Abstract
Paracentric chromosomal inversions are major architects of organismal evolution and have been associated with adaptations relevant to malaria transmission in anopheline mosquitoes. The processes responsible for their origin and maintenance, still poorly understood, can be illuminated by analysis of inversion breakpoint sequences. Here, we report the breakpoint structure of chromosomal inversion 2La from the principal malaria vector Anopheles gambiae and its relatives in the A. gambiae complex. The distal and proximal breakpoints of the standard (2L+a) arrangement contain gene duplications: full-length genes and their truncated copies at opposite ends. Intact genes without pseudogene copies in the alternative arrangement (2La) imply that 2L+a is derived and was viable despite damage to genes, because duplication preserved gene function. A unique origin for the interspecific 2La inversion was challenged previously by indirect genetic evidence, but breakpoint sequences determined from members of the A. gambiae complex strongly suggest their descent from a single event. The derived position of 2L+a, long considered ancestral in this medically important group, has significant implications for the phylogenetic history and the evolution of vectorial capacity in the A. gambiae complex.
Keywords: genome evolution, transposable elements, malaria vectors, inversion monophyly, sibling species
Chromosomal rearrangements are major architects of evolution in various groups of organisms (1). In anopheline mosquitoes, synteny has been highly conserved, but gene order is extensively shuffled, primarily through paracentric chromosomal inversions (2, 3). By suppressing recombination between alternative arrangements and stabilizing adaptive allelic or regulatory combinations, paracentric inversions play a major role in ecological differentiation within species and in speciation (4–6). For already proficient anopheline vectors of human disease, the abundance of polymorphic inversions confers the ability to adapt rapidly to human-made ecological disturbances such as deforestation and irrigation and leads to more efficient exploitation of heterogeneities in the environment. By expanding opportunities for vector breeding and reducing competition for breeding sites, high chromosomal diversity can have the undesirable public health consequences of increasing the vector's density and longevity (5) and extending the temporal and spatial distribution of vectors into formerly inhospitable environments. This phenomenon is exemplified by the Mopti chromosomal form of the principal African malaria vector Anopheles gambiae. This cytotype exploits semipermanent human-made sites such as rice fields and artificial lakes and can be found in arid northern areas bordering the Sahara that exclude other chromosomal forms of this species (7, 8).
Despite the ecological and epidemiological importance of chromosomal inversions, the mechanisms responsible for their generation are not well understood. Moreover, the traditional view that each inversion has a unique origin has been questioned. Two apparent instances of parallel evolution of cytologically identical inversions have been reported. The first was in the A. gambiae complex with respect to an inversion on the left arm of chromosome 2 designated 2La, and the second was in primates (9, 10). The processes responsible for the origin and maintenance of inversions can be illuminated by comparative analysis of breakpoint sequences from alternative arrangements. This information also reveals the ancestral–descendant relationship of the arrangements, allowing inference of the direction of adaptive evolution and phylogenetic history. These issues are of particular interest for 2La in the A. gambiae complex.
Inversion 2La is highly polymorphic and widespread in A. gambiae sensu stricto (s.s.), but it is nonrandomly distributed with respect to degree of aridity (4). In West Africa along north–south transects spanning hundreds of kilometers, there are strong and stable clines in the frequency of 2La, from absence in humid forest to fixation in arid sahel. Seasonal and microspatial heterogeneities also occur. From rainy to dry seasons, the frequency of 2La cycles from low to high. Similarly, its frequency is higher indoors where there is a nocturnal saturation deficit relative to outdoors, resulting in an increased probability of inversion carriers encountering and biting humans sleeping indoors at night, when blood feeding occurs. The 2La inversion also has been associated with susceptibility to Plasmodium (11). Accordingly, this inversion is expected to influence the vectorial capacity of its carriers.
A. gambiae s.s. is the nominal member of a group of seven morphologically indistinguishable and closely related species that comprise the A. gambiae complex. A. gambiae s.s. is the only member of the complex in which the 2La inversion is polymorphic. Anopheles bwambae, Anopheles melas, and Anopheles quadriannulatus A and B are monomorphic (fixed) for the alternative 2L+a arrangement, which was assumed to be ancestral and was therefore designated as standard (5). The two remaining species, Anopheles arabiensis and Anopheles merus, are fixed for an arrangement that is identical to 2La at the cytological level. Assuming a monophyletic (unique) origin of this arrangement in the A. gambiae complex, its distribution among the component species is not consistent with their phylogenetic relationships and can only be reconciled by invoking genetic introgression and/or multiple origins of 2La. Indeed, a previous study of four gene regions inside or adjacent to the distal breakpoint of 2La (but at least 142 kb away) suggested that A. merus has a cytologically indistinguishable but molecularly independent inversion, 2La′ (10). To elucidate the origin of 2La and to test more precisely for its monophyly in the A. gambiae complex, we cloned and sequenced DNA fragments spanning both breakpoints from two A. gambiae s.s. isolates and two sibling species, all fixed for this arrangement (2La): A. gambiae SUA and Bamako, A. arabiensis, and A. merus. Additionally, we sequenced one or both breakpoints of the alternative arrangement (2L+a) from A. melas and A. quadriannulatus.
Results and Discussion
Integration of the A. gambiae PEST (2L+a) genome sequence (12) and the polytene chromosome complement by means of bacterial artificial chromosome (BAC) clones that were physically mapped and end-sequenced allowed us to identify candidate BAC clones from PEST that were predicted to span the inversion breakpoints. When mapped to A. gambiae s.s. polytene chromosomes by using FISH, BAC clones 146D17 and 160O9 produced one signal corresponding to the proximal or distal breakpoints on 2L+a chromosomes (in divisions 22F-23A and 26D-27A, respectively) and signals at both breakpoints on 2La chromosomes, indicating that they contained distal and proximal breakpoint sequences. Reiteration of this procedure with smaller fragments allowed us to localize the breakpoint region within the BAC and identify probes to screen genomic phage libraries of A. gambiae SUA (2La) and A. merus (2La). Candidate phage were confirmed to span inversion breakpoints by end-sequencing and blast analysis against the PEST (2L+a) genome and by FISH against 2La and 2L+a polytene chromosomes of A. gambiae s.s. Verified phage were subcloned into plasmids and sequenced. We determined 3–4 kb of sequence surrounding each 2La breakpoint and aligned them to PEST 2L+a to identify the exact position of chromosome breakage. Primers designed based on these alignments were used to obtain the proximal and distal 2La breakpoints from A. arabiensis (by PCR from genomic DNA) and from A. gambiae Bamako (by PCR screening of a gridded fosmid library), from which comparable amounts of sequence were determined. Primers also were designed to obtain breakpoints of the alternative 2L+a arrangement from A. quadriannulatus and A. melas by PCR amplification and sequencing of genomic DNA.
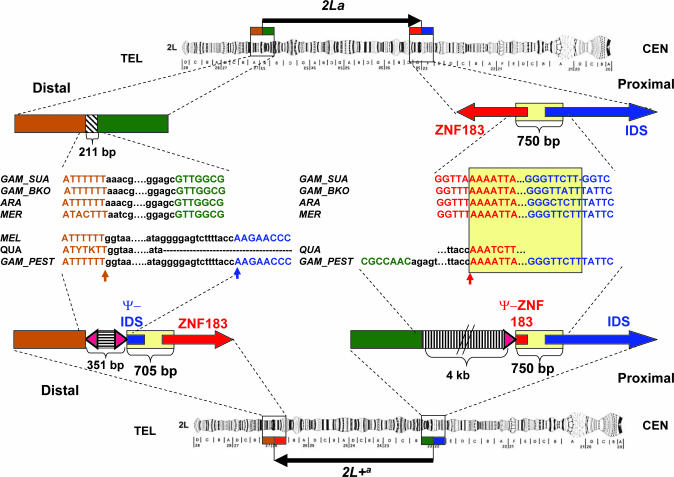
Importantly, the molecular organization of the 2La breakpoints of all three species (A. gambiae SUA and Bamako strains, A. arabiensis, and A. merus) was identical (Fig. 1). Detailed comparison of 2La sequences spanning proximal (4,369 bp) and distal (3,409 bp) breakpoints with the corresponding sequences of A. gambiae PEST 2L+a identified predicted genes, insertions/deletions, and complex assemblies of transposable elements (TEs) and repetitive DNA (Fig. 1; see also Fig. 3, which is published as supporting information on the PNAS web site). The 2La distal breakpoint is flanked by a small segment (≈200 bp) containing noncoding repetitive DNA surrounded by unique sequences with evidence of coding potential (Ensembl transcripts ENSANGT00000031443 and ENSANGT00000031375) but no significant similarity to known proteins. The sequence surrounding the 2La proximal breakpoint contains two predicted full-length and divergently transcribed genes oriented head to head (Ensembl transcripts ENSANGT00000018629 and ENSANGT00000021479) that potentially encode iduronate 2-sulfatase precursor (IDS) and zinc finger protein (ZNF) 183. Surprisingly, in 2L+a, both genes are present at both breakpoints: a full-length copy at one breakpoint (IDS at the proximal breakpoint and ZNF183 at the distal breakpoint) and a truncated copy in reverse orientation at the opposite breakpoint (Ψ-IDS at the distal breakpoint and Ψ-ZNF183 at the proximal breakpoint). Both truncated (pseudogene) copies contain the first exon and a partial intron. Thus, the salient difference between alternative arrangements is a ≈700-bp region present once at the 2La proximal breakpoint or twice in opposite orientations at both 2L+a breakpoints (Fig. 1). These duplicated segments have slightly different lengths, owing to deletions in Ψ-IDS at the distal 2L+a breakpoint. Adjacent to the duplicated segments at both 2L+a breakpoints are assemblies of repetitive DNA and/or TEs (Fig. 3). The distal 2L+a breakpoint has a 351-bp insertion of repetitive DNA framed by short inverted repeats, one of which is immediately adjacent to Ψ-IDS. The proximal 2L+a breakpoint has a 4-kb insertion of clustered and scrambled TE fragments flanked at the end nearest Ψ-ZNF183 by another copy of the same repeat that abuts Ψ-IDS at the distal breakpoint (Fig. 1). Such unexpected complexity at the breakpoints of an inversion has been reported only once previously, for the In(3R)Payne breakpoints of Drosophila melanogaster (13). The remarkably similar organization of these inversion breakpoints in Drosophila and Anopheles suggests a similar and more broadly applicable mechanism governing their generation.
Fig. 1.
Structure of the 2La and 2L+a inversion breakpoints in A. gambiae (not drawn to scale). Vertical arrows indicate the distal and proximal breakpoints in PEST spanning coordinates 42165182–42165532 on scaffold 8807 and 20524058–20528089 on scaffold 8960 (AgamP3 release). Homologous sequences are represented by boxes, and text is colored identically. International Union of Biochemistry codes Y (C or T) and K (G or T) are used at positions that were heterozygous in A. quadriannulatus. Red and blue horizontal arrows and their truncated counterparts correspond to genes (and pseudogenes, Ψ) that potentially encode ZNF183 and IDS proteins. Yellow boxes mark the 750-bp fragment in 2La that is present in opposite orientations at both breakpoints in 2L+a. Hatched boxes show complex assemblies of degenerate TEs and repetitive DNA. Triangles correspond to the terminal fragments of a repetitive element immediately adjacent to the breakpoints. DNA alignments are shown for A. gambiae SUA, Bamako, and PEST strains (GAM_SUA, GAM_BKO, and GAM_PEST); A. arabiensis (ARA); A. merus (MEL); A. melas (MEL); and A. quadriannulatus (QUA). CEN, centromere; TEL, telomere.
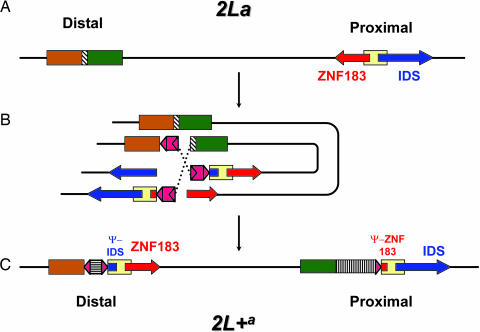
The presence of full-length genes and their pseudogene copies at opposite breakpoints of the 2L+a arrangement strongly suggests that the 2La arrangement is ancestral. To assume otherwise would require the improbable generation of two pseudogene copies before rearrangement, at distant reciprocal locations on the 2L+a chromosome, followed by their exact excision during generation of the inversion. The rearrangement of 2La to 2L+a cannot have been achieved by a simple cut-and-paste mechanism involving only two breaks of the chromosome, as traditionally assumed. We propose a model involving three simultaneous breaks and both homologous chromosomes, whereby breakage in slightly offset segments of the homologs and their recombination with segments of the opposite break can lead directly to an inversion with the characteristics of 2L+a during the same meiotic prophase (Fig. 2). The resulting inversion is viable, despite truncation of two highly conserved and potentially vital genes, because of the preservation of intact copies on the rearranged chromosome.
Fig. 2.
Model for the generation of arrangement 2L+a from 2La. Shapes and color-coding match those in Fig. 1. (A) 2La is the ancestral arrangement. (B) Homologous 2La chromosomes pair in meiotic prophase I. The distal break occurs in a region of noncoding/repetitive DNA (striped box) of one chromosome. Two proximal breaks occur in two homologs at offset positions: one in the ZNF183 gene and the other in the IDS gene. Three copies of a repetitive element (indicated in red) can either generate these breaks or associate with preexisting breaks. Two elements in opposite orientations pair and form a loop, bringing the regions of the distal and proximal breakpoints into close proximity. Dotted lines show recombination that leads to the inversion. (C) 2L+a is the derived arrangement, with intact ZNF183 and IDS genes and insertions of repetitive elements at both breakpoints. DNA sequence homologous to a 750-bp fragment of the 2La chromosome is represented twice in 2L+a, in opposite orientations at both breakpoints.
A mechanism for the proposed asymmetrical exchange is suggested by the presence of homologous repetitive elements immediately flanking each breakpoint (triangles in Fig. 1). The nature of this repetitive element was explored through reiterative blastn searches of the A. gambiae genome, using the element and subsequently its top hits as queries. Alignment of sequences from the blast output plus flanking DNA allowed construction of a consensus sequence ≈10 kb in length. The consensus sequence begins precisely with the repeats flanking each breakpoint, suggesting that they represent terminal remnants of a larger repetitive element, potentially a TE, although no hallmarks are evident. TEs have been implicated in the generation of many, but not all, inversions whose breakpoints have been analyzed, including 2Rd′ of A. arabiensis (13–19). Passive ectopic recombination between homologous repetitive elements already residing at the chromosomal locations of the breakpoints could have generated the rearrangement. However, their apparent absence at the appropriate locations on any of the sampled ancestral 2La chromosomes, and their retention on all sampled 2L+a chromosomes (Fig. 1), suggests a more active mechanism involving simultaneous element activations, chromosome breakage, and rearrangement not unlike the hybrid dysgenesis-induced chromosomal rearrangements described for different families of TEs in Drosophila (20). The accretion of ≈4 kb of additional defective TEs in the 2L+a proximal breakpoint region of A. gambiae most likely postdates the rearrangement and is a reflection of the reduced recombination that is typical of breakpoint regions in inversion heterozygotes.
The traditional view of history in the A. gambiae complex held that its most likely ancestor had the chromosomal features of A. quadriannulatus, which were considered as standard because of their central position relative to other species in the complex (5). Moreover, other features of A. quadriannulatus seemed to support its basal position, including tolerance for temperate climates, disjunct distribution, less specialized host preference, and, importantly, lack of involvement in malaria transmission (5). This view is no longer valid, in light of our data that suggest that the 2L+a arrangement fixed in A. quadriannulatus is derived. Indeed, there is cytogenetic evidence independent from our results that reinforces this conclusion. Based on banding pattern homologies, the 2La arrangement is present in the Oriental Anopheles subpictus complex (6). More conclusive evidence derives from the in situ hybridization of a BAC clone spanning the 2L+a proximal breakpoint to two locations delimiting the 2La arrangement on the chromosomes of Anopheles stephensi; follow-up experiments involving BAC fragments derived from one side of the breakpoint yielded only a single site of hybridization (I.V.S. and M.V.S., unpublished data). The derived nature of the 2L+a arrangement and the ancestral status of 2La has led to a revised history of the A. gambiae complex in which A. arabiensis, fixed for 2La and second only to A. gambiae in importance as a malaria vector, is now considered the most likely ancestral species (6). The implication of the shift from the nonvector A. quadriannulatus to the major vector A. arabiensis as basal in the phylogeny of this species complex is that at least some of the traits that influence the potency of the vectorial system may have been present at its inception and that A. quadriannulatus may have lost vectorial capacity secondarily.
Our breakpoint data leave little doubt that the 2La arrangement is identical in A. arabiensis, A. gambiae, and A. merus. Previous circumstantial evidence to the contrary (10) may be a result of gene conversion events (or crossing over) occurring between alternative arrangements at some distance from the breakpoints. Moreover, available sequence from the 2L+a breakpoints in A. melas and A. quadriannulatus is consistent with a monophyletic origin of this rearrangement in the A. gambiae complex (Fig. 1). Additional sequencing from these species and from more isolates of A. gambiae will help clarify this issue. Assuming that these data reinforce a single origin of 2L+a, the distribution of this rearrangement will remain inconsistent with the phylogenetic hypothesis for the A. gambiae complex; resolution of the conflict requires at least one instance of genetic introgression between nonsister taxa.
Until now, inference of the role of the 2La polymorphism in A. gambiae s.s. ecology and dynamics has relied on cytogenetic analysis of half-gravid females, the only favorable source of polytene chromosomes. The elucidation of the 2La breakpoint sequences opens the possibility of “molecular karyotyping,” an approach that will offer greatly increased efficiency and flexibility to examine all preimaginal stages as well as adult males, where polytene chromosomes cannot be analyzed directly. The presence of repetitive DNA at the breakpoint has complicated development of a robust PCR assay to detect alternative karyotypes. A beta version of this assay provided results largely consistent with cytogenetic analysis when tested on >100 karyotyped specimens from Senegal, Mali, Burkina Faso, and Cameroon (Fig. 4, which is published as supporting information on the PNAS web site), but technical problems remain. Optimization for sensitivity and specificity, as well as large-scale validation, holds promise for an efficient and reliable tool to advance field studies of A. gambiae.
Materials and Methods
FISH.
To obtain polytene chromosome preparations, ovaries of half-gravid females (A. gambiae SUA and 4Arr colonies) were dissected into fresh Carnoy's solution (ethanol:glacial acetic acid at 3:1). Ovaries were gently pressed with a coverslip in 50% propionic acid, dipped in liquid nitrogen, and then dehydrated in 50%, 70%, 95%, and 100% ethanol. The quality of the banding pattern of polytene chromosomes was examined under a BX60 phase-contrast microscope (Olympus, Melville, NY). Probes were prepared from 1 μg of BAC DNA or 100 ng of PCR product labeled with Cy3-AP3-dUTP or Cy5-AP3-dUTP (where AP3 is 5-amino-propargyl) (Amersham Biosciences) by using the GIBCO/BRL BioPrime DNA labeling system (Life Technologies, Gaithersburg, MD) with dNTPs from the nick translation kit (Amersham Pharmacia) in half-volume reactions. The in situ hybridization was performed with the GIBCO/BRL in situ hybridization and detection system, following the manufacturer's recommended protocol. After hybridization, chromosomes were washed in 0.2× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), counterstained with YOYO-1 (Sigma), and mounted in diazabicyclo[2.2.2]octane antifade solution (Sigma). Fluorescent signals were detected by using a Bio-Rad MRC 1024 scanning confocal system (two channel, networked, and using the lasersharp 3.2 program).
Library Screening.
A. gambiae SUA 2La and A. merus V12 Lambda DASH II phage library screening was performed by using the following kits and reagents (Roche Applied Science, Indianapolis) according to protocols supplied by the manufacturer: DIG DNA Labeling Kit, Nylon Membranes for Colony and Plaque Hybridization, DIG Easy Hyb, DIG Wash and Block Buffer Set, CPD-Star, and Anti-Digoxigenin-AP. DNA from positive phage was isolated by using the Lambda Mini Kit (Qiagen, Valencia, CA).
The gridded A. gambiae Bamako 2La phosmid library (Lucigen, Middleton, WI) was screened by PCR amplification of plate, row, and column pools by using primers designed from A. gambiae SUA 2La breakpoint sequences (Table 1, which is published as supporting information on the PNAS web site). Primers for the distal breakpoint were 27A/23A; primers for the proximal breakpoint were BP8960F/BP8807R. PCRs contained 2.5 pmol of each primer, 150 μM dNTP, 3 mM MgCl2, 2.5 units of Taq polymerase, 1 μl of template DNA, and 1× buffer in a 50-μl volume. PCR conditions were 94°C for 5 min; 35 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s; 72°C for 5 min; and 4°C hold.
PCR Amplification of 2La and 2L+a Breakpoints.
Primers designed based on an alignment between available A. gambiae SUA 2La sequence and A. gambiae PEST 2L+a sequence were used to amplify 3–4 kb of sequence surrounding each 2La breakpoint from A. gambiae Bamako fosmids (proximal, 94I12; distal, 55I3) and from A. arabiensis genomic DNA (KGB colony). Multiple primer pairs were used to amplify overlapping fragments of each 2La breakpoint (Table 1). Proximal primer pairs were 8960F1/8960R, 8960F/8807R, and BP8807F1/BP8807R2. Distal primer pairs were 27A0/23A2 and 27A2/23A0. PCRs and conditions matched those used for library screening. PCR amplification of 2L+a breakpoints from the sibling species A. melas (BAL colony and wild specimens from Senegal) and A. quadriannulatus (SKUQUA colony) was attempted by using distal primers BP8807 and 27A2 and proximal primers 750Dupl and PestProx. Amplification of both breakpoints was successful from A. quadriannulatus, but only the distal breakpoint amplified from A. melas.
Sequencing.
Purified PCR products were directly sequenced by using standard T3, T7, or custom (Invitrogen) primers (available on request) and BigDye Terminator v3 chemistry (Applied Biosystems) on the ABI 3700 capillary sequencer (Applied Biosystems) as recommended by the manufacturer. Sequences have been deposited in the GenBank database (accession nos. DQ230889–DQ230901).
Bioinformatic Analysis.
The position of chromosome breakage and repetitive DNA segments within each 3–4 kb of SUA 2La breakpoint sequence was established by blastn searches implemented in the Ensembl A. gambiae genome browser, using SUA 2La sequences as queries. Ensembl gene predictions in the neighborhood of the breakpoints were “blasted” against the nr database at the National Center for Biotechnology Information to identify putative orthologs in other species. Sequences surrounding the breakpoints of 2La and 2L+a chromosomes were aligned by using clustalx 1.81 and seqman (DNASTAR, Madison, WI). Primers were designed based on these alignments by using primer3.
To explore the nature of the repetitive element adjacent to both breakpoints, we used its 163-bp consensus sequence as a query to initiate reiterative blastn searches of the A. gambiae genome, subsequently using a consensus of its top hits (including flanking DNA) as queries. This process was performed both manually and automatically by using the program tealign, which was developed by a team led by Z. Tu (Virginia Polytechnic Institute, Blacksburg). Briefly, tealign is an automated pipeline that links previously published programs (21) to find TEs in a genome, retrieve different TE copies and their flanking sequences, and perform clustal alignment. Of the hits recovered after the initial search, the first 28 had E values ≤4.4 × 10−9; the remainder were ≥0.0044. All 28 hits plus surrounding DNA shared sequence similarity beginning precisely with the sequence corresponding to the breakpoint-flanking repeats, suggesting that we had identified one terminus of a repetitive element in the A. gambiae genome. These 28 hits corresponded to 24 scaffolds, of which only 10 were mapped to chromosomes (5 to 2L, including the three breakpoints; 3 to 2R; and 2 to X). The short, repetitive character of most of these scaffolds hindered the successful identification of the other terminus; most sequences in the multiple alignment apparently terminated prematurely owing to sequencing gaps (Ns). The longest possible consensus sequence that could be reconstructed manually from scaffolds without sequencing gaps was ≈10 kb, after which sequence similarity ended. This consensus lacked evident long terminal or inverted repeats. Screening the consensus sequence against a reference repeat database (22) identified only fragments of an AMPLICON_AA repeat region, partial Merlin1_CB and HARBINGER1_AG transposons, and a nonautonomous SINEX-1_AG non-LTR retrotransposon. blastx searches at the National Center for Biotechnology Information and Ensembl revealed similarity to Haemophilus somnus 2336 transposase (GenBank accession no. ZP_00132108) in the sequence corresponding to Merlin1_CB transposon.
Supplementary Material
Acknowledgments
This paper is dedicated to Mario Coluzzi. We thank L. Goeddel, M. Fitzpatrick, and D. Williams for technical help; Z. Tu for sharing the tealign program; Z. Tu and Jim Hogan (University of Notre Dame) for sharing data from their TE databases; and Kathy Campbell for bioinformatics advice. This work was supported by National Institutes of Health Grants R01 AI44003 (to N.J.B.), R01 AI63508 (to N.J.B.), and U01 AI48846 (to F.H.C.).
Abbreviations
- s.s.
sensu stricto
- BAC
bacterial artificial chromosome
- TE
transposable element
- IDS
iduronate 2-sulfatase precursor
- ZNF
zinc finger protein.
Footnotes
References
- 1.Coghlan A., Eichler E. E., Oliver S. G., Paterson A. H., Stein L. Trends Genet. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Cornel A. J., Collins F. H. J. Hered. 2000;91:364–370. doi: 10.1093/jhered/91.5.364. [DOI] [PubMed] [Google Scholar]
- 3.Sharakhov I. V., Serazin A. C., Grushko O. G., Dana A., Lobo N., Hillenmeyer M. E., Westerman R., Romero-Severson J., Costantini C., Sagnon N., et al. Science. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- 4.Powell J. R., Petrarca V., della Torre A., Caccone A., Coluzzi M. Parassitologia (Rome) 1999;41:101–113. [PubMed] [Google Scholar]
- 5.Coluzzi M., Sabatini A., Della Torre A., Di Deco M. A., Petrarca V. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 6.Ayala F. J., Coluzzi M. Proc. Natl. Acad. Sci. USA. 2005;102(Suppl. 1):6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toure Y. T., Petrarca V., Traore S. F., Coulibaly A., Maiga H. M., Sankare O., Sow M., DiDeco M. A., Coluzzi M. Parassitologia (Rome) 1998;40:477–511. [PubMed] [Google Scholar]
- 8.Della Torre A., Tu Z., Petrarca V. Insect Biochem. Mol. Biol. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Goidts V., Szamalek J. M., de Jong P. J., Cooper D. N., Chuzhanova N., Hameister H., Kehrer-Sawatzki H. Genome Res. 2005;15:1232–1242. doi: 10.1101/gr.3732505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caccone A., Min G. S., Powell J. R. Genetics. 1998;150:807–814. doi: 10.1093/genetics/150.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrarca V., Beier J. C. Am. J. Trop. Med. Hyg. 1992;46:229–237. doi: 10.4269/ajtmh.1992.46.229. [DOI] [PubMed] [Google Scholar]
- 12.Holt R. A., Subramanian G. M., Halpern A., Sutton G. G., Charlab R., Nusskern D. R., Wincker P., Clark A. G., Ribeiro J. M., Wides R., et al. Science. 2002;298:129–149. [Google Scholar]
- 13.Matzkin L. M., Merritt T., Zhu C. T., Eanes W. F. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wesley C. S., Eanes W. F. Proc. Natl. Acad. Sci. USA. 1994;91:3132–3136. doi: 10.1073/pnas.91.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cirera S., Martin-Campos J. M., Segarra C., Aguade M. Genetics. 1995;139:321–326. doi: 10.1093/genetics/139.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathiopoulos K. D., della Torre A., Predazzi V., Petrarca V., Coluzzi M. Proc. Natl. Acad. Sci. USA. 1998;95:12444–12449. doi: 10.1073/pnas.95.21.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caceres M., Puig M., Ruiz A. Genome Res. 2001;11:1353–1364. doi: 10.1101/gr.174001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casals F., Caceres M., Ruiz A. Mol. Biol. Evol. 2003;20:674–685. doi: 10.1093/molbev/msg070. [DOI] [PubMed] [Google Scholar]
- 19.Richards S., Liu Y., Bettencourt B. R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M. J., Chen R., Meisel R. P., et al. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidwell M. G., Lisch D. R. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol; 2002. p. 1204. [Google Scholar]
- 21.Biedler J., Tu Z. Mol. Biol. Evol. 2003;20:1811–1825. doi: 10.1093/molbev/msg189. [DOI] [PubMed] [Google Scholar]
- 22.Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.