates (30, 47, 48). Moreover, it has been shown that the initial binding of DnaJ to the protein substrate, by way of its protein-binding domain, is essential for subsequent efficient binding of DnaK to its substrates and for efficient assisted protein refolding (48). Entropic pulling is the first mechanism that can account for this observation: Aggregate-bound DnaJ could entrap free-moving low-affinity DnaK (ATP-bound) molecules in the solution by catalyzing with the J domain, ATP hydrolysis, and the tight locking of DnaK on a nearby misfolded substrate. In the case of mitochondrial-protein import, Tim44 could act as the chaperone-entrapping device, targeting unbound Hsp70 at the pore exit (13, 17, 49), whereas the role of the nearby membrane-bound J domain proteins (PAM18/Tim14 and PAM16/Tim16) would be limited, in this case, to the sole triggering of mtHsp70 locking onto the incoming polypeptides (18).
Nucleotide exchange factors (e.g., GrpE in bacteria and Mge1 in mitochondria) accelerate ADP release from the ATPase domain of Hsp70, a step of the ATPase cycle that is required to allow chaperone unlocking from its substrates and subsequent local native refolding. Whereas the central physical principle of entropic pulling entirely relies on the binding of Hsp70s to their misfolded or translocating substrates, the two cochaperones regulate the time the chaperones must spend in the bound and unbound states, to allow optimal local unfolding and/or translocation and optimal local refolding, respectively.
Conclusions
Entropic pulling describes the thermodynamic basis for a universal mechanism for Hsp70 chaperones, allowing them to apply a pulling, and ultimately unfolding, force to their various protein substrates. Entropic pulling unifies the two Hsp70 functions of protein disaggregation and mitochondrial protein import. In the translocation context, entropic pulling settles decade-long debates between translocation by Brownian ratchet and by power stroke, by naturally integrating the ratcheting property of the former and the active pulling action of the latter in a single framework. The very simple mechanism of entropic pulling suggested here also justifies the ability of antibodies to import specific substrates into endoplasmic reticulum Sec-reconstituted vesicles (8). From a more general point of view, entropic pulling validates on a rigorous basis the definition of Hsp70 chaperones as unfoldases, that is, enzymes catalyzing the (local) unfolding of native-like or otherwise misfolded proteins (27, 28).
Materials and Methods
Chaperone Reactivation Assay.
G6PDH (0.85 μM) from Leuconostoc mesenteroides (Sigma) was incubated for 7.5 min at 52°C in 100 mM Tris, pH 7.5, 100 mM KCl, 10 mM MgAc2, and 10 mM DTT. The residual activity after the inactivation was <2%. For the chaperone-mediated reactivation assays, the stable G6PDH aggregates (0.75 μM, final concentration) were incubated at 30°C with 4 mM ATP, 5 mM phosphoenol pyruvate, and 10 μg/ml pyruvate kinase (Sigma), Escherichia coli DnaJ (0.8 μM), GrpE (0.8 μM), and DnaK (3.5 μM) or recombinant mtHsp70 (3.5 μM) from yeast mitochondria instead of DnaK. The enzymatic activity of G6PDH was measured at 25°C as in ref. 9. MtHsp70 was purified as described in ref. 3.
Theoretical Calculations.
The data for Fig. 3 have been obtained by counting the number N(n) of chaperone-unbound polymer conformations, of a sample of 106, that do not enter any forbidden region (membrane or aggregate volumes) as a function of the number of emerging residues from the translocating polypeptide. Conformations retained in the previous set and whose last residue falls in a shaded region in Fig. 1 were excluded from the count of the number N70(n) of chaperone-bound available conformations. The free energy of the system is thus F = kBTln[N70(n)], which can be rewritten as F = ΔF + kBTln[N(n)], where the term ΔF = −kBTln[N70(n)/N(n)] represents the contribution intrinsically due to the binding of Hsp70, and it is the term that interests us here. We also checked that increasing the polymer sample from 105 to 106 polymers does not change the ratio N70(n)/N(n) and just reduces statistical fluctuations. We thus used the larger sample.
Experimentally, preproteins have been found to interact with matrix proteins in unstructured, flexible conformations (50, 51). Therefore, polypeptides have been described as freely jointed chains (FJC) with a Kuhn length of 1.2 nm (corresponding to 3–4 residues) (33). Modeling proteins as worm-like chains with persistence length varying between 5 and 10 Å, as measured in experiments (33), does not significantly change the results. Hsp70 chaperones have been modeled as spheres with radius of 20 Å attached to the final bead of the FJC. These quantities are difficult to gauge precisely: The ATPase domain alone of Hsp70 could actually be inscribed in an ellipsoid with principal axes of 4, 4, and 6 nm (31). We would expect a more realistic model of Hsp70, including also the protein-binding domain, to lead to a more pronounced entropy reduction and to a stronger pulling. For lack of a three-dimensional structure of the Hsp70 whole molecule, our predictions cannot be more precise, although we expect our estimation to be an underestimation of the real effects because of the conservative nature of the chaperone model.
Supplementary Material
Acknowledgments
We thank G. Dietler and A. Reginelli for discussions. This work was supported by Swiss National Science Foundation Grant 31-65211.01 and German–Israeli Foundation Grant 753-181.13 (to A.A.).
Abbreviations
- G6PDH
glucose-6-phosphate dehydrogenase
- mtHsp70
matrix-localized Hsp70
- PAM
presequence translocase-associated motor
- TIM
translocase of the inner membrane
- TOM
translocase of the outer membrane
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nollen E. A. A., Morimoto R. I. J. Cell. Sci. 2002;115:2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- 2.Sangster T. A., Lindquist S., Queitsch C. BioEssays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 3.Weiss Y. G., Maloyan A., Tazelaar J., Raj N., Deutschman C. S. J. Clin. Invest. 2002;110:801–806. doi: 10.1172/JCI15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picard D. Cell. Mol. Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neupert W., Brunner M. Nat. Rev. Mol. Cell Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 6.Soll J., Schleiff E. Nat. Rev. Mol. Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 7.Theg S. M., Cline K., Finazzi G., Wollman F. A. Trends Plant Sci. 2005;10:153–154. doi: 10.1016/j.tplants.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Rapaport D., Mayer A., Neupert W., Lill R. J. Biol. Chem. 1998;273:8806–8813. doi: 10.1074/jbc.273.15.8806. [DOI] [PubMed] [Google Scholar]
- 9.Diamant S., Ben-Zvi A. P., Bukau B., Goloubinoff P. J. Biol. Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- 10.Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B. Proc. Natl. Acad. Sci. USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClellan A. J., Scott M. D., Frydman J. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Sakahira H., Breuer P., Hayer-Hartl M. K., Hartl F. U. Proc. Natl. Acad. Sci. USA. 2002;99:16412–16418. doi: 10.1073/pnas.182426899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokranjac D., Sichting M., Neupert W., Hell K. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasiljev A., Ahting U., Nargang F. E., Go N. E., Habib S. J., Kozany C., Panneels V., Sinning I., Prokisch H., Neupert W., et al. Mol. Biol. Cell. 2004;15:1445–1458. doi: 10.1091/mbc.E03-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazier A. E., Dudek J., Guiard B., Voos W., Li Y. F., Lind M., Meisinger C., Geissler A., Sickmann A., Meyer H. E., et al. Nat. Struct. Mol. Biol. 2004;11:226–233. doi: 10.1038/nsmb735. [DOI] [PubMed] [Google Scholar]
- 16.Truscott K. N., Voos W., Frazier A. E., Lind M., Li Y. F., Geissler A., Dudek J., Muller H., Sickmann A., Meyer H. E., et al. J. Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Silva P. D., Schilke B., Walter W., Andrew A., Craig E. A. Proc. Natl. Acad. Sci. USA. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer M. P. Nat. Struct. Mol. Biol. 2004;11:6–8. doi: 10.1038/nsmb0104-6. [DOI] [PubMed] [Google Scholar]
- 19.Kozany C., Mokranjac D., Sichting M., Neupert W., Hell K. Nat. Struct. Mol. Biol. 2004;11:234–241. doi: 10.1038/nsmb734. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q. L., D’Silva P., Walter W., Marszalek J., Craig E. A. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- 21.Matouschek A., Pfanner N., Voos W. EMBO Rep. 2000;1:404–410. doi: 10.1093/embo-reports/kvd093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peskin C. S., Odell G. M., Oster G. F. Biophys. J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon S. M., Peskin C. S., Oster G. F. Proc. Natl. Acad. Sci. USA. 1992;89:3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glick B. S. Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- 25.Horst M., Azem A., Schatz G., Glick B. S. Biochim. Biophys. Acta. 1997;1318:71–78. doi: 10.1016/s0005-2728(96)00131-4. [DOI] [PubMed] [Google Scholar]
- 26.Matouschek A., Azem A., Ratliff K., Glick B. S., Schmid K., Schatz G. EMBO J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Zvi A., De Los Rios P., Dietler G., Goloubinoff P. J. Biol. Chem. 2004;279:37298–37303. doi: 10.1074/jbc.M405627200. [DOI] [PubMed] [Google Scholar]
- 28.Slepenkov S. V., Witt S. N. Mol. Microbiol. 2002;45:1197–1206. doi: 10.1046/j.1365-2958.2002.03093.x. [DOI] [PubMed] [Google Scholar]
- 29.Hennessy F., Nicoll W. S., Zimmermann R., Cheetham M. E., Blatch G. L. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer M. P., Bukau B. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Revington M., Zhang Y. B., Yip G. N. B., Kurochkin A. V., Zuiderweg E. R. P. J. Mol. Biol. 2005;349:163–183. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 32.de Gennes P.-G. Scaling Concepts in Polymer Physics. Ithaca, NY and London: Cornell Univ. Press; 1979. p. 29. [Google Scholar]
- 33.Toan N. M., Marenduzzo D., Micheletti C. Biophys. J. 2005;89:80–86. doi: 10.1529/biophysj.104.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer M. P., Schroder H., Rudiger S., Paal K., Laufen T., Bukau B. Nat. Struct. Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 35.Azem A., Oppliger W., Lustig A., Jeno P., Feifel B., Schatz G., Horst M. J. Biol. Chem. 1997;272:20901–20906. doi: 10.1074/jbc.272.33.20901. [DOI] [PubMed] [Google Scholar]
- 36.Thirumalai D., Lorimer G. H. Annu. Rev. Biophys. Biomol. Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 37.Rudiger S., Buchberger A., Bukau B. Nat. Struct. Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 38.Tian P., Andricioaei I. J. Mol. Biol. 2005;350:1017–1034. doi: 10.1016/j.jmb.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 39.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. [Google Scholar]
- 40.Schlierf M., Rief M. Biophys. J. 2006;90:L33–L35. doi: 10.1529/biophysj.105.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell G. I. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 42.Voisine C., Craig E. A., Zufall N., von Ahsen O., Pfanner N., Voos W. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 43.Bomer U., Maarse A. C., Martin F., Geissler A., Merlin A., Schonfisch B., Meijer M., Pfanner N., Rassow J. EMBO J. 1998;17:4226–4237. doi: 10.1093/emboj/17.15.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fersht A. R., Daggett V. Cell. 2002;108:573–582. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto K., Brinker A., Paschen S. A., Moarefi I., Hayer-Hartl M., Neupert W., Brunner M. EMBO J. 2002;21:3659–3671. doi: 10.1093/emboj/cdf358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Germaniuk A., Liberek K., Marszalek J. J. Biol. Chem. 2002;277:27801–27808. doi: 10.1074/jbc.M201756200. [DOI] [PubMed] [Google Scholar]
- 47.Erbse A., Mayer M. P., Bukau B. Biochem. Soc. Trans. 2004;32:617–621. doi: 10.1042/BST0320617. [DOI] [PubMed] [Google Scholar]
- 48.Laufen T., Mayer M. P., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. Proc. Natl. Acad. Sci. USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truscott K. N., Brandner K., Pfanner N. Curr. Biol. 2003;13:R326–R337. doi: 10.1016/s0960-9822(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 50.Taylor A. B., Smith B. S., Kitada S., Kojima K., Miyaura H., Otwinowski Z., Ito A., Deisenhofer J. Structure (London) 2001;9:615–625. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- 51.Pellecchia M., Montgomery D. L., Stevens S. Y., Vander Kooi C. W., Feng H. P., Gierasch L. M., Zuiderweg E. R. P. Nat. Struct. Biol. 2000;7:298–303. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.