Abstract
TRIF-related adaptor molecule (TRAM) is the fourth Toll/IL-1 resistance domain-containing adaptor to be described that participates in Toll-like receptor (TLR) signaling. TRAM functions exclusively in the TLR4 pathway. Here we show by confocal microscopy that TRAM is localized in the plasma membrane and the Golgi apparatus, where it colocalizes with TLR4. Membrane localization of TRAM is the result of myristoylation because mutation of a predicted myristoylation site in TRAM (TRAM-G2A) brought about dissociation of TRAM from the membrane and its relocation to the cytosol. Further, TRAM, but not TRAM-G2A, was radiolabeled with [3H]myristate in vivo. Unlike wild-type TRAM, overexpression of TRAM-G2A failed to elicit either IFN regulatory factor 3 or NF-κB signaling. Moreover, TRAM-G2A was unable to reconstitute LPS responses in bone marrow-derived macrophages from TRAM-deficient mice. These observations provide clear evidence that the myristoylation of TRAM targets it to the plasma membrane, where it is essential for LPS responses through the TLR4 signal transduction pathway, and suggest a hitherto unappreciated manner in which LPS responses can be regulated.
Keywords: innate immunity, lipopolysaccharide
Toll-like receptors (TLRs) recognize microbial products derived from all of the major classes of pathogens and initiate a complex immune response designed to eliminate invading pathogens. A key structural motif involved in the signal transduction of TLRs is the Toll/IL-1 resistance (TIR) domain (for review, see ref. 1). TIR domains can be found in the cytoplasmic portions of all TLRs and the IL-1 receptor family as well as a third subgroup of TIR domain-containing adaptor proteins. The initial events of TLR signal transduction are thought to involve the recruitment of pertinent adaptor molecules, which in turn provide a scaffold to enable the recruitment and activation of additional signaling molecules. To date, four adaptor molecules have been associated with TLR signaling: myeloid differentiation factor 88 (MyD88) (2, 3); MyD88 adaptor-like (Mal) (4), also called TIR domain-containing adaptor protein (TIRAP) (5, 6); TIR domain-containing adaptor-inducing IFN-β (TRIF) (7–9), also called TIR domain-containing adaptor molecule 1 (TICAM-1) (10); and TRIF-related adaptor molecule (TRAM), or TICAM-2 (11–13) (for review, see ref. 14). Exactly how the adaptor molecules initiate signaling to these pathways once a TLR is triggered by its ligand is still unclear. This event is particularly complicated in the case of TLR4, in which all four of the above adaptors are required for a complete LPS response.
LPS (endotoxin) is the major constituent of the outer membrane of Gram-negative bacteria (15, 16). The recognition of LPS by host phagocytes stimulates the release of inflammatory mediators and cytokines from a variety of target cells. In conjunction with CD14 and its coreceptor, MD-2, TLR4 binds LPS and serves to elicit a “danger” signal, thus initiating the host immune response (for a complete review, see ref. 17). The cytoplasmic face of TLR4 is quite unique among TLRs because it utilizes all four TIR domain-containing adaptor molecules: MyD88, Mal, TRIF, and TRAM (18). In recent years, considerable progress has been made delineating the requirement for each of these adaptor molecules in the TLR4-activated immune response. The recruitment of MyD88 to proximal TIR domains of activated TLRs allows for the interaction and activation of the IRAK family members, IRAK1 and IRAK4 (19, 20), and the subsequent activation of TNF receptor-associated factor 6 [TRAF-6 (21)], a RING finger domain-containing protein with ubiquitin ligase activity. TAK1 kinase in turn activates the IKK complex (22, 23), which phosphorylates IκBα. This phosphorylation marks IκB for ubiquitination and degradation by the proteosome. NF-κB is then released, translocates to the nucleus, and regulates κB target genes, including inflammatory cytokines. TLR4 also activates MyD88-independent responses, which lead to the activation of IFN regulatory factor (IRF) 3, the induction of IFN-β and IFN-inducible genes, and the up-regulation of costimulatory molecules (18, 24–26). Mal/TIRAP appears also to regulate inflammatory cytokine genes, suggesting that Mal may cooperate with MyD88 to control these responses (4–6, 27).
Using an RNA interference approach or gene targeting, several reports have suggested that TRAM is uniquely required in the TLR4 signal transduction pathway and together with TRIF coordinates the activation of IRF3 and the MyD88-independent responses outlined above (9, 11, 28, 29). The TRAM–TRIF module also controls MyD88-dependent inflammatory responses, suggesting that TRAM is a master regulator of both arms of the TLR4 signaling pathway. Our earlier studies with overexpression systems suggested that TRAM functioned upstream of TRIF in TLR4 signaling (11). This conclusion was supported by the observation that TRAM binds TRIF directly and recruits it to TLR4 (12) as well as by the observation that TRIF dominant-negative constructs eliminated the direct activation of the MyD88-independent pathway by TRAM, but not vice versa (12). In the case of TLR3 signaling, where TRAM is not required, TRIF binds directly to the TLR3 TIR domain (10).
To characterize further the unique role of TRAM in the initiation of TLR4 signaling, we analyzed the subcellular localization of TRAM and the consequences of TRAM localization for efficient signal transduction. Our study shows that TRAM is localized in the plasma membrane and Golgi apparatus by N-terminal myristoylation, where it colocalizes with TLR4. In fact, TRAM contains a putative N-terminal myristoylation site, similar to that found in mammalian Src kinases. Mutation of this predicted myristoylation site (TRAM-G2A) redistributes TRAM from the plasma membrane to the cytosol. TRAM-G2A did not signal on overexpression and was unable to reconstitute LPS responses in macrophages deficient for wild-type TRAM. These results indicate that myristoylation and plasma membrane localization of TRAM are critical for responses to LPS and also indicate the potential for a hitherto unappreciated mechanism of regulation of LPS responses.
Results
Subcellular Localization of TRAM.
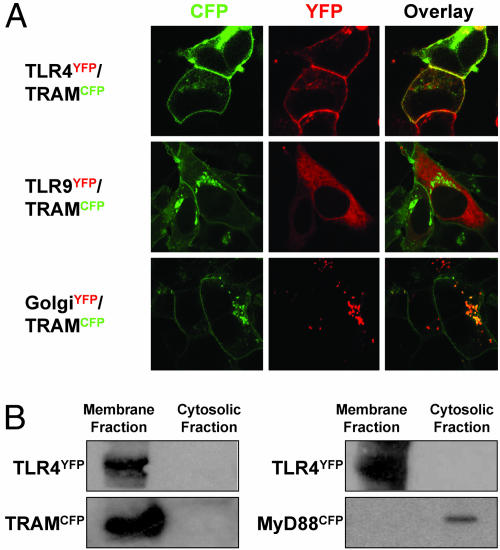
To understand further the role of TRAM in TLR4 signaling, we examined its subcellular localization. By using a C-terminal cyan fluorescent protein (CFP)-tagged version of TRAM (TRAMCFP), we discovered that in resting cells, TRAM resides at the plasma membrane and also in the Golgi apparatus of human embryonic kidney (HEK)293 cells. Similar localization patterns were observed in stable cell lines expressing TLR4 tagged with yellow fluorescent protein (TLR4YFP). TLR4 was both surface-expressed as well as localized in the Golgi apparatus, consistent with previous reports (30). In fact, TRAM and TLR4 appeared to colocalize on the plasma membrane (Fig. 1A, Right, Overlay). There was no colocalization of TRAM with TLR9 (Fig. 1A), which resides in the endoplasmic reticulum (31). In addition to the plasma membrane, TRAM appears to localize in the Golgi apparatus as seen in Fig. 1A (Bottom). Cell fractionation studies in TRAM-expressing HEK293 cells revealed that TRAM is found exclusively in the membrane fraction, in sharp contrast to the localization of MyD88, which resides in the cytosolic fraction (Fig. 1B).
Fig. 1.
TRAM is associated with the cell membrane and Golgi apparatus. (A) Stable TLR4YFP- or TLR9YFP-expressing HEK293 cells were transiently transfected with TRAMCFP. In addition, HEK293 cells were cotransfected with TRAMCFP and β-galactosyltransferaseYFP (GolgiYFP). Twenty-four hours after transfection, cells were visualized by confocal microscopy. (B) Membrane fractionation was carried out on HEK293 cells stably expressing TLR4YFP and either TRAMCFP or MyD88CFP. The fractions were resolved on an SDS/10% polyacrylamide gel before transfer to nitrocellulose and immunoblotting.
TRAM Contains a Putative Myristoylation Site.
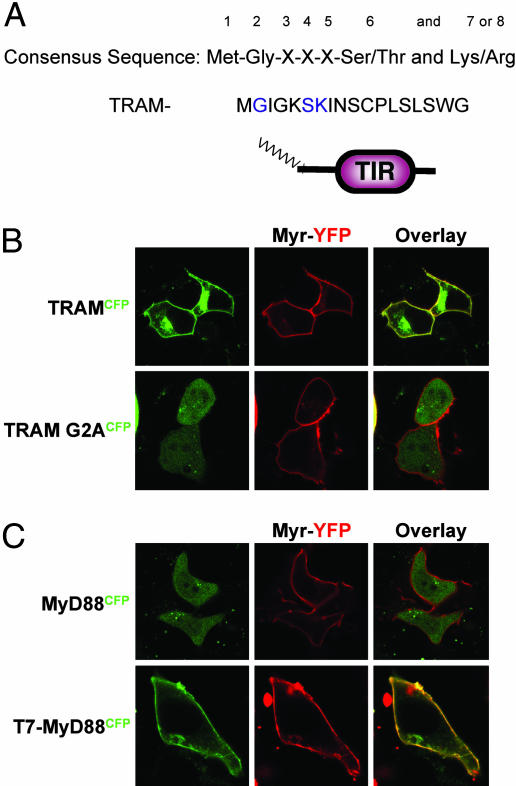
This localization pattern of TRAM in living cells indicates that TRAM is anchored to the membrane by direct interaction with a membrane-associated molecule (such as TLR4), by fatty acid modification, or by both. To examine the possibility that TRAM was anchored by acylation, we subjected the TRAM sequence to N-myristoyltransferase program analysis (http://mendel.imp.univie.ac.at/myristate/SUPLpredictor.html), which revealed that TRAM contained a putative myristoylation consensus sequence consisting of Met-Gly-Xaa-Xaa-Xaa-Ser and Lys (Fig. 2A). Similar myristoylation sequences have been identified previously in members of the Src kinase family.
Fig. 2.
TRAM contains a putative myristoylation sequence. (A) Comparison of the consensus sequence for protein N-terminal myristoylation with amino acids 1–18 of TRAM. Blue characters indicate amino acids that match myristoylation consensus sequences. (B) MyrYFP stable HEK293 cells were transiently transfected with TRAMCFP or TRAM-G2ACFP and visualized by confocal microscopy. (C) MyrYFP stable HEK293 cells were transiently transfected with MyD88CFP or T7-MyD88CFP and visualized by confocal microscopy 24 h later.
To investigate whether TRAM is indeed a myristoylated protein, we took a number of complementary approaches. We generated a mutant form of TRAM in which the glycine at position 2 was mutated to alanine and examined its localization by confocal microscopy. As a comparison, we compared the localization of MyrYFP, a synthetic construct containing an acylation sequence derived from the mammalian Src kinase, Lyn (32). MyrYFP was clearly membrane-localized. In fact, MyrYFP and TRAMCFP colocalized, as indicated by the yellow overlay of the confocal image (Fig. 2B). Mutation of glycine at position 2 to alanine (TRAM-G2A) resulted in the relocalization of TRAM from the cell membrane to the cytosol (Fig. 2B). This change was confirmed by subcellular fractionation, with TRAM-G2A occurring only in the cytosolic fraction (data not shown). As a second approach, we compared the cellular localization of MyD88, a universal TLR adaptor that resides in the cytosol (Fig. 2B). In contrast to TRAM, MyD88CFP did not colocalize with MyrYFP. Next, we performed a gain-of-function experiment and generated a mutant form of MyD88, in which the first seven amino acids of MyD88 were replaced with those from TRAM (T7-MyD88). This experiment resulted in a construct that, in theory, encoded a myristoylated form of MyD88. T7-MyD88 was also capable of localizing in the plasma membrane (Fig. 2C). Together, these results suggest that the putative N-terminal myristoylation site was sufficient for targeting TRAM to the plasma membrane.
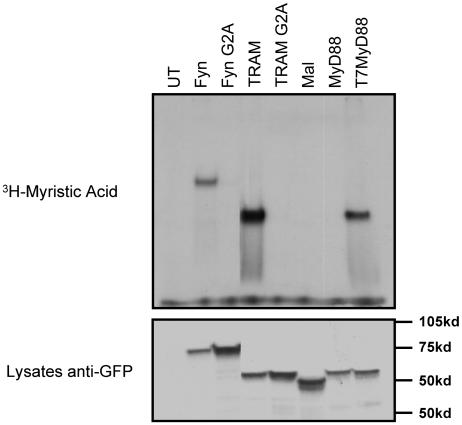
To determine further whether TRAM is a myristoylated protein, we monitored TRAM myristoylation in vivo. 293T cells were transfected with wild-type or G2A mutant versions of TRAM, Mal, MyD88, or T7-MyD88. Cells were metabolically labeled with [3H]myristate. Fyn, an adaptor molecule involved in T cell receptor signal transduction, was used as a positive control for in vivo myristoylation. Likewise, Fyn-G2A served as a negative control for the incorporation of [3H]myristate. The indicated proteins were transfected into HEK293 cells, immunoprecipitated with anti-GFP, run on an SDS/polyacrylamide gel, and examined for incorporation of [3H]myristate by autoradiography (Fig. 3). TRAM, but not TRAM-G2A, was clearly myristoylated in vivo. Furthermore, neither Mal nor MyD88 was labeled with [3H]myristate. In contrast, T7-MyD88 efficiently incorporated [3H]myristate. These results provide clear evidence that TRAM is indeed a myristoylated protein. Even though TRAM and Mal have similar molecular weights, consisting of 232 and 235 aa, respectively, these adaptors resolve differently on SDS/PAGE (Fig. 3 Lower), possibly because of different posttranslational modifications.
Fig. 3.
TRAM is myristoylated in vivo. 293T cells were transiently transfected with CFP-tagged versions of the indicated constructs. Eighteen hours after transfection, cells were incubated in medium containing 250 μCi of [3H]myristate per well for 4 h. Cells were lysed and immunoprecipitated with anti-GFP polyclonal Ab, and immune complexes were resolved by SDS/PAGE followed by autoradiography. Whole-cell lysates were immunoblotted with anti-GFP mAb.
TRAM-G2A Does Not Signal on Overexpression.
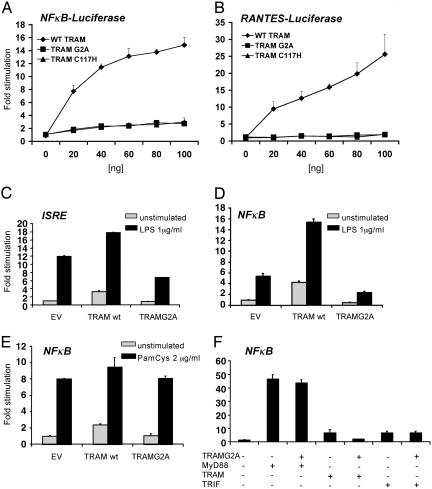
As reported previously, transient expression of TRAM induces both NF-κB- and IRF3-dependent reporter activation in HEK293 cells (11). Furthermore, mutation of the cysteine residue in the predicted TIR domain BB loop of TRAM at position 117 (TRAM-C117H), a mutation equivalent to the C3H/HeJ mutation in the TLR4 TIR domain BB loop, abolished the ability of TRAM to signal (11). To determine whether TRAM-G2A was compromised in its ability to signal on overexpression, we monitored the ability of TRAM-G2A to induce NF-κB- or IRF3-dependent reporter gene activation in HEK293 cells. Wild-type TRAM, but neither the C117H nor the G2A mutant, could drive NF-κB, RANTES, or ISRE (interferon-stimulated response element) reporters (Fig. 4A–C). Moreover, TRAM-G2A inhibited LPS-induced ISRE reporter gene induction modestly, whereas wild-type TRAM potentiated this response (Fig. 4C). Similar results were obtained when LPS-induced NF-κB activation was examined in cells transfected with TRAM or the TRAM mutants (Fig. 4D). As anticipated, TRAM-G2A had no effect on lipopeptide-induced NF-κB activation in TLR2-expressing HEK293 cells (Fig. 4E). We also examined the ability of TRAM-G2A to interfere with signaling by other TIR adaptors. For example, transient expression of MyD88 induced NF-κB reporter gene expression, as was also true in the case of TRAM and TRIF, although to a lesser extent. TRAM-G2A had no effect on either MyD88- or TRIF-induced NF-κB activation, but it inhibited the activation of NF-κB induced by overexpressing wild-type TRAM (Fig. 4F). These results suggest that TRAM-G2A cannot signal efficiently and in fact can act as a dominant-negative interfering mutant. Together, these results suggest that the membrane localization of TRAM is critical for its ability to initiate efficient signaling.
Fig. 4.
TRAM-G2A fails to activate the RANTES promoter and NF-κB. (A and B) HEK293 cells were transiently transfected with the RANTES and NF-κB-luciferase reporters as indicated. FLAG-tagged versions of TRAM, TRAM-G2A, and TRAM-C117H or empty vector were cotransfected as indicated. (C–E) TLR2- or TLR4/MD2-expressing HEK293 cells were transfected as indicated (EV, empty vector) and 24 h later were stimulated with 2 μg/ml LPS or 2 μg/ml tripalmitoyl-Cys-Ser-(Lys)4 (Pam3CSK4) for 6 h. (F) HEK293 cells were transfected with the NF-κB-luciferase reporter and cotransfected with wild-type TRAM, TRIF, or MyD88 in the presence or absence of TRAM-G2A as indicated. In all cases, the thymidine kinase Renilla-luciferase reporter was used to control for transfection efficiency. Twenty-four hours after transfection, cell lysates were generated, and luciferase activity was assayed. All of the data are representative of three independent experiments.
Myristoylation of TRAM Is Critical for LPS Responses in Macrophages.
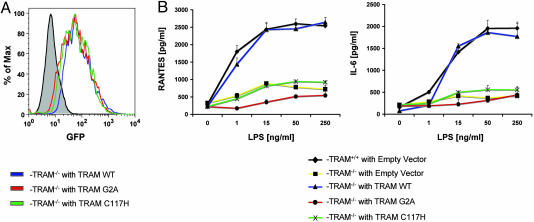
To investigate whether membrane localization of TRAM is critical for LPS responses in vivo, we reconstituted bone marrow-derived macrophages from TRAM-deficient mice. To this end, we generated retroviruses expressing wild-type TRAM, TRAM-G2A, or TRAM-C117H in tandem with an internal ribosomal entry site (IRES)-encoded GFP. This construct was designed to allow the expression of a bicistronic mRNA that would give rise to the translation of TRAM at a 1:1 molar ratio with GFP. Transduced bone marrow-derived macrophages from TRAM-deficient mice were sorted by flow cytometry for GFP fluorescence. As seen in Fig. 5A, the levels of GFP fluorescence, and hence TRAM reconstitution, were comparable in the cells transduced with the wild-type TRAM, the TRAM-G2A, or the TRAM-C117H retroviruses. We were unable to detect TRAM or the mutants in transduced and GFP-sorted cells with Western blotting, presumably because of the low-level expression of TRAM from the construct in these cells (data not shown). However, when we compared the expression level of the three proteins in transfected HEK293 cells, we found no significant difference in expression levels. We therefore believe that the levels of the wild type and TRAM mutants are similar.
Fig. 5.
TRAM myristoylation is critical for optimal LPS responses. Bone marrow-derived macrophages from C57BL/6 or TRAM−/− mice were cultured in 20% conditioned supernatant from L929 cells as a source of macrophage colony-stimulating factor. On day 2, cells were transduced with viral supernatant derived from empty vector, WT TRAM, TRAM-G2A, or TRAM-C117H viral constructs. On day 10, cells were sorted for GFP and seeded in 96-well plates at 40,000 cells per well. (A) Transduced and GFP-sorted cells were examined for GFP fluorescence by FACS analysis. (B) GFP-positive cells were stimulated with LPS as indicated, and cell supernatants were assayed for IL-6 and RANTES by ELISA.
Wild-type or TRAM-knockout macrophages transduced with an IRES vector that encoded only for GFP were also sorted for productive infection (data not shown). The CC-chemokine RANTES is a downstream target gene of the MyD88-independent pathway after LPS stimulation (11). In addition, IL-6 production in response to LPS is also abrogated in TRAM-deficient macrophages (13). We therefore examined IL-6 and RANTES production after LPS stimulation in both wild-type and TRAM-deficient macrophages that had been transduced with either the control vector-expressing retrovirus or the TRAM-expressing retroviruses. As seen in Fig. 5B, GFP-positive, wild-type, empty vector-expressing macrophages produced IL-6 and RANTES after LPS stimulation, whereas GFP-positive, TRAM-deficient, empty vector-expressing macrophages failed to induce either IL-6 or RANTES under identical experimental conditions. Remarkably, TRAM-deficient macrophages transduced with a virus encoding the wild-type TRAM restored responsiveness to LPS challenge completely. As predicted, TRAM-C117H was unable to restore LPS responses, consistent with our earlier in vitro studies. Importantly, TRAM-G2A was also incapable of restoring LPS responses (Fig. 5B). Cells transduced with the TRAM-G2A mutant were unable to produce either RANTES or IL-6 in response to LPS challenge. These results provide clear evidence that the myristoylation and subsequent membrane localization of TRAM are critical for responses to endotoxin.
Discussion
A diverse range of viral and cellular proteins are known to be modified covalently by lipophilic moieties, including protein kinases, guanine nucleotide-binding proteins, transmembrane receptors, and viral structural proteins. The attachment of lipid groups to these molecules influences protein–protein interactions, membrane-binding affinity, and cellular signal transduction. Here, we implicate fatty acid modification as a critical event in TLR4 signaling. We show that the TIR domain-containing adaptor molecule TRAM is colocalized with TLR4 at the plasma membrane and in the Golgi apparatus as a result of myristoylation. Previous studies have demonstrated that LPS traffics rapidly to and from the Golgi apparatus with TLR4, MD2, and CD14; however, these events are distinct from the initiation of signal transduction (30). Therefore, the localization of TRAM with TLR4 in an intact Golgi network is also unlikely to be required for signaling.
Because TRAM does not contain a signal peptide sequence, its presence on the membrane could be the result of protein–protein interactions, prenylation, or fatty acid modification. TRAM does not contain a predicted prenyl modification site because it does not contain a CAAX box at or near its C terminus. There are two distinct types of fatty acid modification: myristoyl and palmitoyl (33–35). In this study, we show that TRAM contains an N-terminal myristoylation sequence. For myristoylation to occur, the initiating methionine is usually removed by methionine aminopeptidase during translation, and the glycine at position 2 becomes the N-terminal amino acid. The requirement for glycine at the N terminus is absolute; no other amino acid can substitute. Such proteins are labeled with myristate on these N-terminal glycines in an irreversible, cotranslational manner by N-myristoyltransferase. This type of covalent modification occurs in many signal transduction components and is central to their function (33). A well characterized myristoylated protein is the Src kinase pp60v-src from Rous sarcoma virus. A myristoylation mutant of this Src kinase (G2A) does not bind to the membrane and is incapable of mediating cellular transformation (33). Myristoylation is also required for membrane association and virion formation by Gag polyproteins of mammalian C- and D-type retroviruses. Nonmyristoylated mutants of murine leukemia C virus Pr65gag and HIV-1 Pr55gag are predominantly cytosolic, and infectious viral particles are not produced (36).
Although myristoylation is clearly necessary for membrane binding of N-myristoylated proteins, myristoylation alone is not sufficient to confer stable membrane binding properties to proteins (37). It is thought that a second signal within the N-myristoylated protein is required for efficient membrane binding. The second signal is most often either palmitoylation or the presence of a polybasic cluster of amino acids. The latter signal is the mechanism used by the Src kinase pp60v-src for membrane anchoring (33, 35). A basic amino acid cluster in Src kinase, downstream from the glycine at position 2, forms electrostatic interactions with acidic phospholipid headgroups in the membrane. It is unclear at present what is the second signal that tethers TRAM to the membrane. TRAM has a number of lysine residues close to the myristoylation site, which may serve as a polybasic cluster. A combination of both myristate and basic motifs in TRAM would allow the hydrophobic and electrostatic forces to synergize and facilitate membrane binding (37). TRAM also has a number of cysteine residues, however, which could potentially become palmitoylated. A third possibility for membrane tethering might involve association with membrane-anchored molecules, which, in the case of TRAM, could be TLR4. This last explanation seems less likely because when TRAMCFP was transfected into HEK293 cells, which ordinarily lack expression of TLR4, TRAM localization in the cell surface and Golgi was nevertheless observed (data not shown). Of course, other cell surface TLRs are theoretically capable of serving this function, but cells from TRAM-deficient animals appear to have normal responses to a variety of TLR ligands, including those for TLR2, TLR5, TLR7, and TLR9 (data not shown), suggesting that TRAM binds the cytoplasmic domain of TLR4 selectively.
Proper localization of signaling molecules to specific cellular membranes is critical for their function. It is tempting to speculate that clustering of myristoylated proteins into specialized membrane microdomains, such as lipid rafts or caveolae, enhances particular protein–protein interactions important for subsequent signal transduction. LPS has been reported to result in the redistribution of TLR4 into lipid rafts (38). The acylation of TRAM might aid in this redistribution. Similarly, in the case of TRAM, colocalization with TLR4 in lipid rafts may be critical for signaling.
A key question, which remains to be answered, is why TRAM must be membrane-localized to allow cells to signal through TLR4. One possible explanation is that membrane-localized TRAM must leave the membrane and dissociate from TLR4 to interact with TRIF and propagate signals further downstream to NF-κB and IRF pathways. Indeed, preliminary experiments suggest that TRAM leaves the membrane after LPS stimulation. In this scenario, the dissociation of TRAM from the membrane could be a regulated event (e.g., demyristoylation, depalmitoylation, or phosphorylation). N-myristoylation is usually a permanent modification; however, there is evidence in particular situations (e.g., brain synaptosomes) that a demyristoylase activity modifies myristoylated alanine-rich C-kinase substrate (MARCKS), promoting its release form the membrane (39). Depalmitoylation is a more common mechanism of protein translocation because the thioester linkage between palmitate and the polypeptide is quite labile. Another possible mechanism involves protein phosphorylation. Electrostatic interactions between polybasic residues and phospholipid headgroups could be destabilized by phosphorylation events adjacent to polybasic clusters. Ligand-induced phosphorylation of TRAM would elicit its dissociation from the membrane. Recent evidence from O’Neill and colleagues suggests that TRAM is indeed phosphorylated by PKCε after LPS signaling. Myristoylated proteins are often phosphorylated by PKC near their N termini, leading to their dissociation from the membrane (40–43). Phosphorylation of TRAM by PKCε is critical for LPS signaling (L.A.O., personal communication), consistent with the possibility that TRAM phosphorylation could facilitate it to leave the membrane after LPS signaling.
In conclusion, the data described in this work show that TRAM is the only adaptor in TLR signaling to be myristoylated, conferring on it membrane localization as a prerequisite for LPS signaling. Defining the functional significance of membrane-localized TRAM in LPS signaling remains to be elucidated completely, but the data strongly suggest that the acylation of TRAM is one means by which cells regulate their responses to bacterial endotoxin.
Materials and Methods
Reagents.
Unless otherwise stated, all reagents were purchased from Sigma. Escherichia coli O111:B4 LPS was subjected to a second phenol extraction to remove contaminating lipopeptides (44). Lipopeptide Pam3CSK4 was purchased from EMC Microcollections (Tuebingen, Germany).
Plasmids.
Most of the constructs described in this work have been described elsewhere. These include: pCDNA3-TRAMCFP, pEF-BOS-TRAMFLAG, pEF-BOS-TRAM-C117HFLAG, pEF-BOS-TRIFFLAG (11); pCDNA3-MyD88CFP (31); MyrYFP (32); β-galactosyltransferaseYFP (GolgiYFP) (30); NF-κB-luciferase, RANTES-luciferase, ISRE-luciferase, and Renilla-luciferase (11); and the retroviral vector pMSCV2.2-ISRE-GFP (45). pCDNA3-TRAM-G2A and pEF-BOS-TRAM-G2A were generated by using a site-directed mutagenesis kit (Stratagene). The plasmid pCDNA3-Fyn was subcloned using PCR cDNA obtained from E. Kurt-Jones (University of Massachusetts, Worcester); pCDNA3-Fyn-G2A was generated by site-directed mutagenesis. pMSCV2.2-IRES-GFP-murine TRAM, murine TRAM-G2A, and murine TRAM-C117H were subcloned from pEF-BOS vectors expressing murine constructs. pCDNA3-T7-MyD88CFP was generated by PCR. The packaging vector for the retroviruses was the proviral clone of Moloney murine sarcoma virus ΨEcu from O. Witte (University of California, Los Angeles).
Cell Lines.
HEK293 cells stably expressing TLR2YFP, TLR4YFP, and TLR9YFP were as described previously (30, 31).
Confocal Microscopy.
HEK293 cells stably expressing TLR4YFP and TLR9YFP or MyrYFP were transfected with TRAMCFP, TRAM-G2ACFP, MyD88CFP, or T7-MyD88CFP where indicated. In addition, HEK293 cells were cotransfected with TRAMCFP and β-galactosyltransferaseYFP (GolgiYFP). Twenty-four hours after transfection, cells were imaged by confocal microscopy with a Leica TCS SP2 AOBS microscope.
Subcellular Fractionation.
HEK293 cells stably expressing TLR4YFP and TRAMCFP, TRAM-G2ACFP, or MyD88CFP were seeded in 10-cm dishes at a density of 1 × 106 cells per dish. After 24 h, the medium was removed, and the cells were washed in PBS and then scraped into fractionation buffer (20 mM Tris·HCl, pH 7.5/10 mM MgCl2/1 mM EDTA/250 μM sucrose/200 μM PMSF). The samples were subjected to 20 strokes of a Dounce homogenizer and spun at 100,000 × gfor 1 h. The supernatant (cytosolic fraction) was removed to a fresh tube, and the pellet (membrane fraction) was resuspended in 50 μl of SDS sample buffer [50 mM Tris·Cl, pH 6.8/10% (vol/vol) glycerol/2% (wt/vol) SDS/0.1% (wt/vol) bromophenol blue/5% (vol/vol) 2-mercaptoethanol]. The fractions were run on an SDS/10% polyacrylamide gel, transferred onto nitrocellulose, and blotted with the appropriate antibody.
Luciferase Reporter Assay.
Cells were seeded into 96-well plates at a density of 40,000 cells per well and transfected 24 h later with 80 ng of the indicated luciferase reporter genes and the indicated amounts of the TRAM constructs, MyD88 or TRIF, by using 0.8 μl of GeneJuice (EMD Biosciences, San Diego) per well. The thymidine kinase Renilla-luciferase reporter was also cotransfected (40 ng) so that the data could be normalized for transfection efficiency. Cells were either left untreated or treated with 1 μg/ml LPS or 2 μg/ml Pam3CSK4 as indicated for 6 h. Cell lysates were then prepared, and reporter gene activity was measured by using the Dual Luciferase Assay System (Promega). Data are expressed as the mean relative stimulation ± SD.
Radiolabeling.
293T cells were transfected by using GeneJuice with the CFP-tagged constructs as indicated. Eighteen hours after transfection, cells were incubated in medium containing 250 μCi (1 Ci = 37 GBq) of [3H]myristate (PerkinElmer) for 4 h. Cells were then lysed in 0.5 ml of lysis buffer [20 mM Tris·HCl/2 mM EDTA/137 mM NaCl/0.5% Triton X-100/10% (vol/vol) glycerol, with protease inhibitors]. Polyclonal anti-GFP (Molecular Probes) was incubated with the cell lysates in protein A–Sepharose for 2 h. The immune complexes were precipitated and subjected to SDS/4–15% PAGE. The gel was then dried and developed by using Amplify (Amersham Pharmacia) according to the manufacturer’s instructions.
Reconstitution of Bone Marrow-Derived Macrophages.
The retroviral vector pMSCV expressing TRAM, TRAM-G2A, TRAM-C117H, and GFP from the IRES element was used to generate high-titer, helper-free retroviral stocks (using the packaging plasmid ΨEcu) by transient cotransfection of 293T cells (45). After 24–72 h, cell supernatants were harvested, filtered, and used to transduce target cells. Bone marrow-derived macrophages were cultured from C57BL/6 mice or age- and sex-matched TRAM−/− mice for 8–10 days. Conditioned supernatant from L929 cells comprised 20% of the total volume as a source of macrophage colony-stimulating factor. Cells were transduced with virus encoding the indicated versions of TRAM or empty vector on day 2 while cells were actively dividing. On day 10, cells were sorted for GFP and seeded into 96-well plates at a density of 35,000 cells per well. Cells were stimulated with LPS overnight after being allowed to recover from cell sorting for 24 h. Cell culture supernatants were analyzed for the presence of IL-6 and RANTES by ELISA (R & D Systems).
Acknowledgments
We thank K. Halmen for helpful discussions and A. Cerny for animal husbandry and care.
Abbreviations
- CFP
cyan fluorescent protein
- HEK
human embryonic kidney
- IRES
internal ribosomal entry site
- IRF
IFN regulatory factor
- Mal
MyD88 adaptor-like
- MyD88
myeloid differentiation factor 88
- Pam3CSK4
tripalmitoyl-Cys-Ser-(Lys)4
- RANTES
regulated on activation, normal T expressed and secreted
- TIR
Toll/IL-1 resistance
- TIRAP
TIR adaptor protein
- TLR
Toll-like receptor
- TRAM
TRIF-related adaptor molecule
- TRIF
TIR domain-containing adaptor-inducing IFN-β
- YFP
yellow fluorescent protein
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dunne A., O’Neill L. A. J. Sci. STKE (2003) 2003;2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 2.Muzio M., Ni J., Feng P., Dixit V. M. Science. 1997;278:1612–1615. [Google Scholar]
- 3.Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., et al. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 5.Horng T., Barton G. M., Flavell R. A., Medzhitov R. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 6.Horng T., Barton G. M., Medzhitov R. Nat. Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. J. Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 9.Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S. O., Goode J., Lin P., Mann N., Mudd S., et al. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 10.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. Nat. Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald K. A., Rowe D. C., Barnes B. J., Caffrey D. R., Visintin A., Latz E., Monks B., Pitha P. M., Golenbock D. T. J. Exp. Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oshiumi H., Sasai M., Shida K., Fujita T., Matsumoto M., Seya T. J. Biol. Chem. 2003;278:4951–4962. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., Akira S. Nat. Immunol. 2003;11:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 14.O’Neill L. A. J., Fitzgerald K. A., Bowie A. G. Trends Immunol. 2003;24:286–290. doi: 10.1016/s1471-4906(03)00115-7. [DOI] [PubMed] [Google Scholar]
- 15.Osborn M. J. Annu. Rev. Biochem. 1969;38:501–538. doi: 10.1146/annurev.bi.38.070169.002441. [DOI] [PubMed] [Google Scholar]
- 16.Raetz C. R. H. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald K. A., Rowe D. C., Golenbock D. T. Microbes Infect. 2004;6:1361–1367. doi: 10.1016/j.micinf.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Vogel S. N., Fenton M. Biochem. Soc. Trans. 2003;31:664–668. doi: 10.1042/bst0310664. [DOI] [PubMed] [Google Scholar]
- 19.Cao Z., Henzel W. J., Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 20.Li S., Strelow A., Fontana E. J., Wesche H. Proc. Natl. Acad. Sci. USA. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D. V. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 22.Sun L., Deng L., Ea C. K., Xia Z. P., Chen Z. J. Mol. Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 23.Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., Chen Z. J. Mol. Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Toshchakov V., Jones B. W., Lentschat A., Silva A., Perera P. Y., Thomas K., Cody M. J., Zhang S., Williams B. R., Major J., et al. J. Endotoxin Res. 2003;9:169–175. doi: 10.1179/096805103125001577. [DOI] [PubMed] [Google Scholar]
- 25.Kawai T., Takeuchi O., Fujita T., Inoue J., Muhlradt P. F., Sato S., Hoshino K., Akira S. J. Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 26.Kaisho T., Takeuchi O., Kawai T., Hoshino K., Akira S. J. Immunol. 2001;166:5688–5694. doi: 10.4049/jimmunol.166.9.5688. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T., et al. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M., Takeda K., Akira S. Mol. Immunol. 2004;40:861–868. doi: 10.1016/j.molimm.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., Akira S. Nat. Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 30.Latz E., Visintin A., Lien E., Fitzgerald K. A., Monks B. G., Kurt-Jones E. A., Golenbock D. T., Espevik T. J. Biol. Chem. 2002;277:47834–47843. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 31.Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. Nat. Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 32.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 33.Resh M. D. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 34.Resh M. D. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 35.Resh M. D. Cell. Signalling. 1996;8:403–412. doi: 10.1016/s0898-6568(96)00088-5. [DOI] [PubMed] [Google Scholar]
- 36.Bryant M., Ratner L. Proc. Natl. Acad. Sci. USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peitzsch R. M., McLaughlin S. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 38.Triantafilou M., Miyake K., Golenbock D. T., Triantafilou K. J. Cell Sci. 2002;115:2603–2611. doi: 10.1242/jcs.115.12.2603. [DOI] [PubMed] [Google Scholar]
- 39.Manenti S., Sorokine O., Van Dorsselaer A., Taniguchi H. J. Biol. Chem. 1994;269:8309–8313. [PubMed] [Google Scholar]
- 40.Rosen A., Keenan K. F., Thelen M., Nairn A. C., Aderem A. J. Exp. Med. 1990;172:1211–1215. doi: 10.1084/jem.172.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thelen M., Rosen A., Nairn A. C., Aderem A. Nature. 1991;351:320–322. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- 42.Aderem A. A., Albert K. A., Keum M. M., Wang J. K., Greengard P., Cohn Z. A. Nature. 1988;332:362–364. doi: 10.1038/332362a0. [DOI] [PubMed] [Google Scholar]
- 43.Wang J. K., Walaas S. I., Sihra T. S., Aderem A., Greengard P. Proc. Natl. Acad. Sci. USA. 1989;86:2253–2256. doi: 10.1073/pnas.86.7.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirschfeld M., Ma Y., Weis J. H., Vogel S. N., Weis J. J. J. Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 45.Hawley R. G., Lieu F. H., Fong A. Z., Hawley T. S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]