Abstract
Understanding the evolution of development in large part relies on the study of phylogenetically old organisms. Cnidarians, such as Hydra, have become attractive model organisms for these studies. However, despite long-term efforts, stably transgenic animals could not be generated, severely limiting the functional analysis of genes. Here we report the efficient generation of transgenic Hydra lines by embryo microinjection. One of these transgenic lines expressing EGFP revealed remarkably high motility of individual endodermal epithelial cells during morphogenesis. We expect that transgenic Hydra will become important tools to dissect the molecular mechanisms of development at the base of the Metazoan tree.
Keywords: EGFP
Cnidarians arose ≈600 million years ago and were first in metazoan evolution to develop a complex body structure composed of specialized tissues. Today, several cnidarian species, including the freshwater polyp Hydra, are important model organisms in both environmental and conservation science (1) as well as in evolutionary developmental biology (2–8). The Hydra body plan consists of two cell layers, with only a limited number of cell types belonging to three distinct cell lineages, the ectodermal and endodermal epithelial cells, and the interstitial cells, which give rise to all nerve cells, gland cells, and nematocytes (2). Cell proliferation takes place continuously in both cell layers along the body column; together with the continuous presence of developmental signals, this gives even adult polyps a remarkable ability to fully regenerate lost body structures (2). Morphogenesis in Hydra does not depend on cell division (9) and is largely driven by epithelial cells (10, 11).
Results and Discussion
Efficient Generation of Transgenic Hydra by Embryo Microinjection.
To study cell behavior during morphogenetic events in vivo, we have developed a system that allows generation of stable transgenic Hydra lines. We have used an EGFP expression construct [homologous transformation vector GFP (hoT G)] based on the Hydra vulgaris β-actin gene (Fig. 1A) for microinjection into H. vulgaris (AEP) embryos at the two- to eight-cell stage. Of 65 embryos injected with hoT G, 18 (27.6%) expressed EGFP as early as 2 days after microinjection, when midblastula transition occurred. Within EGFP-positive cuticle stage embryos, expression was intense and appeared nonuniform within patches of blastomeres. Surprisingly, although the construct did not harbor any sites favoring genomic integration, upon hatching, 6 of 65 (9.2%) polyps stably expressed EGFP in either the endodermal or ectodermal epithelial stem cells. Moreover, 2 of 65 (3%) polyps expressed EGFP permanently in the interstitial cell lineage.
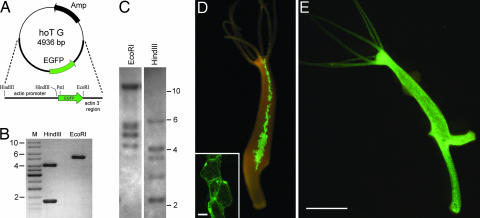
Fig. 1.
Generation and characterization of transgenic H. vulgaris (AEP) line endo-2. (A) The EGFP expression construct hoT G used to generate transgenic H. vulgaris. (B) hoT G construct digested by EcoRI and HindIII showing one band of ≈5 kb and two bands of 4 and 1.5 kb; M, size marker. (C) Southern blot hybridization. DNA from H. vulgaris (AEP) endo-2 line was hybridized with a probe specific for EGFP. Lane 1, EcoRI-digested DNA from transgenic polyps; lane 2, HindIII-digested DNA from transgenic polyps. The differences in fragment size between the restricted genomic DNA and the hoT G vector (shown in B) indicate stable integration of the vector in the Hydra genome. (D) Transgenic endo-2 polyp with a mosaic expression of EGFP in some of its endodermal epithelial cells. (Inset) A patch of EGFP+ endodermal epithelial cells visualized by confocal microscopy. (Scale bar, 20 μm.) (E) Transgenic endo-2 polyp expressing EGFP in all of its endodermal epithelial cells. (Scale bar, 1 mm.)
Transgenic H. vulgaris (AEP) Line endo-2 Expresses the EGFP Marker in All Endodermal Epithelial Stem Cells.
Because in Hydra epithelial cells control morphogenesis (8, 9), here we focus our analysis on line endo-2, which initially showed EGFP expression in patches of endodermal epithelial cells (Fig. 1D; see also Fig. 5 A and B, which is published as supporting information on the PNAS web site). By clonal propagation, we were able to establish lines homogeneously expressing EGFP in all of their endodermal epithelial cells (Fig. 1E), which were then used as founders for a mass culture of endo-2. Transgenic polyps exhibited normal morphology and behavior and proliferated asexually by budding, as well as undergoing frequent sexual reproduction. When we transplanted single transgenic endodermal epithelial cells into wild-type host animals, we observed (data not shown) proliferation of transgenic cells and were ultimately able to generate polyps in which the transgenic cells made up the entire endothelium, demonstrating that all endodermal epithelial cells in Hydra have at least restricted stem cell properties.
Southern blot analyses using an EGFP probe revealed integration of the hoT G construct into the H. vulgaris genome at multiple loci (Fig. 1C). Restriction digests with either EcoRI or HindIII suggested at least five unlinked integration sites. Because a favorable chromosomal site may have been an important factor for integration of the construct, current efforts are directed toward the characterization of flanking sequences. As shown above, genomic integration of the construct into the Hydra genome occurs at a high frequency. One explanation may be that the Hydra genome is much more promiscuous in accepting foreign DNA than previously thought. This view is supported by two recent reports describing the integration of a plant peroxidase (12) and a protist-derived gene (13), most likely via horizontal gene transfer, into the Hydra genome. However, we note that a similar frequency of stable genomic integration was observed previously when foreign DNA was microinjected into fertilized mouse eggs (14, 15).
In Vivo Tracking of Individual Epithelial Cells Within the Transparent Polyp.
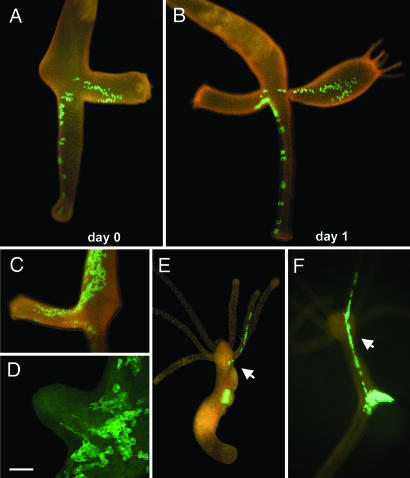
The tissue of adult Hydra polyps is completely transparent, allowing the visualization of individual cells by means of GFP fluorescence and facilitating in vivo tracking of cells within the intact organism. Here we investigated the motility of endodermal epithelial cells and their role in axial tissue expansion. In the absence of zones of localized cell proliferation (16, 17), tissue necessary for bud formation must be recruited from the mother polyp. In vivo tracking of EGFP-labeled cells over time revealed that motility of individual endodermal cells plays a major role during budding. Fig. 2A shows an asexually proliferating polyp containing a small number of EGFP labeled cells, which were located in the basal part of the body axis and in the older bud. One day later, the second bud had grown considerably, and individual EGFP-positive cells were recruited from the parental tissue (Fig. 2B), demonstrating that endodermal cell motility rather than passive tissue displacement is involved in bud formation. Support for this conclusion comes from the analysis of cell behavior both in vivo (Fig. 2C) and by confocal imaging (Fig. 2D), which revealed that recruitment of endodermal epithelial cells into bud tissue is accompanied by dramatic changes in cell shape. At the base of the bud, endodermal epithelial cells elongated and sent out filopodia-like protrusions (Fig. 2D), consistent with active migration of these cells. To independently confirm the motility of individual cells, we used homotopic transplantation experiments. Although in most grafts, transgenic cells remained at the transplantation site, in a significant number of cases (3 of 30), we could observe individual cells or groups of cells migrating to distant sites in the polyp within the first 8 days after transplantation (Fig. 2E). We also detected oriented cell division during the migration process because, 8 days after transplantation, descendants of EGFP-labeled cells formed a string of cells aligned along the oral–aboral axis of the polyp (Fig. 2F). Thus, migration of individual cells, together with oriented cell division, appears to contribute to morphogenetic tissue extension in Hydra. These findings support earlier observations of oriented cell division during Hydra morphogenesis (18). They also fit well with recent in vitro studies, which suggested that the motility of endodermal epithelial cells plays a critical role in setting up the two epithelial layers (19).
Fig. 2.
Individual motility of EGFP-expressing endodermal epithelial cells. (A–D) EGFP+ cells reveal high motility of endodermal epithelial cells during budding. (A–C) Buds recruit EGFP+ endodermal epithelial cells even from distant parts of the parent polyp. (D) Confocal image of evaginating bud showing change in epithelial cell shape during budding. (Scale bar, 80 μm.) (E and F) Homotopic transplantation experiments confirm the motility of individual epithelial cells. Small tissue pieces containing EGFP-expressing cells were transplanted homotopically into the gastric region of a nontransgenic recipient. (E) EGFP-positive epithelial cells migrated away from the transplant 5 days after transplantation; arrow points to emigrated endodermal epithelial cells in the tentacle. (F) String of EGFP-positive cells 8 days after transplantation produced as a result of cell migration and oriented cell divisions; arrow points to emigrated endodermal epithelial cells.
Regeneration in Transgenic Hydra Polyps.
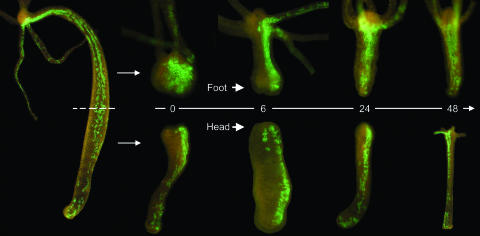
Another process in which cell motility plays an important role is regeneration. The remarkable capacity of Hydra to regenerate lost body structures is considered a morphallactic process (20–22). Consequently, it was shown to involve not localized cell division but rather rapid wound healing initiated by the endoderm (23). Because early studies (reviewed in ref. 19) suggested that endodermal epithelial cells “pile up into multiple layers to seal the injury,” we used transgenic polyps to track individual endodermal epithelial cells in regenerating tissue. As illustrated in Fig. 3, neither localized cell proliferation nor “piling up” of endodermal epithelial cells at the injury site was detected during head or foot regeneration. Thus, our in vivo observations clearly demonstrate that regeneration in Hydra occurs almost exclusively by morphallaxis.
Fig. 3.
Head and foot regeneration occurs via morphallaxis in the absence of local cell proliferation with no evidence for a piling up of endodermal epithelial cells. Numbers indicate hours after amputation.
Response of Transgenic Endodermal Epithelial Cells to Morphogenetic Signals.
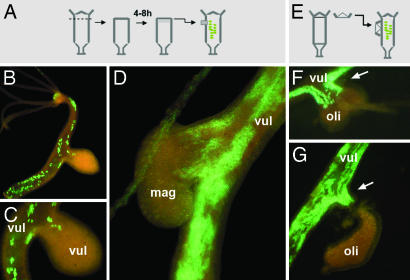
During regeneration, both activating and inhibiting signals are produced to provide Hydra cells with positional information (24, 25). To study in more detail the response of endodermal epithelial cells to morphogenetic signals, regenerating tissue (8 h) from nontransgenic polyps was grafted into the gastric region of EGFP-labeled polyps, as illustrated in Fig. 4A. In most cases (28 of 30 samples), 24 h after grafting, the transplanted tissue had recruited EGFP+ endodermal epithelial cells to form a secondary body axis (Fig. 4 B and C). The observation again demonstrates the high motility of individual epithelial cells and their rapid response to morphogenetic signals. We then extended our analysis to interspecies grafts to investigate whether the signaling circuitry has been conserved during evolution. We found that regenerating tissue from the closely related species Hydra magnipapillata was capable (19 of 22 samples) of recruiting H. vulgaris endodermal epithelial cells to form a secondary body axis very similar to intraspecies control grafts (Fig. 4D). To extend our analysis to more distantly related Hydra species such as Hydra oligactis, we used hypostome contact grafts (Fig. 4E), because head-inducing signals have been shown to be highest in the apical part of the head, which is formed by the hypostome (26). As shown in Fig. 4 F and G, even though H. vulgaris and H. oligactis cannot be stably grafted (27), contact of a H. oligactis head with H. vulgaris gastric tissue efficiently induces H. vulgaris endodermal epithelial cells to form a secondary axis. These results clearly show that the signaling system for head induction has been highly conserved during the several hundred million years of independent evolution in those two species. Furthermore, our data provide direct evidence for the diffusible nature of the head inductive signals.
Fig. 4.
Motility of endodermal cells toward developmental signals from regenerating tissue. (A and E) Experimental procedure. (B and C) Lateral transplantation of regenerating H. vulgaris tissue recruits EGFP+ cells to form a secondary axis. (D) Regenerating tissue from H. magnipapillata induces secondary axis formation and recruits H. vulgaris EGFP+ cells. (F and G) In hypostome-contact grafts, heads from H. oligactis induce H. vulgaris to form a secondary axis by recruiting EGFP+ H. vulgaris endodermal epithelial cells. Arrow points to H. vulgaris EGFP+ endodermal cells, which begin to form a secondary axis. mag, H. magnipapillata; oli, H. oligactis; vul, H. vulgaris.
Conclusion
We have developed a simple and efficient method to generate stable transgenic Hydra lines. Transgenic Hydra will provide a foundation for functional genomics in Cnidaria, and our method may also be extended to other taxa, such as Nematostella and Acropora, which are of considerable interest for evolutionary studies (5, 8). Because Cnidaria are at the base of the Metazoan tree, we hope this research resource will expand our knowledge of the genetic basis for developmental evolution and help to uncover principles that are general to all metazoans.
Materials and Methods
Animals and Culture Conditions.
Experiments were carried out with animals of the AEP strain belonging to the H. vulgaris group. This strain is derived from male and female strains described previously (28).The animals were mass-cultured according to standard procedures at 18°C. To induce gametogenesis, clonally grown polyps were fed daily for 3 weeks, then starved for 5 days, and then fed twice per week.
Preparation of the hoT G.
The hoT G construct was generated by inserting GFP cDNA into plasmid pUC19, which contains 1,386 bp of the H. vulgaris actin 5′ flanking region, transcription start site, native initiator codon, and the first 10 amino acids of H. vulgaris actin (see Fig. 1A; GenBank accession no. DQ369740). In addition, the EGFP reporter gene is flanked by 3′ genomic region of the H. vulgaris actin gene, including the termination/polyadenylation signal. Subsequently, GFP was replaced by EGFP by site-directed mutagenesis, and two additional restriction sites were introduced (29). Plasmid DNA was prepared by using the Qiagen (Valencia, CA) Midi Prep Kit and resuspended in water.
Generation of Transgenic H. vulgaris.
Embryos were removed from H. vulgaris (AEP) females and microinjected with the hoT G expression construct at the two- to eight-cell stage. Microinjection was done by using an inverted microscope (Zeiss Axiovert 100) and two micromanipulators (Leitz, Eppendorf). Embryos were held by a micropipette by using a CellTram Air pump (Eppendorf). The construct was injected by using a CellTram vario pump (Eppendorf). Glass needles for microinjection were produced by using Vertical Pipette Puller 700 C (Kopf Instruments, Tujunga, CA). The construct (0.1 μl; 0.6 μg/μl) was injected into the embryos. Each embryo was injected only once. During the injection procedure, embryos were kept in Hydra medium at room temperature. Microinjected embryos were transferred to 12-well microtiter plates and incubated at 18°C in Hydra medium. Embryos were assessed for reporter gene expression by using a Leica Fluo stereomicroscope. In a pilot experiment, the survival and hatching rates of the injected embryos were assessed. Of 93 injected embryos, 87 (93.5%) survived, and 48 (55.2%) hatched within 49 days. Unexpectedly, some injected embryos hatched as early as 14 days after fertilization. Embryos began to express the reporter gene 2–3 days after injection. Although in some embryos EGFP expression was transient and lasted for 8–12 days, in other embryos, the EGFP construct was stably integrated in the genome. After hatching, EGFP-expressing cells in those polyps became visible as small patches in either the ectoderm or the endoderm. Mass cultures of polyps homogeneously expressing EGFP in all of the endodermal epithelial stem cells were generated by clonal propagation.
Tissue Manipulation/Transplantation.
Homotopic and heterotopic transplantations were done following standard procedures. Hypostome-contact grafts were done as described (26, 30).
Southern Blot Analysis.
Southern blot analysis was performed at high stringency following standard procedures. Twenty micrograms of genomic DNA isolated from transgenic animals was digested with EcoRI and with HindIII, which cut the hoT G construct once or twice, respectively (see Fig. 1B), separated on a 0.7% agarose gel, and hybridized with a radiolabeled EGFP-specific probe. Approximately 1 μg of hoT G was digested with the same restriction enzymes as positive control (see Fig. 1B).
In Vivo Imaging.
Screening for EGFP expression was done by using a stereomicroscope (Leica Fluo). Laser-scanning confocal data were acquired by using Leica TCS SP1. Pictures were taken by using Leica DC300F and AxioCam (Zeiss) digital cameras.
Supplementary Material
Acknowledgments
We thank Prof. Christian Jung (Kiel University, Kiel, Germany) and Dr. Olaf Faustmann (Eppendorf) for support of the microinjection work station, Antje Thomas for excellent assistance with microscopy, and Dr. Angelika Böttger for replacing GFP with EGFP in the hoT G construct. Financial support for this research was provided by the German Research Foundation [Deutsche Forschungsgemeinschaft Grants B0848/13 and SFB617 (to T.C.G.B.)].
Abbreviation
- hoT G
homologous transformation vector GFP.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Orr J. C., Fabry V. J., Aumont O., Bopp L., Doney S. C., Feely R. A., Gnanadesikan A., Gruber N., Ishida A., Joos F., et al. Nature. 2005;437:681–686. doi: 10.1038/nature04095. [DOI] [PubMed] [Google Scholar]
- 2.Gierer A. Sci. Am. 1974;231:44–54. doi: 10.1038/scientificamerican1274-44. [DOI] [PubMed] [Google Scholar]
- 3.Bosch T. C. G., Fujisawa T. BioEssays. 2001;23:420–427. doi: 10.1002/bies.1060. [DOI] [PubMed] [Google Scholar]
- 4.Galliot B., Schmid V. Int. J. Dev. Biol. 2002;46:39–48. [PubMed] [Google Scholar]
- 5.Miller D. J., Ball E. E., Technau U. Trends Genet. 2005;21:536–539. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Hobmayer B., Rentzsch F., Kuhn K., Happel C. M., von Laue C. C., Snyder P., Rothbacher U., Holstein T. W. Nature. 2000;407:186–189. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 7.Meinhardt H. Curr. Opin. Genet. Dev. 2004;14:446–454. doi: 10.1016/j.gde.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Kusserow A., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H. A., Technau U., von Haeseler A., Hobmayer B., Martindale M. Q., et al. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 9.Campbell R. D. Dev. Biol. 1967;15:487–502. doi: 10.1016/0012-1606(67)90039-5. [DOI] [PubMed] [Google Scholar]
- 10.Marcum B. A., Campbell R. D. J. Cell Sci. 1978;29:17–33. doi: 10.1242/jcs.29.1.17. [DOI] [PubMed] [Google Scholar]
- 11.Sugiyama T., Fujisawa T. J. Cell Sci. 1978;29:35–52. doi: 10.1242/jcs.29.1.35. [DOI] [PubMed] [Google Scholar]
- 12.Habetha M., Bosch T. C. G. J. Exp. Biol. 2005;208:2157–2164. doi: 10.1242/jeb.01571. [DOI] [PubMed] [Google Scholar]
- 13.Steele R. E., Hampson S. E., Stover N. A., Kibler D. F., Bode H. R. Curr. Biol. 2004;14:R298–R299. doi: 10.1016/j.cub.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 14.Gordon J. W., Scangos G. A., Plotkin D. J., Barbosa J. A., Ruddle F. H. Proc. Natl. Acad. Sci. USA. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinster R. L., Chen H. Y., Trumbauer M. E., Yagle M. K., Palmiter R. D. Proc. Natl. Acad. Sci. USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David C. N., Campbell R. D. J. Cell Sci. 1972;11:557–568. doi: 10.1242/jcs.11.2.557. [DOI] [PubMed] [Google Scholar]
- 17.Holstein T. W., Hobmayer E., David C. N. Dev. Biol. 1991;148:602–611. doi: 10.1016/0012-1606(91)90277-a. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu H., Bode P. M., Bode H. R. Dev. Dyn. 1995;204:349–357. doi: 10.1002/aja.1002040402. [DOI] [PubMed] [Google Scholar]
- 19.Takaku Y., Hariyama T., Fujisawa T. Mech. Dev. 2005;122:109–122. doi: 10.1016/j.mod.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Cummings S. G., Bode H. R. Roux’s Arch. Dev. Biol. 1984;194:79–86. doi: 10.1007/BF00848347. [DOI] [PubMed] [Google Scholar]
- 21.Holstein T. W., Hobmayer E., Technau U. Dev. Dyn. 2003;226:257–267. doi: 10.1002/dvdy.10227. [DOI] [PubMed] [Google Scholar]
- 22.Bode H. R. Dev. Dyn. 2003;226:225–236. doi: 10.1002/dvdy.10225. [DOI] [PubMed] [Google Scholar]
- 23.Bibb C., Campbell R. D. Tissue Cell. 1973;5:23–35. doi: 10.1016/s0040-8166(73)80003-5. [DOI] [PubMed] [Google Scholar]
- 24.MacWilliams H. K. Dev. Biol. 1983;96:217–238. doi: 10.1016/0012-1606(83)90324-x. [DOI] [PubMed] [Google Scholar]
- 25.MacWilliams H. K. Dev. Biol. 1983;96:239–257. doi: 10.1016/0012-1606(83)90325-1. [DOI] [PubMed] [Google Scholar]
- 26.Broun M., Bode H. R. Development (Cambridge, U.K.) 2002;129:875–884. doi: 10.1242/dev.129.4.875. [DOI] [PubMed] [Google Scholar]
- 27.Kuznetsov S. G., Anton-Erxleben F., Bosch T. C. G. J. Exp. Biol. 2002;205:3809–3817. doi: 10.1242/jeb.205.24.3809. [DOI] [PubMed] [Google Scholar]
- 28.Martin V. J., Littlefield C. L., Archer W. E., Bode H. R. Biol. Bull. 1997;192:345–363. doi: 10.2307/1542745. [DOI] [PubMed] [Google Scholar]
- 29.Bottger A., Alexandrova O., Cikala M., Schade M, Herold M., David C. N. Dev. Genes Evol. 2002;212:302–305. doi: 10.1007/s00427-002-0245-0. [DOI] [PubMed] [Google Scholar]
- 30.Mutz E. Dev. Genes Evol. 1930;121:210–271. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.