Abstract
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. A subset of degradation signals recognized by the N-end rule pathway comprises the signals, called N-degrons, whose determinants include destabilizing N-terminal residues. Our previous work identified a family of at least four mammalian E3 ubiquitin ligases, including UBR1 and UBR2, that share the UBR box and recognize N-degrons. These E3 enzymes mediate the multifunctional N-end rule pathway, but their individual roles are just beginning to emerge. Mutations of UBR1 in humans are the cause of Johanson–Blizzard syndrome. UBR1 and UBR2 are 46% identical and appear to be indistinguishable in their recognition of N-degrons. UBR1−/− mice are viable but have defects that include pancreatic insufficiency, similarly to UBR1−/− human patients with Johanson–Blizzard syndrome. UBR2−/− mice are inviable in some strain backgrounds and are defective in male meiosis. To examine functional relationships between UBR1 and UBR2, we constructed mouse strains lacking both of these E3s. We report here that UBR1−/−UBR2−/− embryos die at midgestation, with defects in neurogenesis and cardiovascular development. These defects included reduced proliferation as well as precocious migration and differentiation of neural progenitor cells. The expression of regulators such as D-type cyclins and Notch1 was also altered in UBR1−/−UBR2−/− embryos. We conclude that the functions of UBR1 and UBR2 are significantly divergent, in part because of differences in their expression patterns and possibly also because of differences in their recognition of protein substrates that contain degradation signals other than N-degrons.
Keywords: ubiquitylation, proteolysis, N-recognin, UBR box, arginylation
A protein substrate of the ubiquitin (Ub)-proteasome system, which controls the levels of many intracellular proteins, is conjugated to Ub through the action of E1, E2, and E3 enzymes (1–5). The substrate’s degradation signal (degron) is recognized by E3. A ubiquitylated protein bears a covalently linked polyUb chain and is degraded by the 26S proteasome (3, 5). The term “Ub ligase” denotes either an E2–E3 holoenzyme or its E3 component. An essential determinant of one class of degrons, called N-degrons, is a substrate’s destabilizing N-terminal residue. The set of destabilizing residues in a given cell type yields a rule, called the N-end rule, that relates the in vivo half-life of a protein to the identity of its N-terminal residue (1, 6). In eukaryotes, the N-degron consists of three determinants: a destabilizing N-terminal residue of a protein substrate, its internal Lys residue(s) (the site of formation of a polyUb chain), and a conformationally flexible region (or regions) in the vicinity of these determinants that is required for the substrate’s ubiquitylation and/or degradation (6–9).
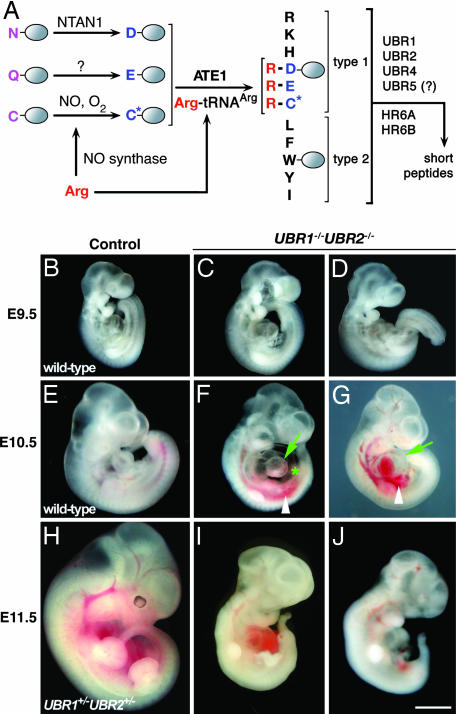
The N-end rule has a hierarchic structure (Fig. 1A). In eukaryotes, N-terminal Asn and Gln are tertiary destabilizing residues in that they function through their enzymatic deamidation (10, 11) to yield the secondary destabilizing N-terminal residues Asp and Glu (Fig. 1A). The activity of Asp and Glu requires their conjugation, by ATE1-encoded isoforms of Arg-tRNA-protein transferase (R-transferase), to Arg, one of the primary destabilizing residues (12–15). The latter are recognized by E3 Ub ligases of the N-end rule pathway (Fig. 1A). In mammals and other eukaryotes that produce nitric oxide (NO), the set of arginylated residues contains not only Asp and Glu but also N-terminal Cys, which is arginylated after its oxidation. The latter requires both NO and oxygen (O2) (Fig. 1A) (14, 15). The functions of the N-end rule pathway include the control of peptide import (through the conditional degradation of import’s repressor) (16, 17), the fidelity of chromosome segregation (through the degradation of a conditionally produced cohesin’s fragment) (18), the regulation of apoptosis (through the degradation of a caspase-processed inhibitor of apoptosis) (19, 20), the regulation of meiosis (21), leaf senescence in plants (22), and cardiovascular development in mammals (13). The last of these processes involves the N-end rule pathway in part through the NO/O2-dependent, arginylation-mediated degradation of regulator of G protein signaling (RGS) proteins RGS4, RGS5, and RGS16, a set of GTPase-activating proteins that bear N-terminal Cys, inhibit the signaling by specific G proteins, and are themselves down-regulated by the N-end rule pathway, at rates controlled by NO and O2 (14, 15).
Fig. 1.
Gross morphology of UBR1−/−UBR2−/− mouse embryos. (A) The mammalian N-end rule pathway (4, 21). N-terminal residues are indicated by single-letter abbreviations for amino acids. The ovals denote the rest of a protein substrate. C*, oxidized Cys residue (14, 15). (B–J) The appearance of control and UBR1−/−UBR2−/− embryos at ED9.5 (B–D), ED10.5 (E–G), and ED11.5 (H–J). Markings are as follows: arrow, heart; asterisk, swollen pericardial sac; arrowhead, a hemorrhage. (Scale bar: 1 mm.)
The E3s that recognize N-degrons are called N-recognins (4, 6, 17). In the yeast Saccharomyces cerevisiae, the N-end rule pathway is mediated by a single N-recognin, UBR1, which contains at least three substrate-binding sites (17). The type-1 and type-2 sites bind, respectively, to basic (Arg, Lys, and His) and bulky hydrophobic (Phe, Leu, Trp, Tyr, and Ile) N-terminal residues of either protein substrates or short peptides (4, 6, 17). The third binding site recognizes internal (non-N-terminal) degrons and is inaccessible to substrates in the autoinhibited UBR1 conformation but can be allosterically activated by the binding of peptides with destabilizing N-terminal residues to the type-1 and type-2 sites of UBR1. The third substrate-binding site of yeast UBR1 targets CUP9, a transcriptional repressor that down-regulates, in particular, the expression of PTR2, a peptide transporter, and thereby mediates the control of peptide import by the N-end rule pathway (16, 17). Our previous work (4, 21, 23, 24) identified mammalian E3s termed UBR1 and UBR2, two sequelogs (25) of S. cerevisiae UBR1. Mouse UBR1 and UBR2 are 46% identical and are apparently indistinguishable in their recognition of N-degrons (21). More recent work expanded the family of (operationally defined) N-recognins to at least four proteins: UBR1, UBR2, UBR4, and UBR5 (4). One common feature of these E3s and of several other E3s, termed UBR3, UBR6, and UBR7, is the presence of an ≈70-residue Cys/His-rich domain termed the UBR box (4).
We constructed mouse strains lacking some components of the N-end rule pathway (Fig. 1A) (4, 11, 13, 21, 24). NTAN1−/− mice, which lacked deamidation of N-terminal Asn, were viable, fertile, and anatomically normal but exhibited abnormalities in learning, memory, and behavior (11). ATE1−/− mice, which lacked N-terminal arginylation, died as embryos, with cardiovascular defects that included ventricular hypoplasia, ventricular septal defects, and delayed angiogenesis (13). UBR1−/− mice, which lacked the founding member of the set of N-end rule’s E3s, were viable and fertile but were leaner than wild-type littermates, exhibited a perturbed regulation of fatty acid synthase, had a mild hypoglycemia, and were behaviorally abnormal (24). Mutations in human UBR1 were recently shown to be the cause of Johanson–Blizzard syndrome, which comprises mental retardation, physical malformations, and severe pancreatitis (26). Further analysis of UBR1−/− mice has revealed that they also exhibit exocrine pancreatic insufficiency, a less severe version of the defect in human Johanson–Blizzard syndrome (UBR1−/−) patients (26). UBR2−/− mice, which lacked the second characterized N-recognin, exhibited both gender- and strain-specific lethality. For example, in the C57/129 hybrid background, UBR2−/− males were viable but infertile, owing to defects in the homologous chromosome pairing during meiotic prophase I, whereas most UBR2−/− females died as embryos (21).
In the present work, we extended genetic analysis of the N-end rule pathway to a double-mutant setting by constructing mouse strains that lacked both UBR1 and UBR2. UBR1−/−UBR2−/− embryos died at midgestation, with defects in both neurogenesis and cardiovascular development. We conclude that the functions of UBR1 and UBR2 are significantly divergent, in part because of differences in their expression patterns and possibly also because of differences in their recognition of substrates that contain degradation signals other than N-degrons.
Results and Discussion
Loss of both UBR1 and UBR2 Results in Embryonic Lethality.
We produced mice of the UBR1−/−UBR2−/− genotype by using compound heterozygous crosses. The UBR1+/− and UBR2+/− mouse strains (21, 24) were intercrossed, yielding (apparently normal) UBR1+/−UBR2+/− mice in the C57BL6/129 background. The progeny of subsequent crosses between UBR1+/−UBR2+/− mice did not contain surviving UBR1−/−UBR2−/− pups, suggesting that the absence of both UBR1 and UBR2 causes embryonic or neonatal lethality. To address these issues, we examined >1,000 embryos at various stages of development, mainly at embryonic days (ED) 7.5–13.5, that were produced in timed intercrosses between UBR1+/−UBR2+/− and/or UBR1−/− UBR2+/− mice. ED9.5 UBR1−/−UBR2−/− embryos were recovered alive and seemed indistinguishable from their control littermates, both anatomically and sizewise (Fig. 1 C and D). ED10.5 UBR1−/−UBR2−/− embryos were also recovered alive and were apparently normal in regard to the neural tube closure, axial rotation, the appearance of branchial arches, and the anterior limb bud development. However, the ED10.5 double-mutant embryos were consistently smaller than littermate controls (Fig. 1 F and G). ED11.5 UBR1−/−UBR2−/− mutants were found to be either dead or still alive but strongly growth-retarded, their growth having ceased at ≈ED10.5 (Fig. 1 I and J). No live UBR1−/−UBR2−/− embryos were recovered at ED12.5.
Impaired Neurogenesis in Embryos Lacking UBR1 and UBR2.
The appearance of neural tubes in ED10.5 UBR1−/−UBR2−/− embryos was apparently normal, but by ED11.5, the tubes became strongly kinked (Fig. 2 G and H, arrowheads). The neuroepithelium of ED10.5 UBR1−/−UBR2−/− embryos was found to be abnormally thin; this defect was more severe in the forebrain than in the spinal cord (Fig. 2 A–D). Neuroepithelial structures in UBR1−/−UBR2−/− embryos did not increase in thickness after ED10.5, in contrast to control embryos (Fig. 2 E and F and data not shown), suggesting the cessation of cell proliferation at ≈ED10.25–10.5. Nevertheless, these UBR1−/−UBR2−/− cells contained neurofilaments, a marker of mature neurons, at levels comparable to those in control embryos (Fig. 2 A and B), suggesting that the development of, for example, the spinal cord was grossly normal in UBR1−/−UBR2−/− embryos until ≈ED10.25. By ED11.5, the morphology of the forebrain in UBR1−/−UBR2−/− embryos became grossly distorted, with serpentine, thin, often disjointed neuroepithelial layers of varying thickness (Fig. 2 E and F). These defects in ED11.5 UBR1−/−UBR2−/− embryos might result from the continuing operation of developmental programs, such as cell differentiation and migration, despite the absence of wild-type amounts of neuronal precursor cells.
Fig. 2.
Abnormal development of the central nervous system in UBR1−/−UBR2−/− embryos. (A and B) Transverse sections of ED10.5 UBR1−/−UBR2−/− (B) and littermate UBR1+/−UBR2+/− control (A) embryos stained for neurofilaments. (C–F) Hematoxylin/eosin-stained transverse sections of UBR1−/−UBR2−/− embryos (D and F) and littermate control embryos (wild type in C; UBR1+/−UBR2+/− in E) at ED10.5 (C and D) and ED11.5 (E and F). (G and H) Dorsal views of ED11.5 UBR1−/−UBR2−/− (H) and control (UBR1+/+UBR2+/−) (G) embryos. drg, dorsal root ganglion; mt, motor neuron; nt, neural tube; SC, spinal cord; FB, forebrain; HB, hindbrain. Asterisk and arrowheads in H mark the swollen pericardial sac and kinked neural tube, respectively. (Scale bars: 200 μm.)
Mammalian neurogenesis begins with a stem cell-like self-renewal of neural progenitor cells (27, 28). In the course of neurogenesis, neural precursor cells undergo several rounds of division at the ventricular zone (VZ) (see Fig. 3E). These cells then migrate from the VZ to the differentiation zone (mantle), which is composed of postmitotic differentiating neurons and glia, and differentiate there, resulting in radially arranged layers of neurons (27, 28). We injected BrdU into pregnant mice and compared the amounts of BrdU-positive cells in transverse sections of ED10.5 UBR1−/−UBR2−/− embryos and littermate controls. In control embryos, BrdU was incorporated, as expected, into cells of the VZ (Fig. 3E). The levels of S-phase cells (BrdU-positive, stained red) were consistently lower in the neural tissues of ED10.5 UBR1−/−UBR2−/− embryos, throughout the anteroposterior axis, than in control embryos (Fig. 3 B, D, F, and H; compare with Fig. 3 A, C, E, and G). Thus, both morphological data (Fig. 2 A–F) and the levels of S-phase cells (Fig. 3 A–H) indicated that the proliferation of neural precursors was impaired in the absence of UBR1 and UBR2. The amounts of neural cells expressing Ki67, a marker for cells active in the cell cycle, were comparable in control and UBR1−/−UBR2−/− embryos (Fig. 3M), suggesting that the impaired proliferation was not caused by cells entering a mitotically quiescent, G0-like state.
Fig. 3.
Abnormal proliferation patterns of UBR1−/−UBR2−/− neural precursors. Transverse sections of ED10.5 UBR1−/− UBR2−/− embryos (B, D, F, H, Ic, Id, K, and M) and littermate control embryos (UBR1+/+UBR2+/− in A, C, E, G, Ia, Ib, and L; UBR1+/−UBR2+/+ in J) that were stained for BrdU (red), pH3 (green), nestin (red), TUNEL (green), and/or Ki67 (red), as indicated above. (E and F) Enlarged views of boxed hindbrain regions in C and D. (I) Representative morphologies of pH3-positive nuclei of cells in ED10.5 UBR1−/−UBR2−/− (c and d) and control (a and b) hindbrains. (Scale bars: A–D, L, and M, 200 μm; E and F, 40 μm; G, H, J, and K, 100 μm.)
We also stained cells in the neural tube with antibody to phosphohistone H3 (pH3), a marker for cells in mitosis. In control embryos, pH3-positive (stained green) mitotic cells were observed at the apical surface of the VZ, a layer closest to the lumen (Fig. 3 A, C, E, and G). However, much higher numbers of pH3-positive mitotic neural precursors were present in similar regions of the forebrain in ED10.5 UBR1−/−UBR2−/− embryos (Fig. 3B). Moreover, the distribution of mutant mitotic cells was overtly disorganized in that they were also found throughout the layer lateral to the VZ (Fig. 3F; see below). In contrast, the pH3 index was similar for control and UBR1−/−UBR2−/− embryos in the hindbrain, and in the spinal cord, the pattern was even partially reversed (Fig. 3 C–H), suggesting that the perturbed proliferation of UBR1−/−UBR2−/− cells was a function of cells’ position along the anteroposterior axis. Furthermore, mitotic cells in control embryos were mainly at prophase or prometaphase (Fig. 3I a and b), whereas many pH3-positive mitotic cells in UBR1−/−UBR2−/− embryos appeared to be between the interphase and prophase (Fig. 3I c and d), suggesting an arrest, possibly a transient one, at the G2–M transition. We also asked, by using the TUNEL assay, whether apoptotic cell death may play a role in the observed deformations of neural tube in UBR1−/− UBR2−/− embryos. Indeed, significantly increased amounts of TUNEL-positive cells were observed throughout the neural tubes of ED10.5 UBR1−/−UBR2−/− embryos (more prominently in the forebrain) (Fig. 3 J and K). We conclude that the thinner and morphologically distorted neuroepithelial layers in UBR1−/−UBR2−/− embryos (Fig. 2 A–F) resulted from both a decrease in cell proliferation and increase in apoptosis.
The neuroepithelium of normal ED10.5 embryos consists of the VZ layer, containing proliferating neural precursors, and the mantle, containing postmitotic neurons (27, 28). In contrast, the neuroepithelium of ED10.5 UBR1−/−UBR2−/− embryos could be subdivided into three layers, BrdU−pH3+ cells, BrdU+pH3+ cells, and BrdU−pH3− cells (Fig. 3F). When progenitor cells in the wild-type VZ become postmitotic, they usually migrate to the mantle. However, at later stages of neurogenesis, some precursor cells that leave the VZ remain mitotically active and enter a secondary proliferative zone called the subventricular zone (SVZ), between the VZ and the mantle (27–29). The nuclei of cells in the VZ migrate up and down (within cells containing them) along the apical–basal axis during the cell cycle, a movement called the interkinetic nuclear migration, whereas the nuclei of cells in the SVZ do not exhibit this effect. A parsimonious interpretation of the three layers observed with UBR1−/− UBR2−/− embryos, vis-a-vis two layers in control embryos (Fig. 3 E and F), is that the former are the VZ (BrdU−pH3+), SVZ (BrdU+pH3+), and mantle (BrdU−pH3−) layers. The SVZ (BrdU+pH3+) layer in UBR1−/− UBR2−/− embryos may be the result of premature migration of mitotically active neural precursors from the VZ. To address this possibility in particular, we used antibodies to nestin (a marker for neural precursors in the VZ), to microtubule-associated protein 2 (a marker for dendrites of postmeiotic neurons in the mantle), and to tubulin-βIII (a marker for tubulin-rich processes of postmitotic neurons in the mantle). In agreement with the above interpretation, the BrdU+pH3+ SVZ-like zone in UBR1−/−UBR2−/− embryos was located between the layer containing nestin-positive cells (Fig. 4F) and the mantle layer containing postmitotic neurons (Fig. 4 H and J). GABA is produced in specific neurons of the forebrain that are derived from the SVZ (30). We asked, by staining for GABA, whether GABA-producing neurons would be precociously generated in the forebrains of ED10.5 UBR1−/−UBR2−/− embryos. That was indeed the case (Fig. 4L): yet another result consistent with the above interpretation.
Fig. 4.
Abnormal differentiation patterns of UBR1−/−UBR2−/− neural precursors. Transverse sections of forebrain regions (boxed in A and B) in ED10.5 UBR1−/−UBR2−/− (B, D, F, H, J, and L) and UBR1+/+UBR2+/− control (A, C, E, G, I, and K) embryos were stained for DNA (blue), BrdU (red), pH3 (green), nestin (red), microtubule-associated protein 2 (MAP2) (green), tubulin-βIII (red), and/or GABA (green), as indicated above. (Scale bars: A and B, 200 μm; C–L, 40 μm.)
Neural progenitor cells in the neural tube normally enter the S phase when their nuclei are located proximally to the outer half of VZ. The subsequent interkinetic nuclear migration brings the nuclei closer to the ventricular surface, followed by mitosis (27, 28). The patterns observed with neural tube cells in UBR1−/−UBR2−/− embryos were different, given the near absence of S-phase cells and the almost completely suppressed cell proliferation in the VZ, normally the main proliferative zone. But cell proliferation was allowed in the SVZ (normally the secondary proliferative zone) of double-mutant embryos (see above). Our findings (Figs. 2–4) suggest that neural progenitor cells in UBR1−/−UBR2−/− embryos prematurely migrate from the VZ to the SVZ, possibly as a result of their premature differentiation. If a reduced proliferation of neural precursors is accompanied by their accelerated differentiation, neural precursors would be gradually depleted as neurogenesis progresses. A relative depletion of cells positive for nestin (a marker for neural precursors in the VZ) was indeed observed in ED10.5 UBR1−/−UBR2−/− embryos (Figs. 3K and 4F), and this depletion became more severe by ED11.5 (data not shown). Taken together, our results indicate that the Ub ligases UBR1 and UBR2 are critical for the proper regulation of proliferation and differentiation of neural precursor cells during embryogenesis.
Impaired Cardiovascular Development in Embryos Lacking UBR1 and UBR2.
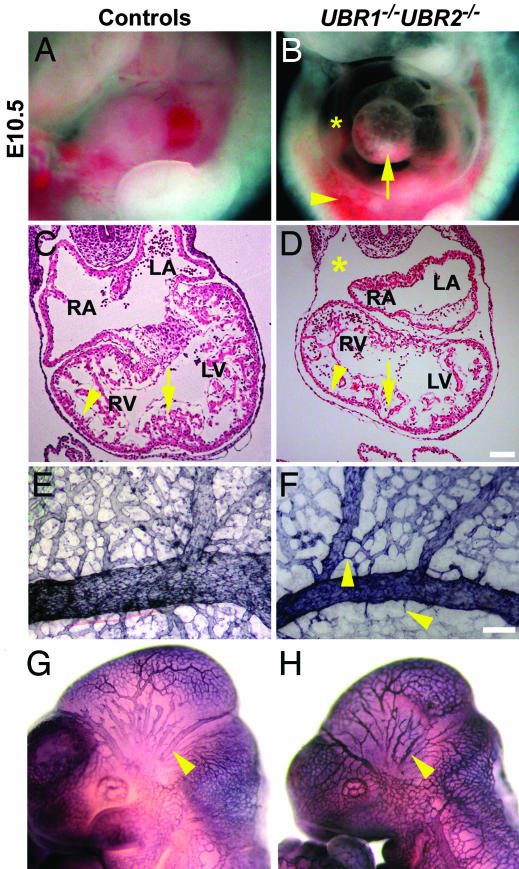
In contrast to single mutants (21, 24), the double-mutant UBR1−/−UBR2−/− embryos had severe cardiovascular defects. The hearts of ED9.5 UBR1−/−UBR2−/− embryos were apparently normal (data not shown), but by ED10.5, the double mutants developed local hemorrhages, a swollen pericardial sac, and/or a pericardial effusion (Figs. 1 F and G, 2H, and 5B), indicating the leakage of blood into the abdominal cavity as well as other defects. Amongst them was a large space between the heart and pericardium in ED10.5 UBR1−/−UBR2−/− embryos, consistent with the accumulation of pericardial fluid (Fig. 5D, asterisk). Development of the atria and ventricles was largely arrested in double mutants by ED10.5. Disorganization of the myocardial wall was also observed, with variable levels of ventricular atrophy (Fig. 5D, arrowhead). Although control hearts at this stage had abundant and thick trabeculae, the trabeculations in UBR1−/−UBR2−/− embryos were thinner and less abundant. Interatrial and interventricular septa began to form in the hearts of control embryos by ED10.5 (Fig. 5C, arrow) but were not observed in double mutants. In the control hearts, the staining for tropomyosin (present in cardiomyocytes) was, as expected, cytoplasmic, delineating well organized cardiomyocyte-based cardiac structures (data not shown). In contrast, the tropomyosin staining in the UBR1−/−UBR2−/− hearts was both cytoplasmic and nuclear, in addition to revealing a morphological disorganization (data not shown). Taken together, these results indicate that proliferation of UBR1−/−UBR2−/− cardiac cells was impaired at ≈ED10.5, resulting in multiple defects.
Fig. 5.
Impaired cardiovascular development in UBR1−/−UBR2−/− embryos. (A and B) Lateral views of ED10.5 UBR1−/−UBR2−/− (B) and littermate UBR1+/+ UBR2+/− control (A) embryos. Markings are as follows: arrow, abnormal heart morphology; asterisk, swollen pericardial sac; arrowhead, hemorrhage. (C and D) Hematoxylin/eosin-stained transverse sections of ED10.5 UBR1−/−UBR2−/− (D) and littermate wild-type control (C) hearts. Markings are as follows: asterisk, swollen pericardial space; arrowheads, ventricular wall; arrows, sites of septum formation in wild-type (but not in UBR1−/−UBR2−/−) embryos. RA, right atrium; LA, left atrium; RV; right ventricle; LV, left ventricle. (E and F) PECAM-stained ED10.5 UBR1−/−UBR2−/− (F) and UBR1+/−UBR2+/− (E) yolk sacs. Arrowheads in F indicate some of the differences in the branching patterns relative to wild-type embryos. (G and H) PECAM-stained ED10.5 UBR1−/−UBR2−/− (H) and UBR1+/−UBR2+/− (G) whole embryos. Arrowheads mark the intracranial artery. (Scale bars: 100 μm.)
Staining for platelet endothelial cell adhesion molecule (PECAM) 1, a marker for blood vessels, showed that the honeycomb-like network of primary vascular plexus formed normally in the ED9.5 UBR1−/−UBR2−/− yolk sacs and embryos proper (data not shown). In addition, UBR1−/−UBR2−/− yolk sacs had apparently normal endothelial cells as well as blood islands that contained nucleated fetal erythrocytes. However, by ED10.5, the UBR1−/−UBR2−/− yolk sacs appeared pale, and their blood vessels were thinner and less branched than their wild-type counterparts. Staining for PECAM-1 showed that the growth, remodeling, and branching of both small and large vessels were impaired in double-mutant embryos (Fig. 5F, arrowhead). Similarly, larger vessels such as the intracranial artery (Fig. 5H, arrowhead) were thin and poorly developed in ED10.5 UBR1−/−UBR2−/− embryos. These findings and related results (Fig. 5 and data shown) indicated that UBR1 and UBR2 were required for normal cardiovascular development.
Molecular Analysis of Mouse Embryos Lacking UBR1 and UBR2.
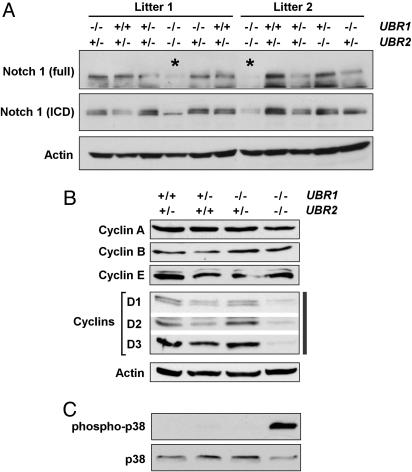
We focused, in the present study, on molecular circuits known to underlie cell proliferation, neurogenesis, and cardiovascular development. The observed suppression of cell proliferation in both the nervous and cardiovascular systems of UBR1−/−UBR2−/− embryos suggested that this suppression may also take place in other tissues of double-mutant embryos. Therefore, we used whole-embryo extracts to examine the levels of cyclins, specific subunits of cyclin-dependent kinases that control cell growth and division (31). Immunoblotting with antibodies to cyclins D1, D2, and D3 revealed their decreased levels in ED10.5 UBR1−/−UBR2−/− embryos, with the level of cyclin D3 being particularly low (Fig. 6B). The levels of other tested cyclins, A, B, and E, were unaffected by the absence of UBR1 and UBR2 (Fig. 6B). Semiquantitative RT-PCR and Northern hybridization showed that the levels of Ccnd3 mRNA were indistinguishable between ED10.5 UBR1−/−UBR2−/− embryos and controls (data not shown). Thus, the strong decrease of cyclin D3 in double-mutant embryos (Fig. 6B) could not be caused by effects on transcription or mRNA stability, suggesting either a shorter half-life of this regulator and/or a less efficient translation of its mRNA. Because D-type cyclins are critical for the conversion of prereplication complex into an active replication fork at the G1–S transition (31), our findings with cyclin D3 (Fig. 6B) may be relevant to the observed decrease in the levels of S-phase neural precursor cells in UBR1−/−UBR2−/− embryos (Fig. 3).
Fig. 6.
Immunoblot analysis of UBR1−/−UBR2−/− embryos. Whole-embryo extracts from ED10.5 UBR1−/−UBR2−/− and littermate control embryos were immunoblotted for the proteins indicated in A–C.
The defects of UBR1−/−UBR2−/− embryos, a deformed neural tube and the impairment of neurogenesis and cardiovascular development, are also a major feature of mouse embryos lacking components of the Notch signaling pathway. The functions of this pathway include the inhibition of neuronal differentiation, a pattern of control that maintains the required cell-type diversity of neural precursors (32–34). A ligand-activated Notch1 is cleaved by the γ-secretase complex, and the released intracellular domain of Notch1 translocates to the nucleus, where it forms a complex with RBP-Jκ (recombination signal sequence-binding protein Jκ) and thereby activates target genes of the Notch pathway (32–34). ED10.5 UBR1−/−UBR2−/− embryos contained a significantly decreased level of Notch1 in comparison with littermate controls (Fig. 6A), whereas the level of Notch1 mRNA was not significantly affected (data not shown). Thus, a suppression of the Notch pathway in UBR1−/−UBR2−/− embryos may account, in part, for the impaired neurogenesis and cardiovascular development in this mutant background.
We also asked whether the absence of both UBR1 and UBR2 would affect the mitogen-activated protein kinase (MAPK) pathway, which regulates, in particular, cell proliferation, differentiation, and apoptosis. The level of phosphorylated (active) p38 MAPK was substantially increased in ED10.5 UBR1−/−UBR2−/− embryos in comparison with wild-type or single-mutant embryos (Fig. 6C and data not shown), in contrast to the levels of phosphorylated or unphosphorylated extracellular signal-regulated kinase and c-Jun N-terminal kinase MAPKs (data not shown). Activated p38 MAPK causes the exit from the cell cycle and differentiation in many cell types (35). Taken together, our biochemical findings (Fig. 6) suggest that decreased levels of D-type cyclins and Notch1, as well as the enhanced phosphorylation of p38 MAPK, are amongst the causes of multiple phenotypic defects (Figs. 1–5) of UBR1−/−UBR2−/− embryos. Because both D3 and Notch1 decrease in concentration in UBR1−/−UBR2−/− embryos (Fig. 6), UBR1 and UBR2 apparently down-regulate specific (currently unknown) proteins that are a part of circuits that influence the levels of Notch1 and/or D3.
Concluding Remarks.
The multiple and severe defects of the double-mutant UBR1−/−UBR2−/− mice, including their embryonic lethality (Figs. 1–5), are in contrast to the viability of UBR1−/− mice and the conditional (genetic background-dependent) viability of UBR2−/− mice (21, 24). Given both strong sequelogy similarity (25) between UBR1 and UBR2 and their previously demonstrated indistinguishable patterns of binding to destabilizing N-terminal residues (21), our results can be formally accounted for by presuming that the substrate recognition and other properties of UBR1 and UBR2 are identical or nearly so. In this interpretation, the main reason for greater severity of the double-mutant phenotype would be significant differences in the expression patterns of UBR1 and UBR2 in specific cell types and tissues. Some of these differences were detected previously (21). According to this model, the previously observed apoptosis of meiotic spermatocytes in UBR2−/− mice (21) is caused by functionally insufficient levels of UBR1 in these cells, which, in wild-type mice, would be “rescued” by UBR2. In a converse example, the same model would predict that defects (and eventual apoptosis) of the acinar pancreatic cells in UBR1−/− human patients with Johanson–Blizzard syndrome (26) are caused by insufficient levels of UBR2 in these cells, which are normally “rescued” by UBR1.
Although no evidence at present directly contradicts this interpretation, the actual disposition is likely to be more complex. For example, S. cerevisiae UBR1, the sole E3 Ub ligase of the yeast N-end rule pathway, contains at least three substrate-binding sites, one of which recognizes the transcriptional repressor CUP9 through its internal (non-N-terminal) degron (see the Introduction) (16, 17). Because S. cerevisiae UBR1 and mouse UBR1 and UBR2 are sequelogous (25) throughout their lengths (4, 23), and because mouse UBR1 and UBR2 do contain the first two substrate-binding sites (21, 24), one would expect either of them to contain a third substrate-binding site as well. In contrast to S. cerevisiae UBR1, such third sites in mammalian UBR1 and UBR2 are still conjectural. [Although human UBR1 and UBR2 bind to RECQL4, a putative helicase that is absent or damaged in patients with the Rothmund–Thomson syndrome (36), it is unclear whether this binding is relevant to the targeting that results in substrates’ ubiquitylation and degradation.]
If UBR1 and UBR2 contain the third substrate-binding sites, such sites may be similar in their recognition specificity, in which case the above “differential expression” model might suffice to account for the entire gamut of phenotypic differences between UBR1−/−UBR2−/− mice and their single-mutant counterparts. However, if the (presumed) third substrate-binding sites of UBR1 and UBR2 are significantly divergent in regard to substrates they recognize, the relative severity of double-mutant phenotype may also stem from this divergence. The mammalian N-end rule pathway contains at least four, and possibly as many as seven, distinct N-recognins, including UBR1 and UBR2 (4). Although all of these E3 Ub ligases share the UBR-box domain, they are largely dissimilar otherwise, with the exception of UBR1 and UBR2 (4). The remarkable range of Ub ligases that mediate the mammalian N-end rule pathway is yet another complexity that the eventual mechanistic (circuits-based) understanding of defects in UBR1−/−UBR2−/− mice (Figs. 1–5) would have to illuminate.
Materials and Methods
UBR1−/−UBR2−/− Mice.
The construction and characterization of single-mutant UBR1−/− and UBR2−/− mice are described in refs. 21 and 24. UBR1−/−UBR2−/− mice were produced through compound heterozygous crosses in the mixed 129SvImJ/C57BL/6 genetic background (4).
Histology, Immunohistochemistry, and Antibodies.
Embryos were fixed in 4% paraformaldehyde before processing for paraffin sectioning. Paraffin-embedded embryos were serially sectioned and stained with hematoxylin/eosin or antibodies. Antibodies used for immunohistochemistry or immunoblotting were to the following antigens: neurofilaments, tropomyosin, and nestin (Developmental Studies Hybridoma Bank, Iowa City, IA); cyclins A, B, D1, D2, D3, and E (Santa Cruz Biotechnology); p38, phospho-p38, and Notch-ICD (intracellular domain of Notch1) (Cell Signaling Technology, Beverly, MA); Notch1, tubulin-βIII, GABA, and actin (Sigma-Aldrich); MAP2 (microtubule-associated protein 2) (Chemicon); BrdU (Accurate Chemicals); pH3 (Upstate Biotechnology, Lake Placid, NY); and PECAM-1 (Pharmingen). Secondary antibodies were conjugated to Alexa Fluor 555 (Molecular Probes), FITC, Cy3 (Jackson ImmunoResearch), or horseradish peroxidase (Santa Cruz Biotechnology).
Measurements of Cell Proliferation and Apoptosis.
For proliferation assays, pregnant mice were injected i.p. with BrdU (Sigma) (50 μg/g of body weight) and killed 1–5 h later. Embryos were fixed, processed for paraffin sectioning, and stained for BrdU. To visualize apoptotic nuclei, transverse sections of embryos were processed for the TUNEL assay by using the In Situ Cell Death Detection Kit (Roche Applied Science, Indianapolis) and then were counterstained with DAPI.
Acknowledgments
We thank Raphael Kopan (Washington University, St. Louis) for advice about Notch-related experiments, Xiaochun Yu (Mayo Clinic, Rochester, MN) for staging mitotic cells, and Heather Buresch and Zhen Lu (University of Pittsburgh) for care of mice. This work was supported by National Institutes of Health Grants GM069482, GM074000 (to Y.T.K.), GM31530, and DK39520 (to A.V.); the American Heart Association (Y.T.K.); and the Ellison Medical Foundation (A.V.).
Abbreviations
- Ub
ubiquitin
- VZ
ventricular zone
- SVZ
subventricular zone
- EDn
embryonic day(s) n
- pH3
phosphohistone H3
- PECAM
platelet endothelial cell adhesion molecule
- MAPK
mitogen-activated protein kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Varshavsky A. Protein Sci. 2006;15:647–654. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A., Ciechanover A., Varshavsky A. Nat. Med. 2000;10:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 3.Pickart C. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 4.Tasaki T., Mulder L. C. F., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., Kwon Y. T. Mol. Cell. Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochstrasser M. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 6.Varshavsky A. Proc. Natl. Acad. Sci. USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmair A., Varshavsky A. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T., Varshavsky A. EMBO J. 1999;18:6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 10.Baker R. T., Varshavsky A. J. Biol. Chem. 1995;270:12065–12074. doi: 10.1074/jbc.270.20.12065. [DOI] [PubMed] [Google Scholar]
- 11.Kwon Y. T., Balogh S. A., Davydov I. V., Kashina A. S., Yoon J. K., Xie Y., Gaur A., Hyde L., Denenberg V. H., Varshavsky A. Mol. Cell. Biol. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon Y. T., Kashina A. S., Varshavsky A. Mol. Cell. Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon Y. T., Kashina A. S., Davydov I. V., Hu R.-G., An J. Y., Seo J. W., Du F., Varshavsky A. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 14.Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., Kwon Y. T. Proc. Natl. Acad. Sci. USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu R. G., Sheng J., Xin Q., Xu Z., Takahashi T. T., Varshavsky A. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 16.Turner G. C., Du F., Varshavsky A. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 17.Du F., Navarro-Garcia F., Xia Z., Tasaki T., Varshavsky A. Proc. Natl. Acad. Sci. USA. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao H., Uhlmann F., Nasmyth K., Varshavsky A. Nature. 2001;410:955–960. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 19.Ditzel M., Wilson R., Tenev T., Zachariou A., Paul A., Deas E., Meier P. Nat. Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 20.Varshavsky A. Nat. Cell Biol. 2003;5:373–376. doi: 10.1038/ncb0503-373. [DOI] [PubMed] [Google Scholar]
- 21.Kwon Y. T., Xia Z. X., An J. Y., Tasaki T., Davydov I. V., Seo J. W., Xie Y., Varshavsky A. Mol. Cell. Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida S., Ito M., Gallis J., Nishida I., Watanabe A. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- 23.Kwon Y. T., Reiss Y., Fried V. A., Hershko A., Yoon J. K., Gonda D. K., Sangan P., Copeland N. G., Jenkins N. A., Varshavsky A. Proc. Natl. Acad. Sci. USA. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon Y. T., Xia Z., Davydov I. V., Lecker S. H., Varshavsky A. Mol. Cell. Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varshavsky A. Curr. Biol. 2004;14:R181–R183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Zenker M., Mayerle J., Lerch M. M., Tagariello A., Zerres K., Durie P. R., Beier M., Hülskamp G., Guzman C., Rehder H., et al. Nat. Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 27.McConnell S. K. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 28.Götz M., Huttner W. B. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi T. T., Nowakowski R. S., Caviness V. S., Jr J. Neurosci. 1995;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson S. A., Eisenstat D. D., Shi L., Rubenstein J. L. R. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 31.Sherr C. J., Roberts J. M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 32.Yoon K., Gaiano N. Nat. Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 33.Cornell R. A., Eisen J. S. Semin. Cell Dev. Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Louvi A., Artavanis-Tsakonas S. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 35.Engel F. B., Schebesta M., Duong M. T., Lu G., Ren S., Madwell J. B., Jiang H., Wang Y., Keating M. T. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin J., Kwon Y. T., Varshavsky A., Wang W. Hum. Mol. Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]