Abstract
Mutations in KAL1 and FGFR1 cause Kallmann syndrome (KS), whereas mutations in the GNRHR and GPR54 genes cause idiopathic hypogonadotropic hypogonadism with normal olfaction (nIHH). Mixed pedigrees containing both KS and nIHH have also been described; however, the genetic cause of these rare cases is unknown. We examined the FGFR1 gene in seven nIHH subjects who either belonged to a mixed pedigree (n = 5) or who had associated midline defects (n = 2). Heterozygous FGFR1 mutations were found in three of seven unrelated nIHH probands with normal MRI of the olfactory system: (i) G237S in an nIHH female and a KS brother; (ii) (P722H and N724K) in an nIHH male missing two teeth and his mother with isolated hyposmia; and (iii) Q680X in a nIHH male with cleft lip/palate and missing teeth, his brother with nIHH, and his father with delayed puberty. We show that these mutations lead to receptor loss-of-function. The Q680X leads to an inactive FGFR1, which lacks a major portion of the tyrosine kinase domain (TKD). The G237S mutation inhibits proper folding of D2 of the FGFR1 and likely leads to the loss of cell-surface expression of FGFR1. In contrast, the (P722H and N724K) double mutation causes structural perturbations in TKD, reducing the catalytic activity of TKD. We conclude that loss-of-function mutations in FGFR1 cause nIHH with normal MRI of the olfactory system. These mutations also account for some of the mixed pedigrees, thus challenging the current idea that KS and nIHH are distinct entities.
Keywords: normosmia, FGFR1
Idiopathic hypogonadotropic hypogonadism (IHH) occurs either with anosmia [Kallmann syndrome (KS)] (1) or with a normal sense of smell [normosmic IHH (nIHH)] (2). Although KS and nIHH are thought to represent two distinct entities, there are documented pedigrees containing both KS and nIHH (3, 4). Varying degrees of pubertal development exist in association with IHH, although the majority of patients lack evidence of puberty (5). In addition, nonreproductive phenotypes, such as synkinesia, hearing loss, labial or palatine cleft, and dental anomalies, are commonly reported in KS (6, 7) but can also occur in nIHH (8–10). In our experience, 11% of nIHH probands have associated phenotypes.
Genetic heterogeneity also exists in IHH. Approximately one-third of cases are familial, whereas the remainder are sporadic (11). Genetic defects have been mapped in <30% of the IHH population: (i) GNRHR and GPR54 mutations underlie nIHH (12–16), and (ii) KAL1 and FGFR1 mutations cause KS. Mutations in KAL1 cause the X-linked form of KS (17, 18) associated with a high frequency of renal anomalies and synkinesia (6), whereas mutations in FGFR1 are one cause of the autosomal dominant form of KS associated with cleft palate and dental agenesis (19–22).
Human FGFR1 is a member of the receptor tyrosine kinase superfamily. FGFR1 signaling regulates cell proliferation, migration, differentiation, and survival and thus is essential for various stages of human development (23). The prototypical FGFR comprises an extracellular region consisting of three Ig-like domains (D1, D2, and D3), a single transmembrane helix, and an intracellular region containing the tyrosine kinase domain (TKD). D2, D3, and the short D2–D3 linker of FGFR house all of the determinants of ligand (FGF) binding and specificity. In the presence of heparan sulfate (HS), FGF binds with high affinity to FGFR1 and induces receptor dimerization, which allows for the subsequent transautophosphorylation of tyrosine residues in the intracellular domain and activates downstream signaling pathways (24). Heterozygous mutations in FGFR1 have previously been identified within D1, D2, D3, and the TKD of the protein in subjects with KS (19–22). In addition, one KS patient with severe associated phenotypes (cleft palate, hearing loss, and agenesis of the corpus callosum) harbored a homozygous mutation in D2 (19).
To date, no genetic cause has been identified for the mixed pedigrees in which KS and nIHH subjects coexist. Variable expressivity (19, 22) within KS pedigrees harboring the same FGFR1 mutation led us to hypothesize that FGFR1 mutations might also account for both mixed pedigrees and nIHH with concurrent midline defects. Therefore, we expanded our genetic screening for FGFR1 to this cohort and performed genotype–phenotype correlations. Importantly, through structural and biochemical studies using recombinant FGFR1 proteins, we provide evidence that these FGFR1 mutations impair receptor activity.
Results
Molecular Analysis of FGFR1 Gene.
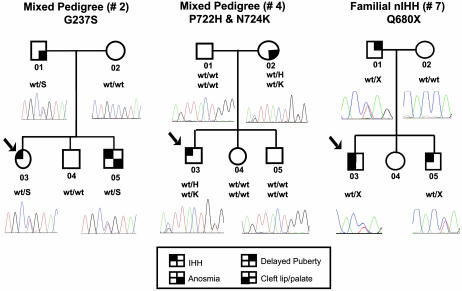
Four rare sequence variants of the FGFR1 gene were found in three unrelated probands (cases 2-03, 4-03, and 7-03) (Table 1 and Fig. 1). A transversion c.709 G → A in exon 6 is predicted to substitute a glycine for serine at position 237 (G237S) in D2 within the extracellular region of FGFR1 (case 2-03; see Figs. 1 and 2). Two rare sequence variants on the same allele, a transversion c.2165 C → A and a transition c.2172 C → G in exon 16, lead to a substitution of proline for histidine at position 722 (P722H) and asparagine for lysine at position 724 (N724K) in the TKD, respectively (case 4-03; see Figs. 1 and 3). Finally, a transversion c.2038 C → T in exon 15 leads to a stop codon (Q680X) in the TKD (case 7-03, Figs. 1 and 3). These changes, segregated appropriately within each pedigree (Fig. 1), were not detected in ethnically matched controls (340 Caucasian chromosomes and 130 Hispanic chromosomes), and were highly conserved both across species and within the family of FGFRs (Figs. 2B and 3B).
Table 1.
Clinical characteristics of the affected family members with an FGFR1 mutation
| Study subjects | G237S pedigree no. 2 |
P722H and N724K pedigree no. 4 |
Q680X pedigree no. 7 |
|||||
|---|---|---|---|---|---|---|---|---|
| Proband 2-03 | Father 2-01 | Brother 2-05 | Proband 4-03 | Mother 4-02 | Proband 7-03 | Father 7-01 | Brother 7-05 | |
| Gender | F | M | M | M | F | M | M | M |
| Diagnosis | nlHH | Anosmia | KS | nlHH | Hyposmia | nlHH | DP | nlHH |
| Sense of smell | Normal | Anosmia | Anosmia | Normal | Hyposmia | Normal | Normal | Normal |
| Formal smell test | 39/40 | 16/40 | 12/40 | 36/40 | 36/40 | 31/35 | ||
| History of puberty | Absent | Normal | Absent | Partial | Normal | Absent | Delayed | Absent |
| Cryptorchidism | None | Bilateral | Unilateral | None | None | None | ||
| Cleft lip/palate | No | No | No | No | No | No | Yes | No |
| Dental agenesis | No | No | No | Yes | No | No | Yes | No |
| MRI of the OB | Normal | Absent | Normal | Normal | ||||
DP, delayed puberty; F, female; M, male; OB, olfactory bulbs.
Fig. 1.
Detection of FGFR1 mutations in pedigree nos. 2, 4, and 7. Pedigree no. 2 reveals a mixed family, with both the proband and her brother carrying a heterozygous missense mutation in FGFR1 (G237S). Pedigree no. 4 reveals a mixed pedigree, with the proband and his mother carrying a heterozygous double mutation in FGFR1 (P722H and N724K). Pedigree no. 7 is consistent with familial nIHH and an autosomal dominant mode of inheritance. The proband, his brother, and his father carry a heterozygous mutation in FGFR1 (Q680X). The proband is identified by the arrow. Circles denote females; squares denote males. Phenotypes are as described in the text.
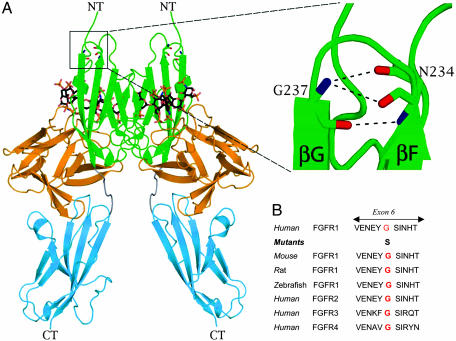
Fig. 2.
Mapping of the G237S mutation onto the crystal structure of the extracellular ligand-binding domain of FGFR1 in complex with FGF ligand and heparin oligosaccharide. (A) The G237S substitution maps to the βF–βG turn within D2 in the extracellular ligand binding region of FGFR1. A close-up view of the Gly-237 locus shows that both backbone nitrogen and carbonyl oxygen of Gly-237 engage in intramolecular hydrogen bonds, which play a critical role in shaping the local fold of the βF–βG turn of D2. Dashed lines represent hydrogen bonds. NT and CT, N terminus and C terminus of FGFR, respectively. FGF is colored orange, and receptor D2, D3, and D2–D3 linker are in green, cyan, and gray, respectively. Heparin oligosaccharides are rendered as sticks. Nitrogen, oxygen, and sulfur atoms are colored blue, red, and yellow, respectively. Carbon atoms of heparin oligosaccharides are colored black, and carbon atoms of the receptor are colored according to the subdomains to which they belong. This figure was made by using pymol (PyMOL Molecular Graphics System; DeLano Scientific, San Carlos, CA). (B) Comparison of the amino acid glycine at position 237 (red) across different species and within the FGFR family.
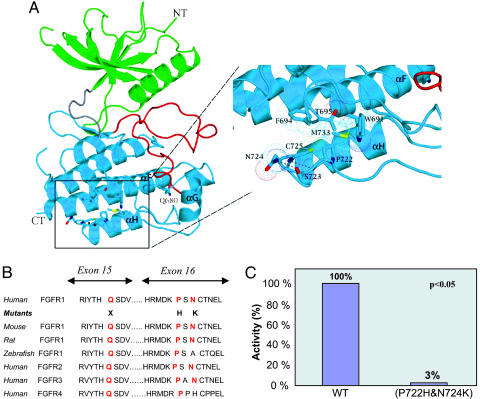
Fig. 3.
Structural and biochemical analysis of the Q680X mutation and the double (P722H and N724K) mutation in the FGFR1 gene. (A) The Q680X nonsense mutation and the (P722H and N724K) missense mutation map to the C-terminal lobe of the TKD. The Q680X mutation will delete a major portion of TKD. The (P722H and N724K) mutation maps to the loop region between αG and αH. These two residues play roles in shaping the conformation of this loop region and effectively preserving the catalytic activity the TKD. Note that the aliphatic side chain of P722 engages in numerous hydrophobic contacts with residues in αF and αH and indirectly contributes to the proper positioning of the residues at the active site of kinase domain. In contrast, Asn-724 is solvent exposed and engages in a single hydrogen bond with S723. Dashed lines represent hydrogen bonds. N-terminal and C-terminal kinase lobes are colored green and cyan, respectively. The hinge region and the regulatory activation loop of the kinase are colored gray and red, respectively. Nitrogen, oxygen, and sulfur atoms are colored blue, red, and yellow, respectively. Dotted spheres represent the van der Waals radius of selected residues. This figure was made by using pymol. (B) Comparison of the amino acid sequences, including arginine at position 680, proline at position 722, and asparagine at position 724 (red) across different species and within the FGFR family. (C) Tyrosine kinase activity of the double mutant FGFR1 (P722H and N724K) as compared with the wild-type FGFR1.
Genotype–Phenotype Correlations in the Pedigrees Found to Have FGFR1 Mutations.
Pedigree no. 2 (G237S).
The Caucasian female proband (case 2-03; see Table 1 and Fig. 1) presented at age 18 with primary amenorrhea, Tanner III breast and pubic hair development, and undetectable serum estradiol. She had no congenital abnormalities and normal olfaction (39/40) and denied eating disorder or excessive exercise. Imaging revealed normal olfactory bulbs and nerves. In response to a gonadotropin-releasing hormone (GnRH)-stimulation test, her luteinizing hormone (LH) increased to 5.2 units/liter and her follicle-stimulating hormone to 4.4 units/liter. We diagnosed her with partial nIHH and initiated hormone-replacement therapy.
The proband’s brother (case 2-05; see Fig. 1 and Table 1) was anosmic (12/40). He had microphallus, bilateral cryptorchidism and failed to go through puberty. At age 18, his testicular volume was prepubertal (2 ml), and imaging revealed absent olfactory bulbs. The patient was also diagnosed with attention-deficit disorder. The proband’s father (case 2-01; see Fig. 1 and Table 1) had congenital anosmia (16/40) with normal puberty. The mother (case 2-02) and brother (case 2-04) had a normal puberty.
Pedigree no. 4 (P722H and N724K).
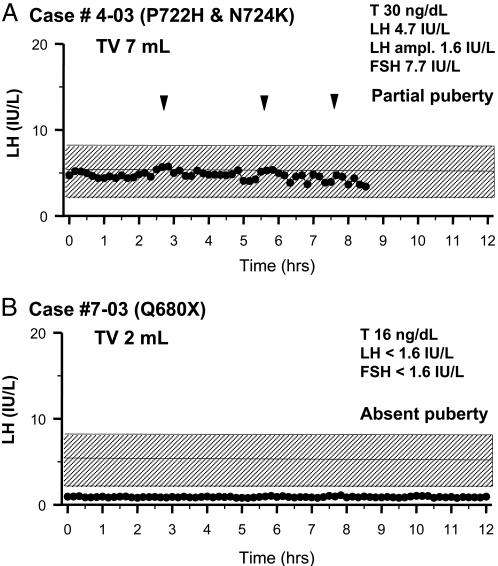
The proband (case 4-05; see Table 1 and Fig. 1) is a 25-yr-old Hispanic male with unilateral cryptorchidism and two congenital missing teeth. By age 14, he reported occasional morning erections, an increased libido, and ejaculations but no virilization. He grew steadily until age 18. At that time, he presented with knee pain. He was unvirilized and eunuchoidal (height of 190 cm; arm span of 201 cm). His testicular volume was 6 ml. He had short fourth metacarpal bones bilaterally and no synkinesia. He had severe bone loss (T score of −3.7 SD at the spine; T score of −3 SD at the hip) in the setting of a low serum testosterone (T) (50 ng/dl) and inappropriately normal gonadotropin levels. We diagnosed him with partial nIHH. The 12-h frequent sampling study revealed an enfeebled pattern of pulsatile LH secretion with mean LH of 4.7 units/liter (Fig. 4A). Additional exams included a formal smell test (36/40), normal MRI of the olfactory system, and renal ultrasound. His mother, who has congenital hyposmia (case 4-02; see Fig. 1 and Table 1), had normal puberty. His father (case 4-01), sister (case 4-04), and brother (case 4-05) had a normal puberty.
Fig. 4.
Neuroendocrine assessment of the proband with a double mutation (P722H and N724K) (A) and the proband with a nonsense mutation (Q680X) (B). An overnight 10-min sampling study for LH depicts endogenous LH secretion. Arrowheads indicate LH pulses, and the normal range of serum LH is hatched.
Pedigree no. 7 (Q680X).
The Caucasian male proband (case 7-03; see Table 1 and Fig. 1) was born with a cleft lip and palate, requiring several surgical procedures. He had three missing teeth and is red/green color blind. At age 18, he presented with a right slipped capital femoral epiphysis. He was eunuchoidal with a height of 181 cm and an arm span of 190 cm. He had absent facial hair, Tanner stage II–III axillary and pubic hair, and a testicular volume of 2 ml bilaterally (normal >12 ml). He had bilateral nystagmus on lateral gaze, and there was no evidence of synkinesia. His serum T was 16 ng/dl with undetectable gonadotropins and otherwise normal pituitary function. He had normal olfaction (36/40) and was diagnosed with a severe form of nIHH. His neuroendocrine evaluation revealed undetectable serum LH and follicle-stimulating hormone (Fig. 4B). A renal ultrasound and MRI of the olfactory system were normal, and a dual energy x-ray absortiometry scan revealed osteopenia of the spine (T score of −1.5) with normal bone density at the femoral neck (T score of −0.6).
The proband’s 19-yr-old brother (case 7-05; see Fig. 1 and Table 1) was diagnosed with absent puberty (prepubertal testes, hypogonadal serum T, and undetectable serum gonadotropins) but no associated phenotypes. He had a normal sense of smell (score of 31), consistent with nIHH. The proband’s father (case 7-01; see Fig. 1 and Table 1) reported having delayed puberty, with initiation of shaving at age 18, and no other associated phenotypes. The proband’s mother (case 7-02; see Fig. 1) and sister (case 7-04; see Fig. 1) experienced menarche at age 12 and both have regular menstrual cycles.
Structural and Biochemical Analysis of the FGFR1 Mutations Responsible for nIHH Show That These Mutations Result in Receptor Loss-of-Function.
The Q680X nonsense mutation maps to the beginning of the α-helix (αF) in the larger, mostly helical C-terminal lobe of the FGFR1 TKD (Fig. 3A). Hence, this mutation will result in the deletion of a catalytically essential portion of the TKD and lead to a “kinase dead” receptor.
The G237S mutation maps to the βF–βG turn in D2 of FGFR1 and therefore is distant from either the ligand or heparin binding sites (Fig. 2A). However, Gly-237 residue plays an important role in determining the conformation of the βF–βG turn and thus contributes to the overall structural integrity of D2. Specifically, both oxygen and nitrogen atoms of Gly-237 make hydrogen bonds that define the end and the beginning of β-strands F and G, respectively. Importantly, because of spatial constraints, only glycine (no side chain) is allowed at this location. Substitution to serine would cause steric conflicts at this region, resulting in the instability of βF–βG turn and ultimately destabilizing the entire D2 fold. The importance of the Gly-237 for D2 fold is underscored by the fact that this residue is invariant among all FGFRs (Fig. 2B). Like the wild-type protein, the G237S mutant extracellular domain was also expressed in large quantities in bacterial inclusion bodies; however, we were unable to refold in vitro the G237S mutant extracellular domain. Importantly, our inability to refold this mutant protein is consistent with our structural analysis, suggesting that the G237S mutation is harmful to D2 folding.
The (P722H and N724K) double mutation maps to the loop connecting the α-helices αG and αH in the FGFR1 TKD (Fig. 3A). Pro-722 is in a cis conformation, and its side chain is engaged in numerous hydrophobic contacts with residues from neighboring αF and αH helices at the bottom corner of the kinase domain. The P722H substitution should weaken these hydrophobic contacts and induce structural perturbations, which at the active site of kinase domain lead to a reduction in tyrosine kinase activity of FGFR1. In contrast, Asn-724 is solvent-exposed and appears to play a subtle role in preserving the loop conformation. Consistent with these structural observations, Pro-722 is invariably conserved among all FGFRs and across species, whereas Asn-724 is highly variable (Fig. 3B). In addition, the double mutant FGFR1 (P722H and N724K) exhibited a marked decreased in the tyrosine kinase activity in vitro as compared with the wild type (Fig. 3C), demonstrating that these mutations result in FGFR1 loss-of-function. It is noteworthy that the expression level of the mutant kinase was only slightly lower than that of the wild type, and the mutant kinase domain had the appropriate elution profiles both in anion-exchange and size-exclusion columns, suggesting that the mutation does not grossly affect the folding of the TKD. Hence, we predict that, unlike the G237S mutant, which likely will be retained in the endoplasmic reticulum because of misfolding, the (P722H and N724K) mutant should be matured appropriately and routed to the cell surface.
Discussion
Herein, we report three FGFR1 mutations in subjects with nIHH either with midline defects or belonging to mixed pedigrees containing both KS and nIHH. This report thus expands the phenotype of FGFR1 mutations beyond anosmic cases of IHH. Importantly, we present in vitro biochemical evidence that these FGFR1 mutations represent loss-of-function mutations.
A nonsense mutation (Q680X) in the catalytic TKD of FGFR1 was identified in pedigree no. 7. This mutation should result in the synthesis of a truncated inactive receptor lacking an essential portion of the catalytic TKD (Fig. 3A), which could act as a dominant negative mutant by forming heterodimers with the normal FGFR1 allele. Alternatively, the FGFR1 mRNA containing this termination codon could be degraded by way of nonsense-mediated decay, leading to haploinsufficiency (25). Pedigree no. 4 harbored the heterozygous double missense mutation (P722H and N724K) in the TKD of FGFR1. Structural and biochemical studies showed that this double mutation leads to loss-of-function of FGFR1 by reducing the tyrosine kinase activity of FGFR1 (Fig. 3 A and C). Finally, a missense mutation (G237S) in D2 of FGFR1 was identified in pedigree no. 2. Structural and in vitro biochemical studies revealed that this mutation leads to receptor loss-of-function by inhibiting the proper folding of D2 (Fig. 2A). Overall, our data, which show that these FGFR1 mutations in nIHH are loss-of-function mutations, agree with a report showing a deletion of one allele of the FGFR1 gene in two KS patients (19).
Deletions and nonsense and missense mutations in FGFR1 cause familial or sporadic KS (19–22). It is proposed that a reduction in FGF signaling abolishes olfactory bulb development, resulting in abnormal GnRH neuronal migration. Consistent with this hypothesis, targeted disruption of FGFR1 expression in mice telencephalon results in aplasia of the olfactory bulbs (26). Furthermore, MRIs have previously identified aplasia or hypoplasia of the olfactory bulb in KS patients carrying an FGFR1 mutation (20, 22).
Herein, we demonstrate that FGFR1 mutations can cause nIHH with apparent normal olfactory bulbs and normal quantitative smell tests. Interestingly, a male IHH patient with cleft lip and palate and without a history of frank anosmia was recently reported to harbor a reciprocal translocation t (7, 8) (p12.3, p11.2), disrupting the FGFR1 gene (27), although no formal smell test or MRI of the olfactory bulbs were performed in this patient. In subjects with nIHH, FGFR1 mutations may lead to an isolated defect in GnRH neuron migration. Indeed, disruptive GnRH neuronal migration in the presence of normal olfactory bulbs and sulci has been demonstrated in the knockout mouse model for another gene, Ebf2 (28). Alternatively, FGFR1 mutations could cause a defect in differentiation or survival of GnRH neurons within the hypothalamus. Notably, FGFR1 expression in the mice can be detected in the nasal placode, in developing olfactory bulbs, and along the GnRH migratory pathways, as well as in mature hypothalamic GnRH neurons and their projection (29). Furthermore, mice expressing dominant negative FGFR1 mutants targeted to GnRH neurons demonstrated a 30% reduction in the GnRH neuronal population of the hypothalamus (30). These mice, although fertile, display subtler reproductive phenotypes, such as delayed puberty and premature ovarian senescence. Thus, it is clear that more IHH subjects with normal olfaction should be screened for mutations in the FGFR1 gene to discover the role of FGFR1 in GnRH neuronal physiology and developmental biology.
This study also identifies previously unrecognized FGFR1 mutations in mixed pedigrees with both KS and nIHH. This finding is informative because it is thought that KS and nIHH are two distinct entities. KS is thought to result from embryonic defects in GnRH neuron migration associated with a disruption in olfactory bulb formation, whereas nIHH is thought to arise from defects in GnRH synthesis, processing, or action. Additionally, previous molecular genetic studies have identified different loci for KS (KAL1 and FGFR1) (17, 19, 31, 32) and nIHH (GNRHR and GPR54) (12–14). Therefore, our finding challenges the absolute nature of this clinical distinction and provides a genetic explanation for some of these mixed pedigrees.
Our data also reveals variable degrees of pubertal development within and across families carrying identical FGFR1 mutations ranging from absent puberty to partial puberty. This wide spectrum of reproductive phenotypes suggests an equally wide range of defects in endogenous GnRH secretion. It is thus tempting to hypothesize that FGFR1 mutations might therefore underlie other milder reproductive phenotypes, such as delayed puberty or perhaps hypothalamic amenorrhea.
Finally, this report confirms the association of IHH with dental anomalies and cleft lip/palate in IHH subjects carrying an FGFR1 mutation (19). FGFRs have been shown to play a key role in dental and palate development. FGFR1 and FGFR2 are differentially expressed in the mesenchyme and epithelium of fusing palatal shelves and in dental follicle development (33). Whereas loss-of-function mutations in FGFR1 cause IHH with a high frequency of cleft palate, somewhat paradoxically, gain-of-function mutations in FGFR2 cause cleft palate associated with craniosynostosis in both humans (33) and mouse models (34). Mice homozygous for a hypomorphic allele for FGFR1 display open palatine shelves in 80% of cases (35).
In summary, we have identified three heterozygous mutations in the FGFR1 gene in nIHH subjects with associated midline defects or belonging to mixed pedigrees. These results expand the phenotypic spectrum of FGFR1 mutations beyond KS and suggest a broader biologic role of FGFR1 in reproduction. Mutations in FGFR1 causing mixed pedigrees suggest that KS and nIHH are part of the same spectrum of disease. Furthermore, we present structural and in vitro biochemical evidences that these FGFR1 missense mutations causing IHH are loss-of-function mutations. Finally, the variable expressivity of the disease within pedigree harboring the same FGFR1 mutation suggests an interaction of FGFR1 with other gene products versus environmental influences on FGFR1 expression. More patients with nIHH should be screened to assess the frequency of heterozygous FGFR1 mutations in nIHH and to further define the role of FGFR1 in human reproduction.
Materials and Methods
Subjects.
IHH population.
Seven probands from unrelated pedigrees (five males and two females) with IHH and normal olfaction were included in this study. All subjects either had associated midline defects (n = 2) or belonged to a mixed pedigree (n = 5). IHH was characterized by (i) absent/incomplete puberty by age 18 yr in otherwise healthy patients; (ii) serum T ≤100 ng/dl in men or estradiol ≤20 pg/ml in women in association with low or normal levels of serum gonadotropins; (iii) otherwise normal pituitary function; (iv) normal serum ferritin concentrations; and (v) normal MRI of the hypothalamic-pituitary region. In addition, six of seven probands were tested for GNRHR mutations and were negative. If an FGFR1 mutation was found, affected and nonaffected parents and siblings of the proband were studied.
Controls.
The Caucasian control population consisted of 170 healthy Caucasian normal controls from Massachusetts General Hospital as assessed by history and clinical examination (n = 340 chromosomes). In addition, 115 healthy Mexican control subjects were selected either from an independently assembled cohort (200 chromosomes) (Coriell Laboratory, Camden, NJ) or from healthy Mexican controls from Massachusetts General Hospital (n = 30 chromosomes).
The study was approved by the Human Research Committee of Massachusetts General Hospital, and all subjects provided written informed consent before participation.
Mutation Analysis of the FGFR1 Gene.
Sequencing of the coding regions of the FGFR1 gene (GenBank accession no. BC018128) was performed as described in ref. 20. Amplified products were sequenced in both directions by using the AmpliTaq Dye Terminator Cycle Sequencing kit and an ABI Prism 377 DNA sequencer (PerkinElmer). Subsequent sequence analysis was performed by using mutation surveyor (Version 2.2; SoftGenetics, State College, PA) and visual inspection. All sequence variations were found on both strands and confirmed in a separate PCR. Nonsense changes resulting in a truncated protein, frameshift, insertion, or deletion were categorized as definite mutations. Nucleotide changes which were (i) absent from the dbSNP and ESTs; (ii) absent in 200 ethnically matched control chromosomes; (iii) evolutionarily conserved across species; and (v) segregated appropriately in the family were also identified as disease-causing mutations. All genes and proteins are described by using standard nomenclature (36).
Clinical and Biochemical Studies.
A detailed personal and family history of pubertal development (5) and associated nonreproductive phenotypes (i.e., dental agenesis, cleft lip/palate, hearing loss, synkinesia, etc.) was preformed. Families including KS or congenital anosmia and nIHH subjects were termed mixed pedigrees. A detailed physical examination, including Tanner staging for pubic and axillary hair and breast development, testicular volume by using a Prader Orchidometer, and mirror movement (37) were obtained. Olfactory acuity was assessed by history and confirmed with formal, quantitative smell testing [the University of Pennsylvania Smell Identification Test (UPSIT); Sensonics, Haddon Heights, NJ]. Thirty or more correct answers indicated a normal sense of smell (>5th percentile of 1,819 men and women between 15 and 50 yrs), 19–29 indicated hyposmia, and ≤18 indicated anosmia (38). In addition, a renal ultrasound and a dual energy x-ray absortiometry scan were performed. Cranial 3D MRI was performed to assess the olfactory system. The examination included routine axial Tesla 2 and fluid-attenuated inversion recovery images of the entire brain. This examination was followed by coronal 3D spoiled gradient recalled echo images with 1.5-mm-thick partitions that included the olfactory system and coronal 3-mm-thick Tesla 2 fast spin echo images focused on the frontal olfactory region from the posterior border of the frontal paranasal sinuses to the optic chiasma.
Five of the seven patients were admitted to the General Clinical Research Center of Massachusetts General Hospital for detailed phenotyping studies, including an overnight 12-h frequent blood-sampling study for LH (q10′ × 12 h) after withdrawal of hormonal therapy for a suitable washout period (typically 8 weeks). Pulsatile hormone secretion was analyzed by using a validated modification of the Santen and Bardin method (39, 40). The pool of the 12-h sampling was also assayed for follicle-stimulating hormone, T, estradiol, and inhibin B. In men able to produce an ejaculate, semen fluid analysis was performed according to World Health Organization guidelines.
Structural Analysis of the Effects of nIHH Mutations on FGFR1 Function.
The crystal structures of the FGFR1 kinase domain (Protein Data Bank entry 1FGK) (41) and the extracellular ligand binding region of FGFR1 in complex with FGF ligand and heparin oligosaccharide (Protein Data Bank entry 1FQ9) (42) were used to explore the effects of the mutations. Crystal structures were visualized by using the program o (43).
Protein Expression and Purification.
The bacterial expression construct for G237S mutant FGFR1c was prepared by introducing the mutation into the bacterial expression construct for the wild-type FGFR1c (residues 142–365). Bacterial expression and in vitro refolding of the mutant protein from inclusion bodies was carried out by using the same protocol as for the wild-type protein. The (P722H and N724K) mutation was introduced into the expression construct for the wild-type minimal kinase domain of FGFR1. Wild-type and mutant minimal kinase domains bearing an N-terminal polyhistidine tag were expressed and purified by using sequential Ni2+-chelating, anion exchange, and size-exclusion chromatographies as described in ref. 44.
Kinase Autophosphorylation Assay.
The tyrosine autophosphorylation activity of the wild-type and the (P722H and N724K) FGFR1 kinase domains were quantified by using a continuous spectrophotometric assay as described in ref. 45. In this assay, hydrolysis of ATP to ADP is coupled to the oxidation of NADH to NAD+ and measured as a reduction in NADH absorbance at 340 nm. Reactions were carried out at 30°C in 50 μl of buffer containing 100 mM Hepes-NaOH (pH 7.5), 1.0 mM ATP, 5 mM MgCl2, 1.5 mM phosphoenolpyruvate, 1.2 mg/ml NADH, 89 units/ml pyruvate kinase, 124 units/ml lactate dehydrogenase, and 4 μM recombinant wild-type or (P722H and N724K) FGFR1 kinase for 1 h.
Hormone Assays.
The hormonal assays for serum LH, follicle-stimulating hormone, T, estradiol, and inhibin B were performed as described in ref. 5.
Acknowledgments
We thank the nurses in the General Clinical Research Center (GCRC) of Massachusetts General Hospital for their excellent care and execution of the detailed neuroendocrine studies. This work was supported by National Institutes of Health Grants R01 HD15788-19, U54HD28138-14, R01 HD42708-03, and R01 DE13686-05 and National Center for Research Resources GCRC Program Grant MO1-RR-01066.
Abbreviations
- IHH
idiopathic hypogonadotropic hypogonadism
- GnRH
gonadotropin-releasing hormone
- KS
Kallmann syndrome
- LH
luteinizing hormone
- nIHH
normosmic IHH
- T
testosterone
- TKD
tyrosine kinase domain.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kallmann F. J., Schoenfeld W. A. Am. J. Ment. Defic. 1944;158:203–236. [Google Scholar]
- 2.Boyar R. M., Wu R. H., Kapen S., Hellman L., Weitzman E. D., Finkelstein J. W. J. Clin. Endocrinol. Metab. 1976;43:1268–1275. doi: 10.1210/jcem-43-6-1268. [DOI] [PubMed] [Google Scholar]
- 3.Seminara S. B., Hayes F. J., Crowley W. F., Jr Endocr. Rev. 1998;19:521–539. doi: 10.1210/edrv.19.5.0344. [DOI] [PubMed] [Google Scholar]
- 4.Quinton R., Duke V. M., de Zoysa P. A., Platts A. D., Valentine A., Kendall B., Pickman S., Kirk J. M. W., Besser G. M., Jacobs H. S., Bouloux P. M. G. J. Clin. Endocrinol. Metab. 1996;81:3010–3017. doi: 10.1210/jcem.81.8.8768867. [DOI] [PubMed] [Google Scholar]
- 5.Pitteloud N., Hayes F. J., Boepple P. A., DeCruz S., Seminara S. B., MacLaughlin D. T., Crowley W. F., Jr J. Clin. Endocrinol. Metab. 2002;87:152–160. doi: 10.1210/jcem.87.1.8131. [DOI] [PubMed] [Google Scholar]
- 6.Hardelin J. P., Levilliers J., Blanchard S., Carel J.-C., Leutenegger M., Pinard-Bertelletto J.-P., Bouloux P., Petit C. Hum. Mol. Genet. 1993;2:373–377. doi: 10.1093/hmg/2.4.373. [DOI] [PubMed] [Google Scholar]
- 7.Quinton R., Duke V. M., Robertson A., Kirk J. M., Matfin G., de Zoysa P. A., Azcona C., MacColl G. S., Jacobs H. S., Conway G. S., et al. Clin. Endocrinol. (Oxford) 2001;55:163–174. doi: 10.1046/j.1365-2265.2001.01277.x. [DOI] [PubMed] [Google Scholar]
- 8.Houang M., Gourmelen M., Moatti L., Le B. Y., Garabedian E. N., Denoyelle F. J. Pediatr. Endocrinol. Metab. 2002;15:219–223. doi: 10.1515/jpem.2002.15.2.219. [DOI] [PubMed] [Google Scholar]
- 9.Abs R., Raes D., Vercruysse H. J. Intern. Med. 1994;236:349–352. doi: 10.1111/j.1365-2796.1994.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 10.Shires R., Whyte M. P., Avioli L. V. Arch. Intern. Med. (Moscow) 1979;139:1187–1189. [PubMed] [Google Scholar]
- 11.Waldstreicher J., Seminara S. B., Jameson J. L., Geyer A., Nachtigall L. B., Boepple P. A., Holmes L. B., Crowley W. F., Jr J. Clin. Endocrinol. Metab. 1996;81:4388–4395. doi: 10.1210/jcem.81.12.8954047. [DOI] [PubMed] [Google Scholar]
- 12.de Roux N., Young J., Misrahi M., Genet R., Chanson P., Schaison G., Milgrom E. N. Engl. J. Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 13.de Roux N., Genin E., Carel J. C., Matsuda F., Chaussain J. L., Milgrom E. Proc. Natl. Acad. Sci. USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seminara S. B., Messager S., Chatzidaki E. E., Thresher R. R., Acierno J. S., Jr, Shagoury J. K., Bo-Abbas Y., Kuohung W., Schwinof K. M., Hendrick A. G., et al. N. Engl. J. Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 15.Messager S., Chatzidaki E. E., Ma D., Hendrick A. G., Zahn D., Dixon J., Thresher R. R., Malinge I., Lomet D., Carlton M. B., et al. Proc. Natl. Acad. Sci. USA. 2005;102:1761–1766. doi: 10.1073/pnas.0409330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahab M., Mastronardi C., Seminara S. B., Crowley W. F., Ojeda S. R., Plant T. M. Proc. Natl. Acad. Sci. USA. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legouis R., Hardelin J. P., Levilliers J., Claverie J. M., Compain S., Wunderle V., Millasseau P., Le Paslier D., Cohen D., Caterina D., et al. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 18.Franco B., Guioli S., Pragliola A., Incerti B., Bardoni B., Tonlorenzi R., Carrozzo R., Maestrini E., Pieretti M., Taillon-Miller P., et al. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 19.Dode C., Levilliers J., Dupont J. M., De Paepe A., Le Du N., Soussi-Yanicostas N., Coimbra R. S., Delmaghani S., Compain-Nouaille S., Baverel F., et al. Nat. Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 20.Sato N., Katsumata N., Kagami M., Hasegawa T., Hori N., Kawakita S., Minowada S., Shimotsuka A., Shishiba Y., Yokozawa M., et al. J. Clin. Endocrinol. Metab. 2004;89:1079–1088. doi: 10.1210/jc.2003-030476. [DOI] [PubMed] [Google Scholar]
- 21.Albuisson J., Pecheux C., Carel J. C., Lacombe D., Leheup B., Lapuzina P., Bouchard P., Legius E., Matthijs G., Wasniewska M., et al. Hum. Mutat. 2005;25:98–99. doi: 10.1002/humu.9298. [DOI] [PubMed] [Google Scholar]
- 22.Pitteloud N., Acierno J. S., Jr, Meysing A. U., Dwyer A. A., Hayes F. J., Crowley W. F., Jr J. Clin. Endocrinol. Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 23.Ford-Perriss M., Abud H., Murphy M. Clin. Exp. Pharmacol. Physiol. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- 24.Mohammadi M., Olsen S. K., Ibrahimi O. A. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Hebert J. M., Lin M., Partanen J., Rossant J., McConnell S. K. Development (Cambridge, U.K.) 2003;130:1101–1111. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- 26.Kim H. G., Herrick S. R., Lemyre E., Kishikawa S., Salisz J. A., Seminara S., MacDonald M. E., Bruns G. A., Morton C. C., Quade B. J., Gusella J. F. J. Med. Genet. 2005;42:666–672. doi: 10.1136/jmg.2004.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corradi A., Croci L., Broccoli V., Zecchini S., Previtali S., Wurst W., Amadio S., Maggi R., Quattrini A., Consalez G. G. Development (Cambridge, U.K.) 2003;130:401–410. doi: 10.1242/dev.00215. [DOI] [PubMed] [Google Scholar]
- 28.Gill J. C., Moenter S. M., Tsai P. S. Endocrinology. 2004;145:3830–3839. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 29.Tsai P. S., Moenter S. M., Postigo H. R., El Majdoubi M., Pak T. R., Gill J. C., Paruthiyil S., Werner S., Weiner R. I. Mol. Endocrinol. 2005;19:225–236. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- 30.Bick D. P., Schorderet D. F., Price P. A., Campbell L., Huff R. W., Shapiro L. J., Moore C. M. Prenatal Diagn. 1992;12:19–29. doi: 10.1002/pd.1970120104. [DOI] [PubMed] [Google Scholar]
- 31.Hardelin J. P., Levilliers J., del Castillo I., Cohen-Salmon M., Legouis R., Blanchard S., Compain S., Bouloux P., Kirk J., Moraine C., et al. Proc. Natl. Acad. Sci. USA. 1992;89:8190–8194. doi: 10.1073/pnas.89.17.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Britto J. A., Evans R. D., Hayward R. D., Jones B. M. Cleft Palate Craniofac. J. 2002;39:332–340. doi: 10.1597/1545-1569_2002_039_0332_tpoacp_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 33.Eswarakumar V. P., Horowitz M. C., Locklin R., Morriss-Kay G. M., Lonai P. Proc. Natl. Acad. Sci. USA. 2004;101:12555–12560. doi: 10.1073/pnas.0405031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trokovic N., Trokovic R., Mai P., Partanen J. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonarakis S. E. Hum. Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 36.Mayston M. J., Harrison L. M., Stephens J. A. Ann. Neurol. 1999;45:583–594. doi: 10.1002/1531-8249(199905)45:5<583::aid-ana6>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 37.Doty R. L., Applebaum S., Zusho H., Settle R. G. Neuropsychologia. 1985;23:667–672. doi: 10.1016/0028-3932(85)90067-3. [DOI] [PubMed] [Google Scholar]
- 38.Santen R. J., Bardin C. W. J. Clin. Invest. 1973;52:2617–2628. doi: 10.1172/JCI107454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes F. J., McNicholl D. J., Schoenfeld D., Marsh E. E., Hall J. E. J. Clin. Endocrinol. Metab. 1999;19:1028–1036. doi: 10.1210/jcem.84.3.5579. [DOI] [PubMed] [Google Scholar]
- 40.Mohammadi M., Schlessinger J., Hubbard S. R. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 41.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. Mol. Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 42.Jones E. Y., Walker N. P., Stuart D. I. Acta Crystallogr. A. 1991;47:753–770. doi: 10.1107/s0108767391006839. [DOI] [PubMed] [Google Scholar]
- 43.Mohammadi M., Dikic I., Sorokin A., Burgess W. H., Jaye M., Schlessinger J. Mol. Cell. Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barker S. C., Kassel D. B., Weigl D., Huang X., Luther M. A., Knight W. B. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 45.Maquat L. E., Carmichael G. G. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]