Abstract
Previous two-hybrid analysis of the 17 soluble class E Vps yeast proteins revealed that Vps46p/Did2p interacts with Vta1p and the AAA (ATPase associated with a variety of cellular activities) ATPase Vps4p. Here we report that the binding of Vps46p to Vps4p and Vta1p is direct and not mediated by additional proteins, and the binding of Vps46p to Vps4p is ATP independent. Vps46p regulates the membrane association of Vps4p and is required for the interaction of Vta1p with Vps32p/Snf7p of the ESCRT-III complex. Vta1p is a potent activator of Vps4p, stimulating the ATPase activity by 6- to 8-fold. These results reveal functional roles for the Vps46p and Vta1p proteins in regulating the ESCRT complex assembly/disassembly cycle in protein sorting at the yeast late endosome.
Keywords: endosome, ESCRT complexes, protein sorting, Vps, class E
Endosomes function to coordinate endocytic and biosynthetic protein trafficking to the yeast vacuole, the equivalent of the mammalian lysosome (1, 2). Newly synthesized proteins destined for vacuolar localization and cell surface receptor proteins destined for vacuolar degradation are first trafficked to endosomes before reaching their final destination (3). The process of protein sorting that occurs at the endosome is characterized by the invagination of the outer membrane, which generates multivesicular bodies (MVBs) (4, 5). On reaching the MVB, proteins are either sorted to the internal vesicle membranes or remain on the outer, limiting membrane. Proteins on the internal vesicle membranes are generally delivered to the lumen of the vacuole, whereas proteins on the outer membrane can be recycled to other compartments or delivered to the limiting membrane of the vacuole (1, 2). MVB sorting is important for a variety of biological processes, such as receptor down-regulation, antigen presentation, and embryonic development (6). In addition, enveloped RNA viruses such as the HIV type 1 (HIV-1) require the MVB sorting machinery to bud from the cell (7–10).
The process of MVB sorting has been studied in a number of systems including the yeast Saccharomyces cerevisiae. Genetic and biochemical studies in yeast have identified four protein complexes that are involved in MVB sorting, the endosomal sorting complexes required for transport (ESCRT)-0, -I, -II, and -III (2, 11–14). In total, these four protein complexes are composed of 12 different class E vacuolar protein sorting (Vps) proteins. The model for sorting at the late endosome includes ubiquitinated protein cargo first binding ESCRT-0 and -I being recruited to the membrane, followed by the recruitment of ESCRT-II and -III to the endosomal membrane. The protein cargo is deubiquitinated by the enzyme Doa4p, whereas the AAA-type ATPase (ATPase associated with a variety of cellular activities) Vps4p binds and disassembles the ESCRT-III complex from the membrane (2, 6, 11, 15). Upon ESCRT-III disassociation, the cargo proteins are found in the membrane invaginations of the MVB. The mechanisms by which endosomal membranes invaginate and the protein cargo is sorted into the invaginating membranes remain poorly understood.
Vps4p belongs to the AAA-type ATPase protein family, whose members are involved in numerous cellular functions such as membrane fusion, proteolysis, and protein folding (16, 17). In MVB sorting, ESCRT-III complex disassembly and release from the membrane requires the ATPase activity of Vps4p (15, 18). The ADP-bound form of Vps4p exists as a dimer and is mainly cytosolic (18). Upon nucleotide exchange, Vps4p assembles into an oligomeric complex that has been shown to contain 10–12 Vps4p monomers (18, 19). The ATP-bound complex of Vps4p is localized to the endosomal membrane where it proceeds to catalyze the release of the ESCRT-III protein complex (15, 16).
In total, there are 18 class E Vps proteins, and the loss of any of these results in similar defects in endosomal protein sorting (2, 12). Deletion or mutation of any class E VPS gene results in the secretion of the vacuolar protein carboxypeptidase Y (CPY) and the formation of an enlarged endosomal compartment in which proteins normally destined for the vacuole accumulate (20–23). We have previously analyzed the interactions of all 17 soluble yeast class E Vps proteins (including the 12 class E Vps proteins that are assigned to the ESCRT complexes), using the yeast two-hybrid system (12). Of particular interest for the present work are interactions identified between Vta1p and Vps46p (also known as Did2p/Fti1p), Vps4p and Vps46p, and Vta1p and Vps32p/Snf7p (ESCRT-III).
In support of our yeast two-hybrid data, recent studies have revealed that Vta1p directly binds Vps4p in vitro (19, 24), and the mammalian Vta1p homologue, SBP1, associates directly with mammalian VPS4B (SKD1) (25). A mammalian orthologue of Vps46p, CHMP1B, interacts by yeast two-hybrid analysis with spastin, another mammalian AAA-type ATPase that when mutated is the most common cause of hereditary spastic paraplegia (26). Recently, the microtubule interacting and transport (MIT) domain of VPS4A, the other mammalian Vps4p orthologue, was solved and found to bind CHMP1B in vitro (27).
Here we report that Vps46p, from yeast extracts, binds to both Vta1p and Vps4p and that these interactions are maintained with recombinantly expressed proteins. The Vps46p interaction with Vps4p is ATP independent. We also report that the membrane association of Vps4p depends on the presence of Vps46p and that Vps46p is required for the interaction of Vta1p with ESCRT-III (Vps32p). Finally, Vta1p is a potent activator of the ATP hydrolysis activity of Vps4p, indicating that both Vta1p and Vps46p are crucial components of the MVB machinery regulating the function of the AAA-ATPase Vps4p.
Results and Discussion
Vta1p and Vps4p Interact with Vps46p.
Data obtained previously from our yeast two-hybrid analysis identified interactions between Vps4p and Vps46p as well as between Vta1p and Vps46p (12). We tested whether these interactions were maintained in various class E vps deletion strains and found that these interactions were in fact maintained in each deletion strain constructed, which included a vps27Δ, vps28Δ, vps36Δ, vps32Δ, and vps60Δ (data not shown). To confirm these interactions biochemically, binding experiments were performed using recombinantly expressed GST fusion proteins and solubilized yeast membrane fractions. For these experiments, we used an epitope-tagged and fully functional version of Vps46p (Vps46p–3XHA) expressed from a low copy plasmid. After incubation of GST-fusion proteins and solubilized membranes, glutathione agarose was used to bind and precipitate the GST-fusion proteins and any associated proteins.
Vps4p and Vta1p were expressed as GST-fusion proteins in bacteria and subsequently purified. Purified proteins were used as described for protein affinity experiments with solubilized yeast membrane fractions. We show that GST–Vta1p, but not GST alone, associates with Vps46p–3XHA from wild-type (WT) solubilized membranes (Fig. 1A). To determine whether this interaction requires the presence of closely associated class E Vps proteins, Vps4p or Vps32p, the experiment was repeated using solubilized yeast membranes from vps4Δ and vps32Δ strains expressing Vps46p–3XHA. Our results show that GST–Vta1p, but not GST alone, associates with Vps46p from solubilized membranes in the absence of Vps4p and Vps32p (Fig. 1A). Although Vta1p and Vps46p associated in the absence of Vps32p, the lower intensity of the band indicated a weaker association (Fig. 1A).
Fig. 1.
Vps46p interacts with Vta1p and Vps4p. (A and B) Protein extracts were prepared from WT, vps4Δ, vps32Δ, and vta1Δ (KEBY88, KEBY92, KEBY112, and JLY03) yeast expressing Vps46p–HA (pJL11). GST pull-downs were performed as described in Materials and Methods. Approximately 10% of input (solubilized P15 fraction) and samples from the GST pull-downs were run on the gel. GST alone was used as a negative control. Blots were probed with anti-HA mAb. GST pull-downs were performed by using GST–Vta1p (A) and GST–Vps4p (B). (C) Vps46p was cloned into a bacterial expression vector to contain a C-terminal 6XHIS tag. Vps46p–6XHIS was recombinantly expressed, purified, and used for in vitro binding analysis. Vps46p–6XHIS was incubated with glutathione Sepharose and GST alone (negative control), GST–Vta1p, GST–Vps4p, or GST–Vma13p (negative control). Unbound protein was recovered from the supernatant and saved. The beads were washed, and bound proteins were eluted by heating in sample buffer. Blots were probed with an anti-6XHIS mAb.
We also examined the interaction between Vps4p and Vps46p in the same manner. We found that GST–Vps4p, but not GST alone, associated with Vps46p using solubilized yeast membranes from WT cells (Fig. 1B). This experiment was again performed using solubilized yeast membranes from class E vps deletion strains (vta1Δ and vps32Δ) expressing Vps46p–3XHA to determine whether this interaction depends on the presence of closely associated proteins. Our results revealed that GST–Vps4p, but not GST alone, associated with Vps46p from solubilized membranes from vta1Δ and vps32Δ yeast (Fig. 1B). Vps4 and Vps46p were able to associate in the absence of Vps32p, but the lower intensity of the band suggested a weaker association (Fig. 1B). Our experiments revealed that the interactions between both Vta1p and Vps46p and the interaction between Vps4p and Vps46p were stable in the absence of other class E Vps proteins, suggesting that both Vps4p and Vta1p interact directly with Vps46p.
To determine whether Vps4p or Vta1p bind Vps46p directly, protein-binding assays were performed using recombinantly expressed and purified proteins. Purified GST–Vps4p, GST–Vta1p (described above), and GST–Vma13p were used in conjunction with Vps46p-6XHIS, which was also recombinantly expressed and purified. We used GST–Vma13p, which is similar in size to Vps4p, as a negative control to test whether the interactions are specific. Glutathione Sepharose was incubated with GST alone, GST–Vps4p, GST–Vta1p, or GST–Vma13p and Vps46p–6XHIS. Vps46p–6XHIS bound to both GST–Vps4p and GST–Vta1p but not GST alone or GST–Vma13p (Fig. 1C), indicating that Vps46p interacts directly with both Vps4p and Vta1p and that no additional proteins were required for these interactions.
Vps46p Interacts with Vps4p Independent of Bound ATP.
To further examine the interaction between Vps4p and Vps46p and the role of Vps4p ATP hydrolysis, we recombinantly expressed and purified the ATP hydrolysis mutant form of Vps4p, GST–Vps4pE233Q. Protein affinity assays were performed using GST–Vps4p and GST–Vps4pE233Q with solubilized membranes from WT cells expressing Vps46p–3XHA. GST–Vps4pE233Q and GST–Vps4p, but not GST alone, bound Vps46p (Fig. 2A). The ATP hydrolysis mutant form of Vps4p has been shown to assemble into a large, oligomeric protein complex that is membrane associated (16, 18, 19). We analyzed the interaction of Vps46p with Vps4pE233Q in both vps32Δ and vta1Δ strain backgrounds to determine whether the absence of these proteins has an effect on the interaction of Vps46p with the ATP hydrolysis mutant form of Vps4p. We found that GST–Vps4pE233Q and GST–Vps4p, but not GST alone, associated with Vps46p from membranes prepared from vps32Δ or vta1Δ yeast cells (Fig. 2A), indicating that the loss of these two proteins does not have an effect on Vps46p association with Vps4pE233Q.
Fig. 2.
Vps46p interaction with Vps4pE233Q is enhanced and ATP independent. (A) Protein extracts were prepared from WT, vps4Δ, vps32Δ, and vta1Δ (KEBY88, KEBY92, KEBY112, and JLY03) yeast expressing Vps46p–HA (pJL11). GST pull-downs were performed as described in Fig. 1 A and B except using GST–Vps4p (positive control) and GST–Vps4pE233Q. (B) In vitro binding analysis was performed as in Fig. 1C except Vps46p–6XHIS was incubated with glutathione Sepharose and GST alone (negative control), GST–Vps4p (positive control), or GST–Vps4pE233Q.
To determine whether Vps46p is able to directly interact with the ATP hydrolysis mutant form of Vps4p, as it does with Vps4p, we used recombinantly expressed and purified GST–Vps4p, GST–Vps4pE233Q, and Vps46p–6XHIS (each described above) in binding assays. Glutathione beads were incubated with GST–Vps4p (positive control), GST–Vps4pE233Q, or GST alone and purified Vps46p–6XHIS. The results show that GST–Vps4pE233Q and GST–Vps4p, but not GST alone, bound Vps46p–6XHIS (Fig. 2B). This assay was performed in the absence of added ATP and revealed that the in vitro binding of Vps46p and Vps4p did not require ATP. In addition, the in vitro binding of Vps46p to Vps4pE233Q illustrates that ATP hydrolysis is not required for the interaction. The binding of Vps46p to Vps4pE233Q was actually enhanced in comparison with WT Vps4p (Fig. 2B). Vps46p and Vta1p interact with Vps4p in vitro, the addition of ATP is not required for either interaction, and they exhibit enhanced binding to the ATP hydrolysis mutant form of Vps4p (this work and ref. 24).
Vps46p Regulates the Membrane Association of Vps4p.
Vps4p exists as a dimer in the cytosol and, upon binding ATP, assembles into an oligomeric complex that associates with endosomes (16, 18, 19). Upon ATP hydrolysis, the Vps4 oligomer is disassembled and dissociates from the endosomal membrane (18, 19). Vps4p localization is mainly cytosolic in a WT background because of the continuous cycling of Vps4p between its two forms, which results in its limited endosomal membrane association. The distribution of the ATP hydrolysis defective form of Vps4p, Vps4pE233Q, shifts from being mainly cytosolic to being more membrane associated (18). To confirm the membrane association of Vps4p, we first constructed an integrated version of vps4E233Q, determined to be correct by the dominant-negative CPY secretion phenotype (data not shown). By using a Vps4p-specific antiserum, we were able to analyze the subcellular fractionation of Vps4p (Fig. 3). Consistent with published localization results for Vps4p and Vps4pE233Q (16), we found that the majority of WT Vps4p localized to the cytosol (S15 fraction) whereas Vps4pE233Q was more membrane associated (P15 fraction) (Fig. 3A). In our analysis we used monoclonal antibodies (mAbs) for Vph1p (a 100-kDa transmembrane protein) and 3-phosphoglycerate kinase (PGK; a cytosolic enzyme) as controls for the fractionation.
Fig. 3.
Vps46p plays a regulatory role in MVB sorting. (A) Differential sedimentation was performed to determine whether Vta1p or Vps46p affects the membrane association of Vps4p. WT, vps4E233Q, vta1Δ vps4E233Q, and vps46Δ vps4E233Q (KEBY88, JVY21, JLY06, and JLY05) yeast were analyzed for the subcellular fractionation of Vps4p. Equivalent amounts of total lysate, membranes from a 15,000 × g spin (P15), or the supernatant from a 15,000 × g spin (S15) were loaded on the gel. Blots were probed for Vps4p using an affinity-purified polyclonal Ab, Vph1p (a 100-kDa vacuolar membrane protein and subunit of the Vo subcomplex), or 3-phosphoglycerate kinase (PGK, a cytosolic enzyme). (B) Protein extracts were prepared from WT, vps4Δ, and vps46Δ (KEBY88, KEBY92, and JLY01) yeast expressing Vps32p–HA (pKEB192). GST pull-downs were performed as described in Fig. 1 A and B using GST–Vta1p or GST alone.
To further examine Vps4p and the role closely associated proteins might be playing, we analyzed Vps4p localization in the absence of Vta1p and Vps46p. Because WT Vps4p cycles rapidly off the membrane, we used the integrated vps4E233Q strain to determine whether Vta1p and/or Vps46p affected the membrane association of Vps4p. Double mutants were constructed (vta1Δ vps4E233Q and vps46Δ vps4E233Q), and the strains were analyzed by subcellular fractionation. The localization of Vps4pE233Q was not affected by the absence of Vta1p (Fig. 3A); however, Vps4pE233Q shifted from an equal distribution between the P15 and S15 fractions to a predominantly S15 localization in the absence of Vps46p (Fig. 3A). This result suggests that Vps46p is regulating the association of Vps4p with the endosomal membrane, possibly by stabilizing the association of Vps4p with the membrane and/or ESCRT-III.
Vps46p Is Required for Vta1p to Interact with ESCRT-III.
Vps4p has been shown to interact with both Vta1p and the ESCRT-III protein Vps20p by in vivo and in vitro analysis (19, 24). ESCRT-III is composed of two subcomplexes, the Vps2p–Vps24p subcomplex and the Vps20p–Vps32p subcomplex (15). Data obtained from our yeast two-hybrid analysis of the class E Vps proteins identified an interaction between Vta1p and Vps32p (ESCRT-III). To test whether Vta1p interacts with ESCRT-III, binding assays were performed from solubilized cell membrane fractions by using GST–Vta1p and GST alone. GST–Vta1p, but not GST alone, associated with Vps32p using solubilized membranes from WT cells (Fig. 3B). This interaction was analyzed in vps4Δ and vps46Δ yeast strain backgrounds to determine whether the Vta1p–Vps32p interaction requires the presence of either protein. We found that the interaction between Vta1p and Vps32p required the presence of Vps46p but not Vps4p (Fig. 3B). Because Vps46p interacts with Vta1p in vitro, this interaction is likely necessary for the association of Vta1p with ESCRT-III (Vps32p).
Vta1p Is a Potent Activator of Vps4p ATPase Activity.
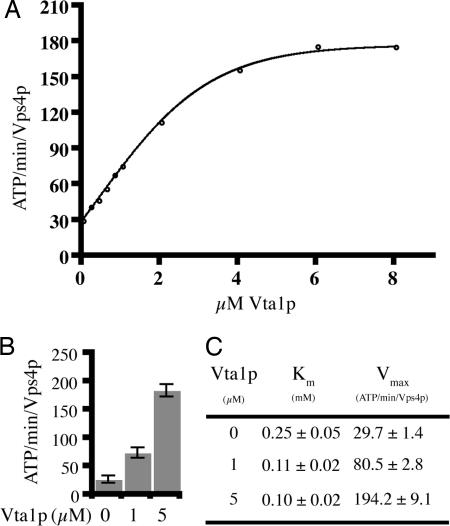
Vps4p oligomers have been shown to directly bind Vta1p (19, 24). Vta1p is a class E Vps protein, which, when mutated, causes cells to accumulate MVB cargo proteins in the endosome (24, 28). To ascertain whether the binding of Vta1p to Vps4p modulates the ATPase activity of Vps4p, Vps4p and Vta1p were recombinantly expressed and purified from E. coli. ATP hydrolysis assays were conducted by using purified Vps4p in the presence and absence of Vta1p. In the absence of Vta1p, the enzyme kinetics of Vps4p can be described by the Michaelis–Menton equation with a maximal activity of 29 ATP/min per Vps4p and a Km of 0.25 mM (Fig. 4C). As expected, the Vps4pE233Q form of the enzyme purified by the same procedure exhibited no ATPase activity, and neither did purified Vta1p alone (data not shown). To determine the effect of Vta1p upon Vps4p activity, Vta1p (after cleavage of the GST portion) was titrated into the reaction at increments from 0 to 8 μM. The presence of Vta1p in the reaction assay resulted in a significant increase of ATP hydrolysis when compared with Vps4p alone and reached a Vmax of 194 ATP/min per Vps4p at 5 μM Vta1p (Fig. 4C). At 2 mM ATP, Vta1p provided a maximal stimulation of 6- to 8-fold (31–186 ATP/min per Vps4p) (Fig. 4B). Scott et al. (19) reported that Vps4p and Vta1p may form a complex at a 1:1 ratio; however, because of the complex equilibria involved, it is not possible to address the issue of stoichiometry from our kinetic data.
Fig. 4.
Vta1p is a potent activator of Vps4p. (A) The rate of ATP hydrolysis by 1 μM Vps4p with increasing amounts of Vta1p (0–8 μM) in the presence of 2 mM ATP. (B) Graph depicting the rate of ATP hydrolysis by 1 μM Vps4p with 0, 1, or 5 μM Vta1p in the presence of 2 mM ATP. (C) The Michaelis–Menton constants, Km and Vmax, of 1 μM Vps4p calculated in the presence of 0, 1, or 5 μM Vta1p by titrating in 0.1–3 mM ATP.
We repeated this experiment using recombinantly expressed and purified Vps46p to determine whether the binding of Vps46p also modulates the activity of Vps4p. We observed that Vps46p had no effect on the ATPase activity of Vps4p, and the ATPase activity observed was consistent with the activity of Vps4p alone (data not shown). We also found that the addition of recombinantly expressed and purified Vps46p had no effect, positive or negative, on the activity of the Vps4p–Vta1p complex (data not shown).
Our results demonstrate that whereas the AAA-ATPase Vps4p possesses a significant endogenous ATPase activity, its activity is stimulated 6- to 8-fold by another class E Vps protein, Vta1p. The importance of Vta1p’s stimulation of Vps4p is reflected in the accumulation of MVB cargo proteins at the endosome in cells lacking Vta1p (24, 28). Because Vps4p exhibits ATPase activity in the absence of Vta1p, the phenotypes associated with the loss of Vta1p are not as severe as for the loss of Vps4p (24, 28).
Model for Vta1p and Vps46p Function in MVB Sorting.
Based on our results, we are able to attribute functionally important regulatory roles for Vta1p and Vps46p, two non-ESCRT class E Vps proteins. Vps46p is regulating the membrane association of Vps4p and is required for the interaction of Vta1p with ESCRT-III. Vta1p has been determined to be a potent stimulator of the ATPase activity of Vps4p. Vta1p might be effecting this stimulation either by stabilizing the oligomeric form of Vps4p or by changing the conformation of the Vps4p active site, resulting in a more efficient enzyme.
An abbreviated model of protein sorting at the late endosome is depicted in Fig. 5. Protein cargo, which in this case is monoubiquitinated, is associated with ESCRT-III and is subsequently sorted into invaginating membranes by an as yet poorly understood mechanism. ESCRT-III is released from the membrane upon hydrolysis of ATP by the AAA-ATPase, Vps4p (Fig. 5B) (15). We propose that Vps46p is functioning to recruit Vps4p dimers to the endosomal membrane where they assemble into an active oligomeric complex (Fig. 5). At this point, Vta1p binds to the Vps4p oligomer increasing the rate of ATP hydrolysis by stabilizing the active form of Vps4p (Fig. 5B). The idea that Vta1p is stabilizing the oligomeric form of Vps4p is an attractive one, because this stabilization by Vta1p would result in multiple rounds of ESCRT-III disassembly without Vps4p oligomer disassembly, thus increasing the overall rate of MVB formation.
Fig. 5.
A model for Vps4p–Vta1p regulation by Vps46p. (A) Monoubiquitination of specific protein cargo can act as a signal for sorting to the endosome and inclusion into invaginating vesicles. Doa4p, a ubiquitin hydrolase, removes the ubiquitin on the cargo protein before its inclusion. ESCRT-III is assembled on the membrane and is required for the sorting of cargo proteins into the invaginating vesicles. Vps46p is localized to the membrane and is functioning to recruit Vps4p, which in turn is assembled into a Vps4p–Vta1p complex. (B) The Vps4p–Vta1p complex directly interacts with Vps46p, and this interaction is required for association with ESCRT-III. Vta1p stimulates the ATPase activity of Vps4p, and this activity catalyzes the release of ESCRT-III proteins from the membrane to be recycled for future rounds of sorting.
Materials and Methods
Supporting Information.
Further details of the plasmids used in this study can be found in Table 2, which is published as supporting information on the PNAS web site.
Yeast Strains.
Yeast strains used in this study are shown in Table 1. Strains were constructed by standard genetic techniques and grown in rich medium [yeast extract/peptone/dextrose (YEPD) (12)] or synthetic dextrose minimal medium with appropriate amino acid supplements [SD (12)]. Amino acid dropout mixtures were from Formedium Ltd. (Norwich, U.K.). vps46Δ::Kanr (JLY01), and vta1Δ::Kanr (JLY03) yeast strains were generated by transformation of PCR-amplified DNA into KEBY88. The knockout plasmid constructs described above were used as PCR templates, with primers 500 bp up and downstream of the ORF of each gene. Transformants were selected on YEPD with 0.2 mg/ml geneticin sulfate (G418) and screened for CPY secretion using a colony overlay assay (29) and then for complementation of the CPY secretion phenotype after transformation of a plasmid containing the WT gene. The vps46Δ::Kanr (JLY01) and vta1Δ::Kanr (JLY03) strains were checked by PCR from genomic DNA. The CPY mAb used for colony overlay assays was obtained from Molecular Probes.
Table 1.
Yeast strains used in this work
| Strain | Genotype | Source |
|---|---|---|
| KEBY88 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 | Ref. 12 |
| KEBY92 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 vps4Δ::Kanr | Ref. 12 |
| KEBY112 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 vps32Δ::Kanr | This work |
| JLY01 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 pep4-3 vps46Δ::Kanr | This work |
| JLY03 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 pep4-3 vta1Δ::Kanr | This work |
| JVY21 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 vps4E233Q | This work |
| JLY05 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 vps4E233Q vps46Δ::Kanr | This work |
| JLY06 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pep4-3 vps4E233Q vta1Δ::Kanr | This work |
An integrated vps4E233Q (JVY21) strain was generated after transformation of KEBY88 with BglII digested pJL18. After selection of transformants on SD-ura, colonies were plated onto 5-fluoroorotic acid-containing minimal medium to select for Ura− loop outs. vps4E233Q dominant-negative mutant strains were verified by assaying for CPY secretion. PCR-amplified DNA was transformed into vps4E233Q (JVY21) to generate vps4E233Q vps46Δ::Kanr (JLY05) and vps4E233Q vta1Δ::Kanr (JLY06). Transformants were selected on YEPD with 0.2 mg/ml geneticin sulfate (G418) and the strains were checked by PCR from genomic DNA because of the dominant-negative CPY secretion phenotype of the vps4E233Q strain.
Plasmid Construction.
The plasmids used in this study are listed in Table 2. Enzymes used in DNA manipulations were purchased from New England Biolabs, and Pfu turbo DNA polymerase was obtained from Stratagene Europe. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) or Qiagen (Valencia, CA). VPS4, VPS46, and VTA1 were amplified by PCR from yeast genomic DNA including 500 bp up and downstream of the ORF, with appropriate restriction sites in the oligonucleotides. The PCR products were subcloned into pRS316 (30) to generate pKEB83, pKEB125, and pJL19. The VPS plasmid-borne gene fully complements the CPY secretion phenotype of the appropriate deletion strain. An in-frame BglII site was added just before the stop codon of VPS46 in pKEB125 by QuikChange (Stratagene) mutagenesis and a 123-bp DNA fragment containing three copies of the hemagglutinin (HA) epitope tag sequence with BglII ends was subcloned into the BglII site, creating pJL11. QuikChange mutagenesis was used to change the amino acid codon at position 233 in pKEB83 from glutamic acid (GAA) to glutamine (CAA) to create vps4E233Q (pJL14). BamHI and XhoI digested pJL14 was used to subclone vps4E233Q from pRS316 into pRS306 to create pJL18.
For protein expression in E. coli, the ORF sequences of VTA1, VPS46, and vps4E233Q were amplified by PCR from pJL19, pKEB125, and pJL14 with appropriate restriction enzyme sites in the primers. The PCR products were subcloned into pGEX-5X-3 and pGEX-4T-1 (GE Healthcare) to generate in-frame fusions to GST creating pJL12, pJL21, and pJL15. The ORF sequences of VPS46 and VTA1 were amplified by genomic PCR from pJL19 and pKEB125, respectively, with a 5′ NcoI site and a 3′ XhoI site in the primers. The PCR products were subcloned into pET28a (EMD Biosciences, San Diego) to generate in-frame fusions to the 6XHIS tag sequence creating pJL13 and pJL17, respectively.
Purification of GST-Fusion and HIS-Fusion Proteins from Bacteria.
pKEB244, pJL12,, pJL13, pJL15, pJL17, pJL21, and GST–Vma13p were transformed into E. coli strain Rosettatm (DE3) pLysS (Novagen) and expressed in the following manner. A single colony was grown in LB medium containing 25 μg/ml chloramphenicol and either 100 μg/ml ampicillin for cells containing plasmids pKEB244, pJL12, pJL15, pJL21, or GST–Vma13p or 50 μg/ml kanamycin for plasmids pJL13 or pJL17 to an A600 = 0.5 at 37°C. Once A600 = 0.5, protein expression was induced with the addition of 2 mM IPTG. After cultures were incubated at 25°C for 3 h, the cells were harvested. For cells expressing GST fusion proteins, the cell pellets were resuspended in PBS lysis buffer (140 mM NaCl/2.7 mM KCL/10 mM Na2HPO4/1.8 mM KH2PO4, pH 7.3) and lysed using a French press. The lysates were clarified by centrifugation at 100,000 × g for 45 min, and the soluble proteins were purified by glutathione Sepharose chromatography (GE Healthcare). Fractions containing the eluted proteins were analyzed by SDS/PAGE and pooled. Proteins destined for GST pulldown experiments were dialyzed into Vps protein buffer (250 mM NaCl/1 mM DTT/10 mM Hepes, pH 7.6) and concentrated to ≈1–1.5 mg/ml. Proteins destined for ATPase assays were dialyzed into thrombin cleavage buffer (10 mM CaCl2/50 mM Tris·HCl, pH 8.0) and concentrated to 1.5 mg/ml. The GST moieties were cleaved from the purified proteins by using the thrombin CleanCleave kit (Sigma). The cleaved proteins were dialyzed into PBS lysis buffer, and GST was removed by passing the protein solution over a glutathione column. The flowthrough was dialyzed into Vps protein buffer and concentrated to 1.5 mg/ml.
For cells expressing proteins fused to a histidine tag, the cell pellets were resuspended in extraction buffer (300 mM NaCl/5 mM imidazole/50 mM Na2HPO4, pH 7.0) and lysed by using a French press. Lysates were clarified by centrifugation at 100,000 × g and purified by Talon metal affinity chromatography (Clontech). Purified proteins were dialyzed into Vps protein buffer and concentrated to 1–1.5 mg/ml.
Preparation of Antiserum to Vps4p.
Polyclonal Abs were raised against recombinantly expressed and purified GST–Vps4p (pKEB244). Rabbits were injected with purified protein as described in ref. 31. Purified GST–Vps4p (pKEB244) protein also was used for affinity purification of the anti-Vps4p Abs.
GST Pull-Down Experiments.
GST pull-down experiments were performed as described in ref. 12, with the following exceptions. Spheroplasts (from 150 ODs) were resuspended in 4 ml of ice-cold PBS with protease inhibitor mixture (1 mM PMSF/2 μg/ml pepstatin/2 μg/ml leupeptin) and incubated for 30 min on ice to lyse the cells. Lysates were centrifuged to generate P15 and S15 fractions. The lysate pellet fractions (P15) were solublized in 800 μl of PBS with 1% Triton X-100 and incubated on ice for 15 min. An additional 3.2 ml of PBS was added, and insoluble material was spun out at 15000 × g for 5 min. One milliliter of solubilized membranes (P15 fraction) was used per pull-down, with ≈50 μg of GST-fusion protein and 70 μl of a 50% glutathione Sepharose slurry. After 3 h at 4°C with constant mixing, the glutathione Sepharose and bound proteins were washed three times with PBS plus 0.2% Triton X-100 before eluting in sample buffer (188 mM Tris, pH 6.8/6% SDS/30% glycerol/0.03% bromophenol blue) with 5% 2-mercaptoethanol (BME). The samples were boiled for 5 min at 100°C and loaded onto a SDS/PAGE gel.
In Vitro Binding Experiments.
Purified Vps46–6XHIS in Vps protein buffer (250 mM NaCl/1 mM DTT/10 mM Hepes, pH 7.6) with protease inhibitors was used for each binding assay at a final concentration of 2 μM. GST, GST–Vma13p GST–Vta1p, or GST–Vps4p was added to each assay at a final concentration of 1.5 μM with 100 μl of a 50% glutathione Sepharose slurry. The total volume was brought to 1 ml with Vps protein buffer (made fresh) containing protease inhibitors and 0.1% Triton X-100. The samples were incubated at 4°C for 12 h and processed as for GST pull-down experiments. Vps46p–6XHIS was detected by using 6XHIS mAb (Boehringer Mannheim).
Differential Sedimentation Experiments.
P15 and S15 fractions were prepared as for GST pull-down experiments (12), with a fraction of the total lysate reserved. Equal volumes of each fraction were loaded on a gel, and immunoblotting was carried out as described above. mAbs 22C5D8 against 3-phosphoglycerate kinase and 10D7A against Vph1p were obtained from Molecular Probes.
ATPase Assays.
Rates of ATP hydrolysis at 30°C were determined by the coupled enzyme assay of Lotscher et al. (32). Briefly, 1 μM Vps4p and 0–8 μM Vta1p or 0–8 μM Vps46p were incubated in 200 μl of reaction buffer (25 mM Mes/Mops, pH 7.6/30 units/ml lactate dehydrogenase/30 units/ml pyruvate kinase/0.6 mM NaDH/2 mM phosphoenolpyruvate/0–3 mM ATP). The reactions were initiated with the addition of MgCl2 to a final concentration of 5 mM. The A340 was monitored in a Versamax tunable plate reader (Molecular Devices). Kinetic data were analyzed with the software application softmaxpro (Molecular Devices) and Michaelis–Menton variables were fitted with kaleidagraph (Synergy Software, Reading, PA).
Supplementary Material
Acknowledgments
We thank Laurie Graham and Katherine Bowers for critical reading of the manuscript and helpful discussions and Leslie Schwarcz for help purifying proteins. This work was supported by National Institutes of Health (NIH) Training Grants HD07348 (to J.M.L.) and GM07759 (to A.R.F.) and NIH Grant GM32448 (to T.H.S.).
Abbreviations
- AAA
ATPase associated with a variety of cellular activities
- HA
hemagglutinin
- MVB
multivesicular body
- ESCRT
endosomal sorting complexes required for transport
- Vps
vacuolar protein sorting.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lemmon S. K., Traub L. M. Curr. Opin. Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- 2.Bowers K., Stevens T. H. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Prescianotto-Baschong C., Riezman H. Traffic. 2002;3:37–49. doi: 10.1034/j.1600-0854.2002.30106.x. [DOI] [PubMed] [Google Scholar]
- 4.Piper R. C., Luzio J. P. Traffic. 2001;2:612–621. doi: 10.1034/j.1600-0854.2001.20904.x. [DOI] [PubMed] [Google Scholar]
- 5.Raiborg C., Rusten T. E., Stenmark H. Curr. Opin. Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 6.Katzmann D. J., Odorizzi G., Emr S. D. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 7.Luban J. Nat. Med. 2001;7:1278–1280. doi: 10.1038/nm1201-1278. [DOI] [PubMed] [Google Scholar]
- 8.Pelchen-Matthews A., Kramer B., Marsh M. J. Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Schwedler U. K., Stuchell M., Muller B., Ward D. M., Chung H. Y., Morita E., Wang H. E., Davis T., He G. P., Cimbora D. M., et al. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 10.Pornillos O., Garrus J. E., Sundquist W. I. Trends Cell Biol. 2002;12:569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 11.Babst M. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 12.Bowers K., Lottridge J., Helliwell S. B., Goldthwaite L. M., Luzio J. P., Stevens T. H. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 13.Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Dev. Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 14.Katzmann D. J., Babst M., Emr S. D. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 15.Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 16.Babst M., Sato T. K., Banta L. M., Emr S. D. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finken-Eigen M., Rohricht R. A., Kohrer K. Curr. Genet. 1997;31:469–480. doi: 10.1007/s002940050232. [DOI] [PubMed] [Google Scholar]
- 18.Babst M., Wendland B., Estepa E. J., Emr S. D. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott A., Chung H. Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., Sundquist W. I. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothman J. H., Howald I., Stevens T. H. EMBO J. 1989;8:2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankaitis V. A., Johnson L. M., Emr S. D. Proc. Natl. Acad. Sci. USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piper R. C., Cooper A. A., Yang H., Stevens T. H. J. Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeo S. C., Xu L., Ren J., Boulton V. J., Wagle M. D., Liu C., Ren G., Wong P., Zahn R., Sasajala P., et al. J. Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- 25.Fujita H., Umezuki Y., Imamura K., Ishikawa D., Uchimura S., Nara A., Yoshimori T., Hayashizaki Y., Kawai J., Ishidoh K., et al. J. Cell Sci. 2004;117:2997–3009. doi: 10.1242/jcs.01170. [DOI] [PubMed] [Google Scholar]
- 26.Reid E., Connell J., Edwards T. L., Duley S., Brown S. E., Sanderson C. M. Hum. Mol. Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 27.Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. Proc. Natl. Acad. Sci. USA. 2005;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiflett S. L., Ward D. M., Huynh D., Vaughn M. B., Simmons J. C., Kaplan J. J. Biol. Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- 29.Roberts C. J., Raymond C. K., Yamashiro C. T., Stevens T. H. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 30.Sikorski R. S., Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaitukaitis J. L. Methods Enzymol. 1981;73:46–52. doi: 10.1016/0076-6879(81)73055-6. [DOI] [PubMed] [Google Scholar]
- 32.Lotscher H. R., deJong C., Capaldi R. A. Biochemistry. 1984;23:4128–4134. doi: 10.1021/bi00313a018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.