Abstract
The identification of mutations in genes that cause human diseases has largely been accomplished through the use of positional cloning, which relies on linkage mapping. In studies of rare diseases, the resolution of linkage mapping is limited by the number of available meioses and informative marker density. One recent advance is the development of high-density SNP microarrays for genotyping. The SNP arrays overcome low marker informativity by using a large number of markers to achieve greater coverage at finer resolution. We used SNP microarray genotyping for homozygosity mapping in a small consanguineous Israeli Bedouin family with autosomal recessive Bardet–Biedl syndrome (BBS; obesity, pigmentary retinopathy, polydactyly, hypogonadism, renal and cardiac abnormalities, and cognitive impairment) in which previous linkage studies using short tandem repeat polymorphisms failed to identify a disease locus. SNP genotyping revealed a homozygous candidate region. Mutation analysis in the region of homozygosity identified a conserved homozygous missense mutation in the TRIM32 gene, a gene coding for an E3 ubiquitin ligase. Functional analysis of this gene in zebrafish and expression correlation analyses among other BBS genes in an expression quantitative trait loci data set demonstrate that TRIM32 is a BBS gene. This study shows the value of high-density SNP genotyping for homozygosity mapping and the use of expression correlation data for evaluation of candidate genes and identifies the proteasome degradation pathway as a pathway involved in BBS.
Keywords: genetic mapping, obesity, SNP genotyping, zebrafish model
A continuing goal of the Human Genome Project is to determine the function of all human genes. Of particular interest is the identification of human phenotypes associated with the mutation of each gene. Although considerable progress has been made, the majority of genes causing complex and Mendelian disorders have yet to be identified. The discovery of most human disease-causing genes has relied on using large and/or multiple human pedigrees for genetic linkage mapping. The identification of additional disease-causing genes is hindered by the paucity of pedigrees that are suitable for traditional linkage studies. Therefore, opportunities to identify novel disease genes that take advantage of smaller pedigrees and genomic resources must be sought. One recent development that can aid in the identification of disease genes are high-density SNP arrays for genotyping (1). In this study, we used SNP genotyping of a small nuclear family to aid in the identification of a gene causing an extremely heterogeneous human obesity syndrome known as Bardet–Biedl syndrome (BBS; MIM 209900).
BBS is a pleiotropic, autosomal recessive disorder characterized by obesity, pigmentary retinopathy, polydactyly, renal abnormalities, learning disabilities, and hypogenitalism (2–4). The disorder is also associated with diabetes mellitus, hypertension, and congenital heart disease (2, 5, 6). The disorder displays extensive genetic heterogeneity. To date, nine BBS genes have been mapped and identified (7–21). Mutation screening of the known genes indicates that additional BBS genes and mutations have yet to be identified (21–23).
The initial identification of BBS genes relied on positional cloning (11–14). Subsequently, bioinformatic comparisons of protein sequences aided in the identification of additional BBS genes (15–17, 21). Further identification of BBS genes is hindered by the extensive genetic heterogeneity and the paucity of additional large multiplex families for genetic mapping. We used high-density SNP genotyping to identify the disease-causing gene in a single small BBS family in which previous linkage studies failed to identify a disease locus.
Results
Genomewide SNP Genotyping.
We performed short tandem repeat polymorphism (STRP) genotyping of an inbred Bedouin Arab family (Fig. 1A) by using 400 highly informative STRP markers. We assumed that the disease locus would be inherited homozygously by descent in affected individuals. No informative STRPs were homozygous in all four affected individuals.
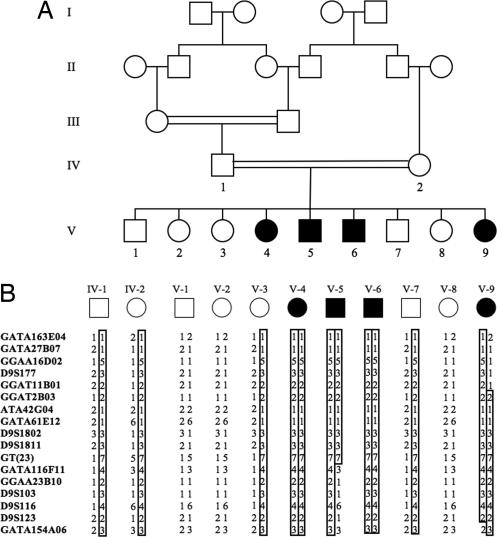
Fig. 1.
BBS11 pedigree and shared haplotype. (A) Pedigree of BBS family. Filled symbols indicate affected individuals. (B) Segregation of STRP haplotype in parents (IV-1 and IV-2) and offspring. The disease haplotype is indicated by the boxed genotypes. Recombinant events observed in affected individuals V-5 and V-9 define the interval.
To search further for homozygous regions consistent with linkage, the four affected members of the BBS family were genotyped with an Affymetrix (Santa Clara, CA) GeneChip probe array containing 57,244 SNPs. The SNP genotype call rate was >96%, and 32,631 (≈57%) SNP genotypes were homozygous in all four affected individuals, a finding reflecting the relative lack of informativity of SNP markers and the inbred nature of the pedigree. Fourteen autosomal regions were consistent with linkage based on homozygosity of 25 consecutive SNPs in the four affected siblings (Table 1).
Table 1.
Regions that are homozygous in all four affected Bedouin Arab siblings
| Chromosome band | Consecutive SNPs | Interval size, Mb |
|---|---|---|
| 9q33.1 | 83 | 2.40 |
| 16q16.3 | 50 | 0.96 |
| 10q23.1 | 42 | 0.90 |
| 2p22.1 | 34 | 1.20 |
| 8q13.3 | 34 | 0.69 |
| 3p26.3 | 32 | 0.53 |
| 2q21.1 | 30 | 2.96 |
| 2q24.3 | 30 | 1.00 |
| 4q21.22 | 30 | 0.67 |
| 9q31.1 | 29 | 0.95 |
| 6q16.1 | 28 | 0.69 |
| 4p15.1 | 26 | 1.24 |
| 9q31.1 | 26 | 1.14 |
| 7q11.22 | 25 | 1.25 |
Shown are the cytogenetic location, the number of consecutive homozygous SNPs, and the physical interval size.
We next genotyped the four affected patients, their unaffected siblings, and their parents with STRP markers that mapped within the 14 regions of apparent homozygosity identified by the SNP genotyping. Genotyping with informative STRPs excluded all but one region as being linked to the disease phenotype, a 2.4-Mb region containing 83 consecutive homozygous SNPs on chromosome 9q33.1 (Fig. 1B). Of interest, this region contained no STRPs from the original 400 STRPs that were used for linkage analysis. Logarithm of the odds score analysis using completely informative markers within the 2.4-Mb region reveals highly significant linkage with a maximum logarithm of the odds score of 3.7 (θ = 0).
Candidate Gene Analysis and Mutation Screening.
Analysis of the 2.4-Mb homozygous region on chromosome 9 reveals four RefSeq genes [pregnancy-associated plasma protein-A (PAPPA, Hs.13067), astrotactin 2 isoform a (ASTN2, Hs.209217), tripartite motif (TRIM)-containing protein 32 (TRIM32, Hs.195633), and Toll-like receptor 4 precursor (TLR4, Hs.174312)] and two placental-specific genes (DIPLA and DIPLAS). No gene within the linked interval stood out as the single-best candidate based on bioinformatic comparison with known BBS genes (18–21).
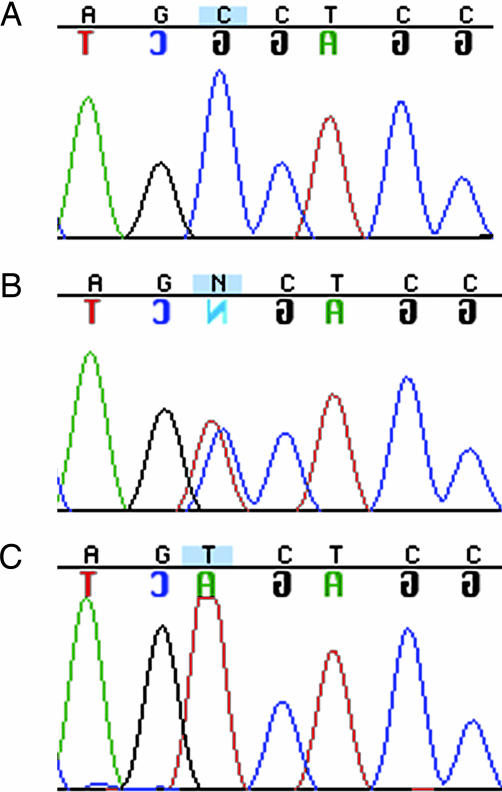
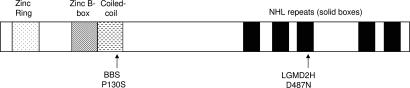
DNA sequencing of the entire coding sequence and consensus splice sites of the six genes within the 2.4-Mb interval revealed a single potential disease-causing variant in the four affected siblings, a homozygous transition (C388T) resulting in a proline to serine substitution at codon 130 (P130S) in TRIM32 (Fig. 2). The parents were heterozygous for the P130S allele, and all five unaffected siblings were either heterozygous for P130S or homozygous for the normal allele. No P130S alleles were detected in 184 control individuals, including 94 Bedouin Arab control individuals and 90 ethnic diversity controls. The proline residue at position 130 was found in a conserved B-box domain of TRIM32 (Fig. 3).
Fig. 2.
Representative TRIM32 sequence. (A) Normal proline homozygote at position 130 (CCT). (B) Heterozygous sequence. (C) Mutant serine homozygote (TCT).
Fig. 3.
Schematic diagram of TRIM32 (653 residues). N-terminal tripartite motif (zinc RING finger, zinc B-box, and coiled-coil domains) and five NHL repeats (solid boxes) are shown.
We also performed mutation screening by using single-strand conformational polymorphism analysis of the coding sequence in a panel of 90 BBS probands. No additional mutant alleles were found. Additional studies, described below, were performed to validate TRIM32 as a BBS gene.
TRIM32 Expression Is Strongly Correlated with Expression of Other BBS Genes.
The tissue expression pattern of TRIM32 has been reported (24–26) and is similar to the pattern of expression of other BBS genes. Expression of TRIM32 in the mammalian eye and hypothalamus has not been previously evaluated to our knowledge. We performed Northern blot analysis on RNA isolated from multiple mouse tissues including whole eye and hypothalamus with a 3′ UTR Trim32 probe. Our Northern blot results confirm an expression pattern similar to other BBS genes, including expression in the eye and hypothalamus (data not shown).
Recent studies in humans and animal models have used microarray expression data from thousands of genes in combination with genomewide polymorphism data to search for loci controlling variation in gene expression (27–29). This approach, known as expression quantitative trait loci (eQTL) mapping, demonstrates the correlation of expression of specific genes with specific genetic loci. We have recently performed a large-scale eQTL mapping study with a cross of 120 F2 rats genotyped with 400 STRPs across the rat genome to identify loci involved in regulation of thousands of genes expressed in the eye. In addition to eQTL mapping analysis, we performed a pairwise gene expression correlation analysis of the microarray expression data to identify genes whose expression levels are highly correlated among the 120 F2 animals. We hypothesized that the genetic permutations created by the mapping cross would allow the detection of functional relationships among genes because the regulatory mechanisms shared by related genes would likely cause their expression to respond to biological variations in a coordinated fashion.
The Affymetrix rat 230.20 chip containing ≈31,000 probe sets was used for the experiments, and ≈19,000 probe sets, including the nine known BBS genes and Trim32, were shown to be expressed in the eye and exhibit enough expression variation among the 120 F2 animals to allow for detection of significantly correlated expression. Evaluation of pairwise gene expression correlations in the eyes from the 120 F2 rats revealed that the expression levels of the nine known BBS genes were positively correlated with one another. Specifically, of the 36 possible pairwise comparisons of expression correlations among the nine BBS genes, all displayed positive correlation and 21 of the 36 comparisons were individually statistically significant (Table 2). The correlation among the nine known BBS genes was determined by comparing the mean multiple correlation coefficient of each gene individually to the other eight, and the significance of this value was assessed by comparing it to 10,000 randomly selected sets of nine genes. The result is highly significant (P = 0.0027). This finding leads to the hypothesis that expression of novel BBS genes should be positively correlated with the known BBS genes and suggests an approach for prioritizing candidate BBS genes. We then examined the pairwise gene expression variation correlation of each gene in the 2.4-Mb 9q candidate interval with the nine known BBS genes. The only gene demonstrating significant positive correlation with multiple BBS genes was Trim32 (Table 2). The significance of the correlation of Trim32 was determined to be P < 0.0001 based on a multiple correlation coefficient of 0.72 between Trim32 and the nine known BBS genes and after correcting for assessment of the multiple genes in the interval.
Table 2.
Pairwise (Pearson's) correlation expression values (among the 120 F2 rats analyzed with Affymetrix expression arrays) between the nine known BBS genes and four genes in the 9q33.1 candidate interval
| Gene name | Gene name |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBS1 | BBS2 | BBS3 | BBS4 | BBS5 | BBS6 | BBS7 | BBS8 | BBS9 | TRIM32 | PAPPA | ASTN2 | TLR4 | |
| BBS1 | 1 | 0.59 | 0.44 | 0.41 | 0.47 | 0.43 | 0.53 | 0.40 | 0.47 | 0.40 | −0.36 | −0.29 | 0.22 |
| BBS2 | 1 | 0.71 | 0.41 | 0.69 | 0.55 | 0.73 | 0.72 | 0.68 | 0.58 | −0.30 | −0.38 | 0.35 | |
| BBS3 | 1 | 0.31 | 0.82 | 0.34 | 0.78 | 0.77 | 0.57 | 0.60 | −0.17 | −0.18 | 0.28 | ||
| BBS4 | 1 | 0.54 | 0.25 | 0.62 | 0.23 | 0.31 | 0.23 | −0.08 | −0.25 | 0.23 | |||
| BBS5 | 1 | 0.34 | 0.79 | 0.65 | 0.52 | 0.63 | −0.22 | −0.28 | 0.30 | ||||
| BBS6 | 1 | 0.46 | 0.35 | 0.30 | 0.40 | −0.24 | −0.35 | 0.52 | |||||
| BBS7 | 1 | 0.65 | 0.57 | 0.53 | −0.16 | −0.32 | 0.38 | ||||||
| BBS8 | 1 | 0.58 | 0.62 | −0.25 | −0.15 | 0.24 | |||||||
| BBS9 | 1 | 0.49 | −0.37 | −0.30 | 0.10 | ||||||||
| TRIM32 | 1 | −0.44 | −0.34 | 0.43 | |||||||||
| PAPPA | 1 | 0.27 | −0.29 | ||||||||||
| ASTN2 | 1 | −0.50 | |||||||||||
| TLR4 | 1 | ||||||||||||
Empirically, correlation values >0.48 are significant at P < 0.05, and correlation values >0.64 are significant at P < 0.01.
Knockdown of TRIM32 in Zebrafish Reveals BBS Phenotypes.
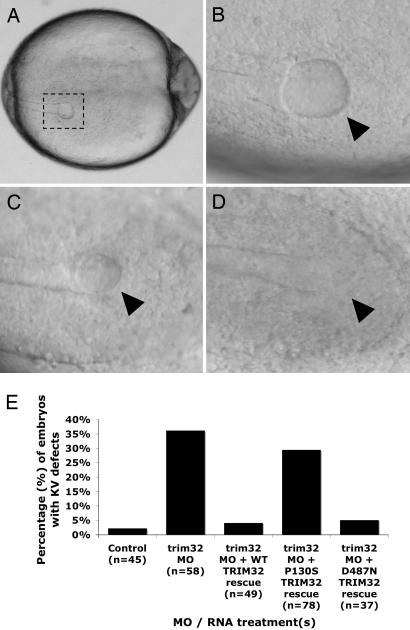
We have recently developed zebrafish models of BBS by using antisense morpholino oligonucleotides (MOs) to knock down the expression of BBS genes in developing zebrafish embryos (30). Two specific phenotypes were observed in common with individual knockdown of known BBS zebrafish orthologues (bbs1–bbs8): (i) disruption of Kupffer's vesicle (KV), a transient ciliated organ involved in left–right patterning, and (ii) delay of intracellular transport as determined by measuring the intracellular rate of retrograde melanosome transport (30). To determine whether knockdown of zebrafish trim32 results in similar defects, we identified and sequenced the zebrafish orthologue. The zebrafish trim32 is 62% identical and 75% similar to the human protein. Knockdown of zebrafish trim32 with an antisense MO flanking the initiator methionine resulted in 36% of fish having abnormal KV as defined by a reduced KV diameter compared with control-injected embryos (P < 0.0001) (Fig. 4). This finding is consistent with those observed with knockdown of other zebrafish BBS orthologues (range 25–40%) (30). In addition, similar to knockdown of other BBS genes, trim32-MO injected fish showed a delay in melanosome transport compared with controls (P < 0.0001) (Fig. 5). Both the KV and melanosome transport phenotypes were rescued when MOs were coinjected with normal human TRIM32 mRNA (P < 0.0001) (Figs. 4 and 5).
Fig. 4.
Representative KV phenotypes and summary of zebrafish trim32 knockdown. (A–D) Photographs of live zebrafish embryos at the 10- to 13-somite stage. (A) KV (dashed box) located in the posterior tailbud in a representative control-injected embryo. (B) Control KV (arrowhead). (C) trim32 MO-injected embryo with a reduced KV (arrowhead). (D) trim32 MO-injected embryo with no morphologically visible KV (arrowhead). (Magnifications: A, ×5; B–D, ×10.) (E) Percentage of zebrafish with altered KV (reduced or absent). MO refers to zebrafish trim32 antisense MO-injected embryos. In rescue experiments, WT, P130S, or D487N containing full-length trim32 mRNA was coinjected with the trim32 MO. Controls were injected with an MO containing mismatched bases to the trim32 sequence. Thirty-six percent of trim32 MO-injected embryos displayed KV defects, whereas only 2% of control-injected embryos exhibited KV defects (P < 0.0001). Both WT human TRIM32 (4%) and the D487N allele (11%) rescued the KV phenotype (not significantly different from controls); however, the P130S allele (30%) failed to rescue the KV phenotype (P < 0.0001 compared with controls).
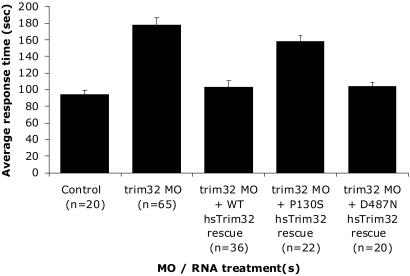
Fig. 5.
Summary of the melanosome transport assay in 5-day zebrafish embryos injected with trim32 MO with and without mRNA rescue. Control MO- and trim32 MO-injected embryos were observed for melanosome transport response time after epinephrine treatment. Embryos treated with trim32 MO alone showed an average response time of 178 s compared with an average 94-s response time for embryos treated with the control MO (P < 0.0001). Both WT human TRIM32 (103 s) and the D487N allele mRNA (103 s) rescued the melanosome transport defect (not significantly different from controls). The P130S allele (158 s) failed to rescue the transport defect (P < 0.0001 compared with controls).
Of interest, a single TRIM32 missense variant (D487N) has been reported to cause limb-girdle muscular dystrophy (LGMD) type 2H (LGMD2H) (Fig. 3) (26). To evaluate the known human TRIM32 variants as BBS-causing mutations, we generated expression constructs individually containing the BBS P130S allele and the LGMD2H D487N allele (Fig. 3A). Coinjection of the variant human mRNAs with the trim32 MO was performed to determine whether mutant variants could functionally rescue both the KV defects and the melanosome transport delay. Human TRIM32 mRNA containing the P130S variant failed to rescue both the KV defect and melanosome transport, indicating that the P130S variant results in an abnormal protein. Human TRIM32 mRNA containing the D487N variant successfully rescued both phenotypes (Figs. 4 and 5).
Discussion
The nine previously identified BBS genes account for approximately half of the known BBS cases, indicating that multiple additional BBS genes remain to be identified (21–23). The paucity of additional large BBS pedigrees and the extensive genetic heterogeneity make identification of additional BBS genes challenging. We used genomewide SNP genotyping to identify regions of homozygosity in a small consanguineous family in which lower-density genomewide STRP genotyping had failed to identify a linked locus. Genotyping of informative STRP markers within the key regions of homozygosity provided statistically significant (logarithm of the odds > 3) evidence of linkage to a single 2.4-Mb interval on chromosome 9q. Analysis of the genomic sequence of this interval revealed only six genes.
Independent of the data generated in this study, three separate lines of evidence suggest TRIM32 as the best BBS candidate gene in the 2.4-Mb interval. First, the expression pattern of TRIM32 is similar to the other known BBS genes (24–26). Second, there are three relevant knockout mouse models for genes within the linked interval: Pappa (31), Astn1 (paralogue of Astn2) (32), and Tlr4 (33). These models do not have phenotypes that resemble BBS mouse models (34–36). Finally, functional characterization of other TRIM proteins indicates involvement with components of the cytoskeleton, a finding consistent with the function of other BBS proteins (37–39). Nevertheless, we sequenced the entire coding regions and splice sites of the six genes in the 2.4-Mb interval. Sequencing revealed only one potential disease-causing mutation, P130S in TRIM32. The P130S allele was not detected in a screen of 184 control individuals. These data strongly suggest, but do not prove, that TRIM32 is a BBS gene. A screen of the TRIM32 coding sequence in 90 additional BBS probands failed to identify additional disease-causing mutations. This latter finding is not unexpected because of the extensive genetic heterogeneity of BBS and the fact that other recently discovered BBS genes each account for <2% of cases (16–20).
We provide additional evidence that demonstrates that TRIM32 is a BBS gene. First, expression variation of TRIM32 shows significant positive correlations with expression of the other known BBS genes. Second, knockdown of trim32 expression in zebrafish embryos exhibits phenotypes identical to those resulting from knockdown of the other known BBS genes (30). Human TRIM32 mRNA harboring the P130S variant fails to rescue the knockdown phenotype, indicating that the P130S variant is a disease-causing mutation. Collectively, the linkage data, mutation data in the linked family, gene expression correlation data, and functional data in the zebrafish model demonstrate that TRIM32 is a BBS gene (BBS11).
TRIM32 was first characterized in a yeast two-hybrid study screening for proteins that bind to the Tat protein, a protein that activates the transcription of lentiviruses (40). TRIM32 is a member of the TRIM family that is characterized by a common domain structure composed of a RING finger, a B-box, and a coiled-coil motif. TRIM32 also contains five C-terminal NHL repeats. The TRIM protein family participates in a variety of cellular processes, including apoptosis, cell growth, differentiation, transcriptional regulation, and ubiquitination. Recent studies show that TRIM32 has E3 ubiquitin ligase activity and binds to the head and neck region of myosin and ubiquitinates actin (41), implicating TRIM32 in regulating components of the cytoskeleton, a function that fits well with the observed zebrafish knockdown phenotypes (30).
Of note is a previous report that a single TRIM32 missense variant (D487N) is associated with autosomal recessive LGMD (28). There are many examples where different mutations in the same gene can result in different disorders (42–46). The TRIM32 LGMD mutation lies in a different domain (C-terminal NHL domain) than the BBS mutation (N-terminal B-box domain). A study of 37 members of the TRIM protein family has shown that ablation or disruption of N-terminal domains have differential subcellular localization effects than those observed with disruption of C-terminal domains (26). A recent study determined that the LGMD2H allele D487N did not affect the E3 ubiquitin ligase activity, whereas disruption of TRIM32 coiled-coil domain reduced the binding affinity to myosin (41). The hypothesis that different domains of TRIM32 may be involved in different processes is supported by our study of the two different mutations in the zebrafish model system. Although the LGMD2H D487N mRNA is able to rescue the zebrafish trim32 knockdown phenotypes, the P130S mRNA does not rescue the zebrafish knockdown phenotypes, indicating that the P130S mutation disrupts aspects of the protein function that are not affected by the D487N variant.
To our knowledge, TRIM32 is the first BBS gene identified to be involved in the ubiquitin/proteasome system. This system of protein degradation is a multistep cascade that relies on a series of enzymes to tag substrates with multiubiquitin for degradation (47–50). The third enzyme in this series, an E3 ubiquitin-protein ligase, of which there are many in the human genome, is involved in the recognition and transfer of ubiquitin to the protein substrate. Determination of substrate specificity provided by TRIM32 may help to explain the multiorgan system defects observed in BBS patients. Additional BBS genes may be either direct or downstream targets of TRIM32.
Besides resulting in the identification of a BBS gene, this study has demonstrated the effectiveness of higher-density SNP genotyping in identifying linked regions that are missed with lower-density STRP linkage data. There are large numbers of diseases that remain unmapped because of inadequate family resources available for traditional genetic linkage studies using STRP markers at 5- to 10-cM density. In addition, this study has demonstrated the utility of using expression correlation data generated from large-scale gene expression studies to aid in the identification and verification of disease genes.
Materials and Methods
Subjects.
Signed informed-consent forms, approved by the Institutional Review Board at the University of Iowa and collaborating institutions, were obtained from all study participants. The diagnosis of BBS was based on the presence of at least three of the following: obesity, polydactyly, renal anomalies, retinopathy, hypogonadism, and learning disabilities.
Genotyping.
STRP genotyping was performed as described (14). SNP genotyping was performed with the HindIII array of the Affymetrix GeneChip Mapping 100K set array. This array consists of 57,244 SNP markers with an average intermarker distance of 47.2 kb. Sample processing and labeling were performed by using the manufacturer's instructions. The arrays were hybridized, washed, and scanned in the University of Iowa DNA facility. Array images were processed with GeneChip DNA Analysis Software (gdas).
DNA Sequencing and Mutation Screening.
PCR products for sequencing were gel-purified with the QIAquick gel extraction kit (Qiagen, Valencia, CA). Sequencing was performed bidirectionally by using dye-terminator chemistry on an ABI 3730 DNA sequencer (Applied Biosystems). TRIM32 primer sequences are available on request.
For some patient DNA samples, the coding sequence of the TRIM32 gene was screened by single-strand conformational polymorphism analysis as described (14).
Zebrafish BBS Orthologues.
We performed blast analysis of human TRIM32 against the European Molecular Biology Laboratory/GenBank and genome project (Sanger Center, Cambridge, U.K.) databases to detect the homologous zebrafish sequence. Gene-specific primers were designed and used to amplify a full-length zebrafish trim32 cDNA sequence. PCR products were cloned into the pSTBlue vector (Novagen) and sequence-verified. Point mutations were introduced into full-length human TRIM32 cDNA clones by using targeted mutagenesis. The sequence of mutations and entire cDNA inserts was verified by sequencing and subcloned into the pCs2+ expression vector. mRNA was in vitro-transcribed with a mMessage kit (Ambion, Austin, TX) and coinjected into zebrafish embryos.
Antisense MOs.
Antisense MOs were designed and purchased from Gene Tools (Philomath, OR). MOs were microinjected into one to eight cell-staged embryos at a variety of concentrations (250, 100, and 50 μM). trim32 MO sequence is CAACATGGTTTAGGTTTAACTCCAT, and control MO sequence is GCTTTATTTGAGATCTCACTGCATCC.
Zebrafish Functional Assays.
Live somite staged embryos were photographed with a Zeiss Axiocam camera as described (30). KVs with a diameter less than or equal to half the WT mean diameter were considered abnormal. Day-5 fish were exposed to epinephrine added to embryo medium for a final concentration of 500 μg/ml. Melanosome transport was continuously monitored under the microscope, and the endpoint was scored when all melanosomes in the head and trunk were perinuclear.
Gene Expression Correlation Among Known BBS Genes and Trim32.
Two inbred strains of laboratory rats (SR/JrHsd and SHRSP) were crossed, and the resultant F1 animals were intercrossed. At 12 weeks of age, 120 healthy males of the resulting F2 generation were killed. Total RNA was extracted from both eyes by using the guanidinium isothiocyanate method (TRIzol reagent; Life Technologies, Gaithersburg, MD), followed by purification with an RNeasy column (Qiagen). Double-stranded cDNA was synthesized from 5 μg of total RNA with the Affymetrix GeneChip one-cycle target labeling kit. The resultant biotinylated cRNA was fragmented and hybridized to the GeneChip Rat Genome 230 2.0 Array containing 31,099 probes (Affymetrix). The arrays were washed, stained, and scanned with the Affymetrix model 450 fluidics station and model 3000 scanner by using the manufacturer's protocols at the University of Iowa DNA Core Facility. Values were generated by using the microarray suite (mas) Version 5.0 software (Affymetrix). The hybridizations were normalized by using the robust multichip averaging method to obtain summary expression values for each probe set (51). Regression, ANOVA (including t tests), and the Mann–Whitney–Wilcoxon rank test were used to identify differentially expressed genes. Cluster analysis was used to find coregulated genes with similar expression profiles.
Acknowledgments
We thank the families for participating in this study; T. Bair, G. Beck, K. Bugge, A. L. Ferguson, T. Kucaba, and C. Searby for technical assistance; and Drs. J. Wei and K. Wang for helpful discussions. This work was supported by National Institutes of Health Grants P50-HL-55006 (to V.C.S.) and R01-EY-11298 (to E.M.S. and V.C.S.), the Carver Endowment for Molecular Ophthalmology (E.M.S. and V.C.S.), and Research to Prevent Blindness, New York (Department of Ophthalmology, University of Iowa). T.E.S. was supported by a career development award from Research to Prevent Blindness. E.M.S. and V.C.S. are Investigators of the Howard Hughes Medical Institute.
Abbreviations
- BBS
Bardet–Biedl syndrome
- STRP
short tandem repeat polymorphism
- TRIM
tripartite motif
- TRIM32
TRIM-containing protein 32
- MO
morpholino oligonucleotide
- KV
Kupffer's vesicle
- LGMD
limb-girdle muscular dystrophy
- LGMD2H
LGMD type 2H.
Note Added in Proof.
A report of a 10th BBS (52) appeared while this article was going to press, hence we refer to the TRIM32 as BBS11.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Syvanen A. C. Nat. Genet. 2005;37(Suppl.):S5–S10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 2.Green J. S., Parfrey P. S., Harnett J. D., Farid N. R., Cramer B. C., Johnson G., Heath O., McManamon P. J., O'Leary E., Pryse-Phillips W. N. Engl. J. Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- 3.Bardet G. Ph.D. thesis. Paris: University of Paris; 1920. [Google Scholar]
- 4.Biedl A. Dtsch. Med. Wschr. 1922;48:1630. [Google Scholar]
- 5.Harnett J. D., Green J. S., Cramer B. C., Johnson G., Chafe L., McManamon P., Farid N. R., Pryse-Phillips W., Parfrey P. S. N. Engl. J. Med. 1988;319:615–618. doi: 10.1056/NEJM198809083191005. [DOI] [PubMed] [Google Scholar]
- 6.Elbedour K., Zucker N., Zalzstein E., Barki Y., Carmi R. Am. J. Med. Genet. 1994;52:164–169. doi: 10.1002/ajmg.1320520208. [DOI] [PubMed] [Google Scholar]
- 7.Kwitek-Black A. E., Carmi R., Duyk G. M., Buetow K. H., Elbedour K., Parvari R., Yandava C. N., Stone E. M., Sheffield V. C. Nat. Genet. 1993;5:392–396. doi: 10.1038/ng1293-392. [DOI] [PubMed] [Google Scholar]
- 8.Sheffield V. C., Carmi R., Kwitek-Black A., Rokhlina T., Nishimura D., Duyk G. M., Elbedour K., Sunden S. L., Stone E. M. Hum. Mol. Genet. 1994;3:1331–1335. doi: 10.1093/hmg/3.8.1331. [DOI] [PubMed] [Google Scholar]
- 9.Carmi R., Elbedour K., Stone E. M., Sheffield V. C. Am. J. Med. Genet. 1995;59:199–203. doi: 10.1002/ajmg.1320590216. [DOI] [PubMed] [Google Scholar]
- 10.Young T. L., Penney L., Woods M. O., Parfrey P. S., Green J. S., Hefferton D., Davidson W. S. Am. J. Hum. Genet. 1999;64:900–904. doi: 10.1086/302301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsanis N., Beales P. L., Woods M. O., Lewis R. A., Green J. S., Parfrey P. S., Ansley S. J., Davidson W. S., Lupski J. R. Nat. Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- 12.Slavotinek A. M., Biesecker L. G. Am. J. Med. Genet. 2000;95:208–215. [PubMed] [Google Scholar]
- 13.Mykytyn K., Braun T., Carmi R., Haider N. B., Searby C. C., Shastri M., Beck G., Wright A. F., Iannaccone A., Elbedour K., et al. Nat. Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura D. Y., Searby C. C., Carmi R., Elbedour K., Van Maldergem L., Fulton A. B., Lam B. L., Powell B. R., Swiderski R. E., Bugge K. E., et al. Hum. Mol. Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 15.Mykytyn K., Nishimura D. Y., Searby C. C., Shastri M., Yen H. J., Beck J. S., Braun T., Streb L. M., Cornier A. S., Cox G. F., et al. Nat. Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 16.Ansley S. J., Badano J. L., Blacque O. E., Hill J., Hoskins B. E., Leitch C. C., Kim J. C., Ross A. J., Eichers E. R., Teslovich T. M., et al. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 17.Badano J. L., Ansley S. J., Leitch C. C., Lewis R. A., Lupski J. R., Katsanis N. Am. J. Hum. Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang A. P., Nishimura D., Searby C., Elbedour K., Carmi R., Ferguson A. L., Secrist J., Braun T., Casavant T., Stone E. M., Sheffield V. C. Am. J. Hum. Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y., Esmail M. A., Ansley S. J., Blacque O. E., Boroevich K., Ross A. J., Moore S. J., Badano J. L., May-Simera H., Compton D. S., et al. Nat. Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 20.Li J. B., Gerdes J. M., Haycraft C. J., Fan Y., Teslovich T. M., May-Simera H., Li H., Blacque O. E., Li L., Leitch C. C., et al. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura D. Y., Swiderski R. E., Searby C. C., Berg E. M., Ferguson A. L., Hennekam R., Merin S., Weleber R. G., Biesecker L. G., Stone E. M., Sheffield V. C. Am. J. Hum. Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsanis N. Hum. Mol. Genet. 2004;13:R65–R71. doi: 10.1093/hmg/ddh092. Spec. No. 1. [DOI] [PubMed] [Google Scholar]
- 23.Hichri H., Stoetzel C., Laurier V., Caron S., Sigaudy S., Sarda P., Hamel C., Martin-Coignard D., Gilles M., Leheup B., et al. Eur. J. Hum. Genet. 2005;13:607–616. doi: 10.1038/sj.ejhg.5201372. [DOI] [PubMed] [Google Scholar]
- 24.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horn E. J., Albor A., Liu Y., El-Hizawi S., Vanderbeek G. E., Babcock M., Bowden G. T., Hennings H., Lozano G., Weinberg W. C., Kulesz-Martin M. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 26.Frosk P., Weiler T., Nylen E., Sudha T., Greenberg C. R., Morgan K., Fujiwara T. M., Wrogemann K. Am. J. Hum. Genet. 2002;70:663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brem R. B., Yvert G., Clinton R., Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 28.Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N., Colinayo V., Ruff T. G., Milligan S. B., Lamb J. R., Cavet G., et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 29.Morley M., Molony C. M., Weber T. M., Devlin J. L., Ewens K. G., Spielman R. S., Cheung V. G. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yen H. J., Tayeh M. K., Mullins R. F., Stone E. M., Sheffield V. C., Slusarski D. C. Hum. Mol. Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 31.Conover C. A., Bale L. K., Overgaard M. T., Johnstone E. W., Laursen U. H., Fuchtbauer E. M., Oxvig C., van Deursen J. Development (Cambridge, U.K.) 2004;131:1187–1194. doi: 10.1242/dev.00997. [DOI] [PubMed] [Google Scholar]
- 32.Adams N. C., Tomoda T., Cooper M., Dietz G., Hatten M. E. Development (Cambridge, U.K.) 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- 33.Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 34.Mykytyn K., Mullins R. F., Andrews M., Chiang A. P., Swiderski R. E., Yang B., Braun T., Casavant T., Stone E. M., Sheffield V. C. Proc. Natl. Acad. Sci. USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura D. Y., Fath M., Mullins R. F., Searby C., Andrews M., Davis R., Andorf J. L., Mykytyn K., Swiderski R. E., Yang B., et al. Proc. Natl. Acad. Sci. USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fath M. A., Mullins R. F., Searby C., Nishimura D. Y., Wei J., Rahmouni K., Davis R. E., Tayeh M. K., Andrews M., Yang B., et al. Hum. Mol. Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 37.Kim J. C., Badano J. L., Sibold S., Esmail M. A., Hill J., Hoskins B. E., Leitch C. C., Venner K., Ansley S. J., Ross A. J., et al. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 38.Kulaga H. M., Leitch C. C., Eichers E. R., Badano J. L., Lesemann A., Hoskins B. E., Lupski J. R., Beales P. L., Reed R. R., Katsanis N. Nat. Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 39.Blacque O. E., Reardon M. J., Li C., McCarthy J., Mahjoub M. R., Ansley S. J., Badano J. L., Mah A. K., Beales P. L., Davidson W. S., et al. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fridell R. A., Harding L. S., Bogerd H. P., Cullen B. R. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 41.Kudryashova E., Kudryashov D., Kramerova I., Spencer M. J. J. Mol. Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 42.Bonne G., Di Barletta M. R., Varnous S., Becane H. M., Hammouda E. H., Merlini L., Muntoni F., Greenberg C. R., Gary F., Urtizberea J. A., et al. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 43.Cao H., Hegele R. A. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 44.Eriksson M., Brown W. T., Gordon L. B., Glynn M. W., Singer J., Scott L., Erdos M. R., Robbins C. M., Moses T. Y., Berglund P., et al. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muchir A., Bonne G., van der Kooi A. J., van Meegen M., Baas F., Bolhuis P. A., de Visser M., Schwartz K. Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 46.Speckman R. A., Garg A., Du F., Bennett L., Veile R., Arioglu E., Taylor S. I., Lovett M., Bowcock A. M. Am. J. Hum. Genet. 2000;66:1192–1198. doi: 10.1086/302836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciechanover A. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 48.Ciechanover A. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 49.Hershko A. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 50.Rose I. Cell Death Differ. 2005;12:1198–1201. doi: 10.1038/sj.cdd.4401710. [DOI] [PubMed] [Google Scholar]
- 51.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K. J., Scherf U., Speed T. P. Biostatstics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 52.Stoetzel C., Laurier V., Davis E. E., Muller J., Rix S., Badano J. L., Leitch C. C., Salem N., Chouery E., Corbani S., et al. Nat. Genet. 2006 Apr 2; doi: 10.1038/ng1771. 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]