Abstract
Cysts of Azotobacter vinelandii are resting cells that are surrounded by a protective coat, conferring resistance to various chemical and physical agents. The major chemical components of the cyst coat are alkylresorcinols, which are amphiphilic molecules possessing an aromatic ring with a long aliphatic carbon chain. Although alkylresorcinols are widely distributed in bacteria, fungi, plants, and animals, no enzyme systems for their biosynthesis are known. We report here an ars operon in A. vinelandii that is responsible for the biosynthesis of the alkylresorcinols in the cysts. The ars operon consisted of four genes, two of which encoded a type III polyketide synthase, ArsB and ArsC. In vitro experiments revealed that ArsB and ArsC, sharing 71% amino acid sequence identity, were an alkylresorcinol synthase and an alkylpyrone synthase, respectively, indicating that ArsB and ArsC are not isozymes but enzymatically distinct polyketide synthases. In addition, ArsB and ArsC accepted several acyl-CoAs with various lengths of the side chain as a starter substrate and gave corresponding alkylresorcinols and alkylpyrones, respectively, which suggests that the mode of the ring folding is uninfluenced by the structure of the starter substrates. The importance of the alkylresorcinols for encystment was confirmed by gene inactivation experiments; the lack of alkylresorcinols synthesis caused by ars mutations resulted in the formation of severely impaired cysts, as observed by electron microscopy.
The genus Azotobacter, comprising Gram-negative, nitrogen-fixing soil bacteria, produces phenolic lipids, which are a mixture of alkylresorcinols and alkylpyrones with various lengths of their side chain (Fig. 1A). Alkylresorcinols, distributed widely in bacteria, fungi, plants, and animals, play several roles that are concerned with cellular biochemistry and membrane chemistry (1). For example, 5-alkylresorcinols are readily incorporated into phospholipid bilayers and biological membranes, thereby causing considerable changes in their structure and properties (1). They also act as modulators of oxidation of liposomal membranes and fatty acids (1). When Azotobacter vinelandii differentiates to form metabolically dormant cysts, the phospholipids in the membrane are replaced by 5-alkylresorcinols (2). This unique membrane matrix is presumed to contribute to the physiology and desiccation resistance of the cyst (2) and, therefore, is sometimes called a “cyst coat.” Because of high incorporation of 14C-labeled acetate into alkylresorcinols in A. vinelandii (3), they were thought to be synthesized through a polyketide biosynthetic pathway. However, no enzyme systems for the biosynthesis of alkylresorcinols, not only in Azotobacter but also in other organisms, have been elucidated.
Fig. 1.
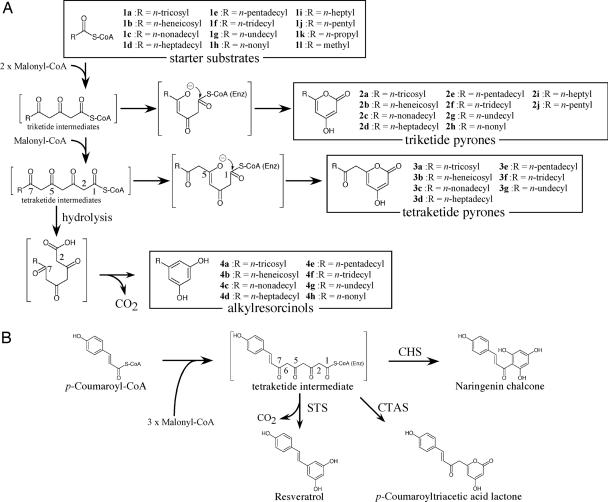
Reactions catalyzed by type III PKSs. (A) Synthesis of alkylresorcinol by ArsB and alkylpyrone by ArsC. (B) Reactions of plant type III PKSs. CHS, STS, and p-coumaroyltriacetic acid synthase (CTAS) use common tetraketide intermediates, which are derived from p-coumaroyl-CoA and malonyl-CoA.
Polyketides are synthesized by polyketide synthases (PKSs) that fall into three groups, type I to type III. All three bacterial PKSs possess a ketosynthase activity that catalyzes the sequential head-to-tail incorporation of acetate building blocks into a growing chain. Type I PKSs are multifunctional enzymes that are organized into modules, each of which harbors a set of distinct catalytic domains (4). Modular type I PKSs assemble polyketides by using each ketosynthase domain once to add many extender substrates onto a “starter substrate,” whereas iterative type I PKSs use a single ketosynthase domain repeatedly for several times of extension (4). Type II PKSs consist of a “minimal PKS,” and their auxiliary subunits contain a single set of iteratively used active sites that are carried on separate proteins (5). Type III PKSs are ketosynthases with a homodimeric form that act iteratively for polyketide chain extension (6). Chalcone synthase (CHS) (Fig. 1B), responsible for flavonoid synthesis in plants, is a member of type III PKSs (6). Stilbene synthase (STS) and p-coumaroyltriacetic acid synthase (CTAS), both sharing >70% amino acid sequence identity to CHS, catalyze the same iterative condensation to yield tetraketide intermediates in the same manner as CHS (Fig. 1B). However, the mode of ring folding differs among these three type III PKSs. STS cyclizes tetraketide intermediates via intramolecular C2-to-C7 aldol condensation, and CTAS catalyzes intramolecular C5 oxygen-to-C1 lactonization (Fig. 1B). Because the products of STS differ from alkylresorcinols only in their moiety derived from the starter substrate and because type III PKSs often produce monocyclic polyketides, we expected that a type III PKS(s) would be involved in the formation of the alkylresorcinols in the cysts of A. vinelandii.
This report deals with the enzyme system responsible for the biosynthesis of the alkylresorcinols and alkylpyrones localized in the cysts of A. vinelandii, together with the biological importance of these phenolic lipids for cyst formation. As we expected, the phenolic lipids were shown to be synthesized by type III PKSs. We named the operon encoding the type III PKSs alkylresorcinol synthesis (ars) (Fig. 2A). In vitro analysis of these type III PKSs revealed that ArsB and ArsC encoded in the operon were an alkylresorcinol synthase and an alkylpyrone synthase, respectively. To our knowledge, this is the first report on the enzymes responsible for alkylresorcinol synthesis. An additional important finding in this study is that the phenolic lipids are essential for the encystment in A. vinelandii.
Fig. 2.
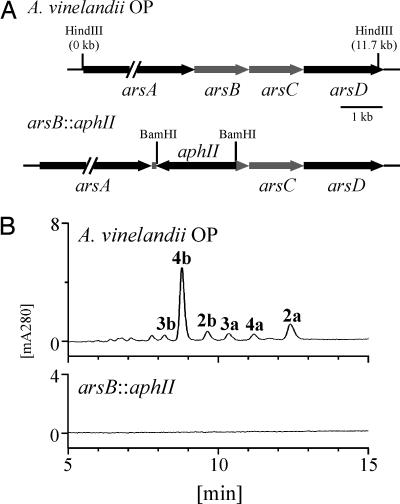
Requirement of the ars operon for phenolic lipid synthesis. (A) Gene organization of the ars operon in A. vinelandii OP. ArsA (accession no. ZP_00418324) and ArsD (accession no. ZP_00418327) are similar in amino acid sequence to type I PKSs. ArsB (accession no. ZP_00418325) and ArsC (accession no. ZP_00418326) are homologous to type III PKSs. The arsB::aphII mutant was constructed by replacing the arsB sequence with the kanamycin-resistance gene, aphII. (B) HPLC chromatograms of lipid extracts prepared from β-hydroxybutyrate-induced cells of A. vinelandii and the arsB::aphII mutant.
Results and Discussion
Isolation and Identification of Phenolic Lipids from Cysts of A. vinelandii.
The phospholipids in the membrane of the vegetative cells of A. vinelandii are replaced by alkylresorcinols and alkylpyrones when it forms metabolically dormant cysts (2). We prepared a lipid fraction from mature cysts of A. vinelandii OP and analyzed it by HPLC (Fig. 2B). The major compound migrating at a retention time of 8.9 min was identified to be 5-heneicosylresorcinol (4b) by proton and carbon NMR spectrometry and liquid chromatography-atmospheric pressure chemical ionization mass spectrometry (LC-APCIMS) analyses. The molecular masses of 3b and 2b were 448 and 406, respectively, on the basis of the [M − H]− ions on LC-APCIMS analysis, suggesting that 3b and 2b were α-pyrones. Their structures were verified as 6-heneicosyl-4-hydroxy-2-pyrone (2b) and 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone (3b) by their comigration with the synthetic standards (data not shown). Although 2b (a triketide pyrone), 3b (a tetraketide pyrone), and 4b (a resorcinol) had different structures from one another, their alkyl chains were presumed to be derived from a common starter substrate, n-behenyl-CoA (1b), for a PKS(s). In addition, we also assumed that 2a, 3a, and 4a were polyketides that were derived from n-heneicosyl-CoA (1a) as a starter, because the molecular masses of 2a, 3a, and 4a were 434, 476, and 432, respectively. Comparison of their fragmentation patterns with those from n-behenyl-CoA in LC-APCI tandem mass analysis predicted that their carbon skeletons were a triketide pyrone (2a), a tetraketide pyrone (3a), and a resorcinol (4a). These data show that A. vinelandii OP predominantly produces phenolic lipids as a mixture of alkylresorcinols and alkylpyrones with saturated side chains ranging from C21 to C23, as was found for other Azotobacter species (2, 7). There were no significant differences in the amount or composition of phenolic lipids when the lipid extract was prepared from the β-hydroxybutyrate-induced cells and the aging cells that were harvested from an 8-day culture without an induction of encystment (data not shown).
Gene Cluster Responsible for Biosynthesis of the Phenolic Lipids.
A blast search in the protein database of A. vinelandii OP using the alfalfa CHS sequence (8) as a query revealed the presence of two type III PKSs: one (named ArsB) sharing 23% amino acid sequence identity to the CHS and the other (ArsC) sharing 25% identity (Fig. 2A). arsB and arsC appeared to form an operon, together with a putative type I fatty acid synthase (arsA) and a functionally unknown protein (arsD). ArsD contains a 4′-phosphopantetheinyl transferase domain that would catalyze the posttranslational modification of acyl carrier protein (ACP) by the covalent attachment of the 4′-phosphopantetheine moiety of CoA to a conserved serine (9), in addition to a functionally unknown N-terminal domain of ≈400 aa in length.
We disrupted arsB by replacing the arsB sequence by a kanamycin resistance gene to determine possible involvement of this gene cluster in the biosynthesis of the phenolic lipids (Fig. 2A). It seemed likely that the arsB::aphII mutation was polar on the arcCD expression. This polar effect might result in the inactivation of the entire gene cluster, because the 4′-phosphopantetheinyl transferase domain of ArsD is presumed to be crucial for the modification of the ACP domain of ArsA. As expected, no phenolic lipids were produced by the arsB::aphII mutant (Fig. 2B), indicating that the ars operon is responsible for the biosynthesis of the phenolic lipids.
In Vitro Analysis of ArsB and ArsC Reactions.
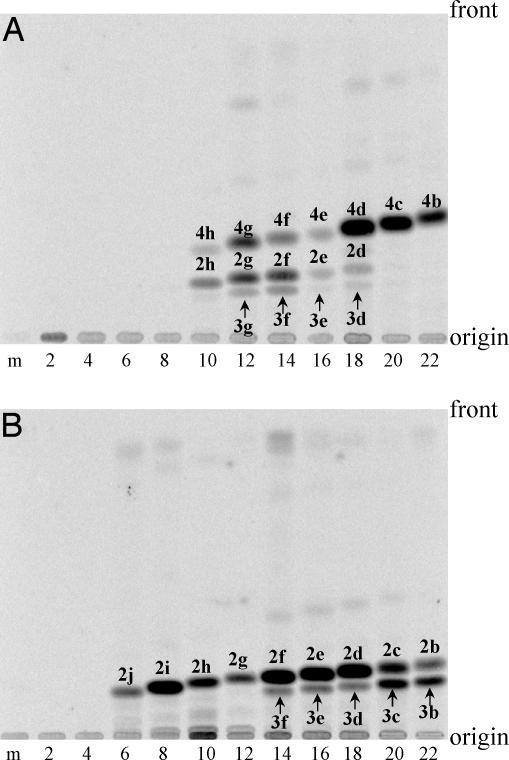
It was particularly interesting to determine whether ArsB and ArsC are isozymes or not, because ArsB and ArsC shared 71% amino acid sequence identity. We prepared ArsB and ArsC with a His-tag at their N-termini from Escherichia coli carrying these genes under the control of the T7 promoter in pET vector. Both ArsB and ArsC migrated at a position of ≈43 kDa on SDS/polyacrylamide gel electrophoresis (data not shown). We first assayed ArsB by using n-behenyl-CoA (1b) as a starter substrate, because the major component of the phenolic lipids of A. vinelandii was 5-heneicosylresorcinol (4b), as described above. Incubation of n-behenyl-CoA (1b) as a starter substrate and [2-14C]malonyl-CoA as an extender substrate gave a single radioactive product (Fig. 3A, lane 22), which was identified as 5-heneicosylresorcinol (4b) by LC-APCIMS analysis of the product similarly synthesized from nonradiolabeled substrates (Table 2, which is published as supporting information on the PNAS web site). On the other hand, under the same reaction conditions, ArsC gave two radioactive compounds different from 5-heneicosylresorcinol (4b) in the Rf values (Fig. 3B, lane 22). We identified them as 6-heneicosyl-4-hydroxy-2-pyrone (2b) and 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone (3b) by comparing their LC-APCIMS with those of the synthetic standards. These results clearly show that ArsB and ArsC are type III PKSs with different catalytic properties and are responsible for the biosynthesis of alkylresorcinols and alkylpyrones, respectively.
Fig. 3.
Radio-TLC analysis of products synthesized by ArsB (A) and ArsC (B) from various acyl-CoA starter substrates and [2-14C]malonyl-CoA. The starter substrates used were acetyl-CoA [1l] (lane 2), butyryl-CoA [1k] (lane 4), hexanoyl-CoA [1j] (lane 6), octanoyl-CoA [1i] (lane 8), decanoyl-CoA [1h] (lane 10), lauroyl-CoA [1g] (lane 12), myristoyl-CoA [1f] (lane 14), palmitoyl-CoA [1e] (lane 16), stearoyl-CoA [1d] (lane 18), arachidoyl-CoA [1c] (lane 20), and behenyl-CoA [1b] (lane 22). Lane m, control incubation without a starter substrate.
We measured the steady state kinetics parameters for 4b, 2b and 3b synthesis by varying the concentration of n-behenyl-CoA (1b) (Table 1). ArsB and ArsC exhibited saturation kinetics in response to the increasing concentration of n-behenyl-CoA (1b). Interestingly, the kcat/Km value of ArsB for n-behenyl-CoA consumption is similar to that of ArsC, although A. vinelandii OP predominantly produced alkylresorcinols in vivo. Therefore, the in vitro catalytic abilities of ArsB and ArsC do not reflect the amounts of alkylresorcinol and alkylpyrones in vivo. This difference may result from a decrease of alkylpyrones due to further metabolism or the instability of the compound itself.
Table 1.
Steady-state kinetic parameters∗ of the ArsB or ArsC reactions
| Enzyme |
n-Behenyl-CoA (1b) |
Triketide pyrone (2b) |
Tetraketide pyrone (3b) |
Resorcinol (4b) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (× 10−3·sec−1) | kcat/Km (× sec−1·M−1) | Km (μM) | kcat (× 10−3·sec−1) | kcat/Km (× sec−1·M−1) | Km (μM) | kcat (× 10−3·sec−1) | kcat/Km (× sec−1·M−1) | Km (μM) | kcat (× 10−3 ·sec−1) | kcat/Km (× sec−1·M−1) | |
| ArsB | 4.86 ± 0.50 | 15.6 ± 0.73 | 3,202 | — | — | — | — | — | — | 4.86 ± 0.50 | 15.6 ± 0.73 | 3,202 |
| ArsC | 1.47 ± 0.18 | 6.80 ± 0.07 | 4,626 | 1.34 ± 0.33 | 2.77 ± 0.13 | 2,069 | 1.59 ± 0.06 | 4.04 ± 0.07 | 2,541 | — | — | — |
∗Results are mean ± SE (n = 3).
Substrate Specificity of ArsB and ArsC.
Type III PKSs of bacterial and plant origins show rather promiscuous starter substrate specificity (6). We examined the reactions of ArsB and ArsC toward CoA esters of C2 to C22 straight chain fatty acids. The radio-TLC (Fig. 3), as well as the LC-APCIMS analyses (Table 2) of the reaction products, revealed broad starter substrate specificities of ArsB and ArsC. ArsB accepted the C10 to C22 esters, whereas ArsC exhibited broader substrate specificity than ArsB. Neither the substrate specificities of the enzymes nor the product profiles of the reactions were affected by the change from pH 6 to pH 9 (data not shown). In contrast, the optimal pH was different depending on the starter substrates (results not shown).
Although ArsB and ArsC accepted a broad range of starter substrates in vitro, the corresponding phenolic lipids were hardly detected in the lipid fraction of A. vinelandii (data not shown). This strict selectivity is perhaps because of the availability of starter CoA esters, which is presumably synthesized by the coaction of ArsA and ArsD. Very recently, Sankaranarayanan et al. (10) found a unique tunnel extending from the active site to the surface of the protein in the crystal structure of PKS18, a type III PKS from Mycobacterium tuberculosis, which accounts for the binding of long-chain aliphatic substrates. PKS18 exhibits a broad specificity for C6 to C20 esters to produce tri- and tetraketide pyrones (11). The catalysis of PKS18 is assumed to be essentially the same as that of ArsC that accepts C6 to C22 esters (Fig. 3B), although they share only 23% identity in amino acid sequence. We speculate that a tunnel similar to that in PKS18 is also present in both ArsB and ArsC.
A vivid contrast between ArsB and ArsC was that the former produced alkylresorcinols in all of the reactions, but the latter produced pyrones. Therefore, the mode of ring folding is characteristic of the enzymes and is independent of the starter substrates incorporated in the polyketide intermediates. Although both enzymes use the same tetraketide intermediate, the mechanism of ring folding is different between ArsB and ArsC; ArsB catalyzes intramolecular C2-to-C7 aldol condensation of STS type to yield 5-heneicosylresorcinol (4b), whereas ArsC catalyzes intramolecular C5 oxygen-to-C1 lactonization of p-coumaroyltriacetic acid synthase type to yield 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone (3b) (Fig. 1 A and B).
Another difference in their reactions from n-behenyl-CoA (1b) is that the size of the ArsB product is strictly confined to the tetraketide, whereas ArsC produces 6-heneicosyl-4-hydroxy-2-pyrone (2b) that is synthesized by lactonization of the triketide intermediate (Fig. 1A). We assume that the hydrolysis of the thioester, which is linked to CoA or ArsB, takes place before the aldol reaction (Fig. 1A), because no alkylresorcinolic acids were detected by the LC-APCIMS analysis of the reaction mixture of ArsB. It is unclear whether decarboxylation is accompanied by the aldol reaction or occurs during the dehydration/aromatization step after the aldol reaction. A similar catalytic property was found for the STS reaction in which stilbene carboxylic acid was not detected (12). Furthermore, merulinic acid, an alkylresorcinolic acid supplemented to the culture of Azotobacter chroococcum, was not incorporated into the corresponding alkylresorcinol. Therefore, enzymatic decarboxylation of alkylresorcinolic acids seems unlikely (1). Recently, a novel mechanism leading to hydrolysis of the thioester, which in turn results in an STS-type ring folding, has been proposed for the STS reaction on the basis of the crystal structure (12). The difference between ArsB and ArsC could be explained by the absence of the STS-like thioesterase activity in ArsC and the presence of this activity in ArsB.
A Proposed Route for Phenolic Lipid Synthesis.
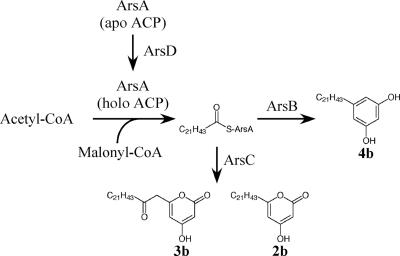
The de novo products by bacterial type II fatty acid synthases are released as ACP esters (13), and the fatty acids are directly transacylated from acyl-ACP to the membrane phospholipids by specific glycerol phosphate transacylases (14). In addition, the chain lengths of fatty acids in phospholipids of A. vinelandii are mainly C16 and C18 (15), which suggests the absence of n-behenyl-CoA (1b) in the primary metabolism. We therefore suppose that ArsA, which is similar in amino acid sequence to type I fatty acid synthases, is responsible for the synthesis of a starter substrate of ArsB and ArsC. Because ArsA contains no thioesterase domain, which cleaves a thioester linkage between fatty acids and ACP (4), docosanoate, a plausible product of ArsA, perhaps remains attached on ACP of ArsA. Alternatively, ArsA may contain a domain that is responsible for transacylation of docosanoate from the enzyme to CoA, like the palmitoyl transferase domain of the yeast type I system (14). Taken together, we hypothesize that the docosanoate bound to ACP of ArsA could be transferred hand-to-hand toward ArsB and ArsC, as illustrated in Fig. 4. Using the same tetraketide intermediate, ArsB catalyzes aldol condensation to form alkylresorcinols, whereas ArsC catalyzes lactonization to form alkylpyrones (Fig. 4).
Fig. 4.
A proposed route for biosynthesis of the phenolic lipids in A. vinelandii.
Effect of Inactivation of the ars Operon on Encystment.
When the glucose in the medium for A. vinelandii cultures is replaced by β-hydroxybutyrate or 1-butanol, a metabolic shift from carbohydrate to lipid occurs, leading to encystment (2). Although it is known that >70% of lipids are replaced by alkylresorcinols and other phenolic lipids during encystment (3), there have been no studies concerning the requirement of the alkylresorcinol synthesis for encystment. A. vinelandii OP produced mature cysts when cultured in the presence of β-hydroxybutyrate; the central body, intine, and exine in the cyst were distinct, as observed by transmission electron microscopy (Fig. 5A). This strain accumulated almost no poly-β-hydroxybutyrate, in agreement with the previous observation that poly-β-hydroxybutyrate accumulation is not essential for encystment (16, 17). Concerning phenolic lipid production, the aged cells grown in the absence of β-hydroxybutyrate and harvested from the stationary phase produced the phenolic lipids in similar amounts as those grown in the presence of β-hydroxybutyrate, although they formed no cysts (data not shown). These observations are consistent with the previous observation that the encystment occurs frequently in β-hydroxybutyrate-induced cells, whereas it occurs in <0.01% of the aged, glucose-grown cells (18).
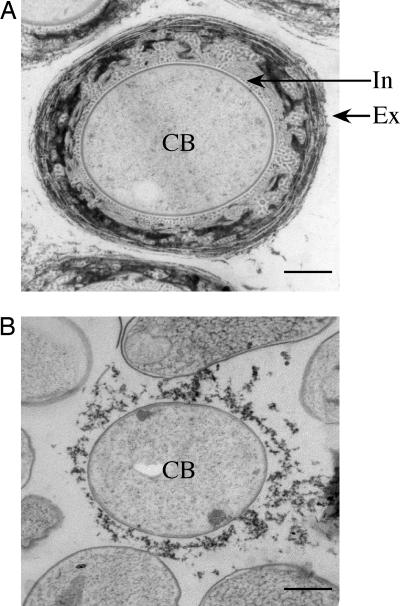
Fig. 5.
Requirement of the phenolic lipid for encystment of A. vinelandii. Electron micrographs of an ultrathin section of the cyst of A. vinelandii OP (A) and a cell of the arsB::aphII mutant (B) are shown. Τhe central body (CB) is surrounded by a multilayered cyst coat (exine, Ex) and an inner amorphous area (intine, In). (Scale bars: 0.5 μm.)
The importance of the phenolic lipids for encystment was confirmed by the failure in encystment of the arsB mutant. As described earlier, the arsB::aphII mutant produced no phenolic lipids. Transmission electron microscopy showed that the mutant formed no mature cysts; the central body with no intine in a cyst-like cell was surrounded by a severely impaired exine (Fig. 5B). Thus, the phenolic lipid synthesis is essential for the mature cyst formation in A. vinelandii. There was no morphological differences between the aged cells of A. vinelandii OP and the arsB::aphII mutant grown in the absence of β-hydroxybutyrate, when observed by optical and transmission electron microscopy (data not shown).
Materials and Methods
Bacterial strains, Plasmids, and Media.
E. coli JM109 and plasmid pUC19, used for DNA manipulation, were purchased from TaKaRa Biochemicals (Shiga, Japan). pET16b (Novagen) was used for expression of His-tagged proteins in E. coli BL21 (DE3). Media and growth conditions for E. coli were described by Sambrook et al. (19).
A. vinelandii strain OP (20), obtained from R. Dixon (John Innes Centre, Norwich, U.K.), was routinely cultured at 30°C for 8 days on Burk's medium (21) containing 2% glucose. For preparation of cysts, the cells collected from a 5-day culture in Burk's medium with 2% glucose were washed once with Burk's buffer (medium without a carbon source) and suspended in the same volume of Burk's buffer containing 0.2% β-hydroxybutyrate for the induction of encystment (18). The culture was incubated at 30°C for an additional 3 days for encystment. For microscopy studies, cells were grown first on 2% agar of Burk's medium containing 2% glucose for 5 days and then on 2% agar of Burk's medium containing 0.2% β-hydroxybutyrate. The agar plates were incubated at 30°C for an additional 3 days for encystment.
Isolation and Identification of 5-Heneicosylresorcinol.
A. vinelandii OP was grown at 30°C for 12 days in Burk's medium containing 2% glucose (2 liters × 4). The cells were harvested by centrifugation, suspended in a small volume of water, and extracted with chloroform/methanol (2:1, vol/vol). After the cell debris had been filtered off, the filtrate was lyophilized to dryness. The crude material was dissolved in chloroform and separated by silica gel chromatography by using chloroform as a solvent. The major compound in the fraction was applied to reversed-phase preparative HPLC equipped with a Pegasil C4 column (10 × 250 mm; Shenshu Scientific, Tokyo) and eluted by 95% CH3CN in water containing 0.1% trifluoroacetic acid at a flow rate of 3 ml/min to provide 2 mg of 5-heneicosylresorcinol as a white solid. 1H NMR (500 MHz, CDCl3) δ 6.22 (m, 2H, C4H, C6H), 6.15 (m, 1H, C2H), 2.46 (t, J = 8 Hz, 2H, C1′H), 1.23 (m, 38H, methylene of C2′ to C21′), 0.86 (t, J = 7 Hz, 3H, C22′H); 13C NMR (125 MHz, CDCl3) δ 156.5 (C-1, C-3), 146.5 (C-5), 108.0 (C-4, C-6), 100.1 (C-2), 35.8 (C-1′), 31.9 (C-20′), 31.1 (C-2′), 29.7 (C4′ to C19′), 29.4 (C3′), 22.7 (C21′), 14.1 (C22′); APCIMS: m/z 405.3 ([M + H]+ ion observed in a positive ion analysis), m/z 403.2 ([M − H]− ion observed in a negative ion analysis).
Synthesis of 6-Heneicosyl-4-hydroxy-2-pyrone and 4-Hydroxy-6-(2′-oxotricosyl)-2-pyrone.
The details of chemical synthesis are in Fig. 6, which is published as supporting information on the PNAS web site. We first prepared 1-eicosanal (22) and 4-benzyloxy-6-methyl-2-pyrone as reported in ref. 23 and used them for synthesis of 4-benzyloxy-6-(2′-hydroxyheneicosyl)-2-pyrone, which was then converted into 4-benzyloxy-6-(heneicos-1′-enyl)-2-pyrone. Hydrogenation of the pyrone by 10% palladium on carbon yielded 6-heneicosyl-4-hydroxy-2-pyrone. For synthesis of 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone, 4-benzyloxy-6-(2′-hydroxytricosyl)-2-pyrone was prepared from 1-docosanal and 4-benzyloxy-6-methyl-2-pyrone. Dess–Martin oxidation of 4-benzyloxy-6-(2′-hydroxytricosyl)-2-pyrone yielded 4-benzyloxy-6-(2′oxotricosyl)-2-pyrone, followed by deprotection to give 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone.
Synthesis of Behenyl-CoA.
Behenyl-CoA was synthesized by a modification of the procedure reported by Blecher (24). The details of chemical synthesis are in Fig. 7, which is published as supporting information on the PNAS web site.
Production and Purification of ArsB and ArsC.
The nucleotide sequence (ATCATG) covering the ATG start codon of arsB was changed to CATATG to create an NdeI site by PCR with primer I: 5′-CGCGAATTCCATATGAGCAGTCCCCACAACGCAGTT-3′ (with the nucleotides to be changed shown by the italic letters and an EcoRI site shown by underlining) and primer II: 5′-CGCGGATCCATCGGCCAGGACCGCGCT-3′ (with a BamHI site shown by underlining). Similarly, the nucleotide sequence (AATATG) covering the ATG start codon of arsC was changed to CATATG to create an NdeI site by PCR with primer III: 5′-CGCGAATTCCATATGAACGACATGGCCCACCCC-3′ (with the nucleotides to be changed shown by the italic letters and an EcoRI site shown by underlining) and primer IV: 5′-CGCGGATCCCGCTGTTGGTATCGAATACC-3′ (with a BamHI site shown by underlining). After amplification by PCR under the standard conditions, the EcoRI–BamHI fragments, excised from amplified arsB and arsC sequences, were separately cloned between the EcoRI and BamHI sites of pUC19, resulting in pUC19-ArsB and pUC19-ArsC, respectively. The absence of undesired alterations was checked by nucleotide sequencing. The NdeI–BamHI fragments, excised from pUC19-ArsB and pUC19-ArsC, were separately cloned between the NdeI and BamHI sites of pET16b, resulting in pET16b-ArsB and pET16b-ArsC, respectively. For production of histidine-tagged ArsB and ArsC, E. coli BL21 (DE3) harboring pET16b-ArsB or pET16b-ArsC was grown overnight in Luria broth containing 100 μg/ml ampicillin. Cells were harvested by centrifugation and resuspended in 10 mM Tris·HCl (pH 8.0) and 145 mM NaCl. After sonication, cell debris was removed by centrifugation, and the cleared lysate was applied to a column with His-bind metal chelation resin (Novagen). The histidine-tagged proteins were purified according to the manual from the manufacturer, except for adding 10% glycerol to each buffer. The purified histidine-tagged protein was dialyzed against 10 mM Tris·HCl (pH 8.0), 500 mM NaCl, and 10% glycerol.
PKS Assay.
The standard reaction mixture contained 100 μM [2-14C]malonyl-CoA, 100 μM starter CoA ester, 100 mM Tris·HCl (pH 8.0), and 61 μg of ArsB or ArsC in a total volume of 100 μl. Reactions were incubated at 30°C for 30 min before being quenched with 20 μl of 6 M HCl. The products were extracted with ethylacetate, and the organic layer was evaporated to dryness. The residual material was dissolved in 15 μl of methanol for TLC and LC-APCIMS analyses. Silica gel 60 WF254 TLC plates (Merck) were developed in benzene/acetone/acetic acid (85:15:1, vol/vol/vol), and the 14C-labeled compounds were detected by using a BAS-MS imaging plate (Fuji). LC-APCIMS analysis was carried out by using the esquire high-capacity trap plus (HCT) system (Bruker Daltonics, Bremen, Germany) equipped with a Pegasil-B C4 reversed-phase HPLC column (4.6 × 250 mm; Shenshu Scientific) and acetonitrile/water/acetic acid [900:100:1, vol/vol/vol (solvent A); 800:200:1, vol/vol/vol (solvent B); or 500:500:1, vol/vol/vol (solvent C)] as an eluant at a flow rate of 1 ml/min. UV spectra were detected on an Agilent 1100 series diode array detector. Spectral data are summarized in Table 2.
Determination of Kinetic Parameters of ArsB and ArsC.
The reactions, containing 100 mM Tris·HCl (pH 8.0), 100 mM NaCl, 100 μM malonyl-CoA, and 9.6 μg (0.674 μM) of ArsB or 9.2 μg (0.646 μM) of ArsC, were performed in a total volume of 300 μl. The concentration of behenyl-CoA was varied between 1 and 5 μM. After the reaction mixture had been preincubated at 30°C for 5 min, the reactions were initiated by adding the substrate and continued for 60 s. The reactions were stopped with 60 μl of 6 M HCl, and the material in the mixture was extracted with ethylacetate. The organic layer was collected and evaporated. The residual material was dissolved in 20 μl of methanol for HPLC analysis. 5-n-heneicosylresorcinol, 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone, and 6-heneicosyl-4-hydroxy-2-pyrone were used to generate the standard curves for the quantification of the products. Steady-state parameters were determined by Hanes plot.
Inactivation of ars Operon in A. vinelandii.
The 1.7-kb DNA fragment containing the region from Ser-1993 of ArsA to Thr-13 of ArsB was amplified with primer V: 5′-CGCAAGCTTCCTCGGCCGGCCTGCCGGTC-3′ (with the Ser-1993 codon of arsA in bold and a HindIII site in italic) and primer VI: 5′-CGCAAGCTTGGATCCTGTGAAGCCGGTGAGAACTG-3′ (with the Thr-13 codon of arsB in bold and a BamHI site in italic). The amplified fragment was cloned between the HindIII and BamHI sites of pUC19, resulting in pUC19-N-BamHI. The 1.6-kb DNA fragment containing the region from Glu-404 of ArsB to His-120 of ArsD was amplified with primer VII: 5′-CGCGGATCCGAGAAAATATGAACGACATG-3′ (with the Glu-404 codon of arsB in bold, the stop codon of arsB shown by the underline, and a BamHI site in italic) and primer VIII: 5′-CGCGAATTCGCTTGACCCCGGGGGCATGG-3′ (with the His-120 codon of arsD in bold and an EcoRI site in italic). The amplified fragment was cloned between the BamHI and HindIII sites of pUC19, resulting in pUC19-C-BamHI. After confirmation of the correct sequence, the HindIII–BamHI fragment from pUC19-N-BamHI and the BamHI–EcoRI fragment from pUC19-C-BamHI were ligated via the common BamHI site and cloned between the HindIII and EcoRI sites of pUC19. The 1.9-kb BamHI fragment containing aphII from Tn5 was inserted into the BamHI site of the resultant plasmid, resulting in pUC19-ΔArsB, in which the sequence encoding from Pro-14 to Leu-403 of ArsB was replaced by the aphII sequence. pUC19-ΔArsB was linearized with DraI, denatured with 0.1 M NaOH, and introduced by transformation into A. vinelandii OP by the method of Page and von Tigerstrom (25). Kanamycin-resistant transformants were selected, and a gene replacement, as a result of double crossover, was confirmed by Southern blot analysis (data not shown).
Electron Microscopy.
A. vinelandii cells were mounted on a copper grid to form a thin layer and immersed in liquid propane cooled with liquid nitrogen (Leica EM CPC and Leica Mikrosystems). The frozen cells were transferred to 2% OsO4 in dry acetone and kept at −80°C for 3 days. The samples were warmed gradually from −80°C to 0°C over 5 h, held for 1 h at 0°C, and warmed from 0°C to 23°C for 2 h (Leica EM AFS and Leica Mikrosystems). After the samples had been washed three times with dry acetone, they were infiltrated with increasing concentrations of Spurr's resin in dry acetone and finally with 100% Spurr's resin. After polymerization, ultrathin sections were cut on an ultramicrotome (Leica Ultracut UCT and Leica Mikrosystems) and stained with uranyl acetate and lead citrate. The sections were viewed on an H-7600 electron microscope (Hitachi, Tokyo) at 100 kV.
Supplementary Material
Acknowledgments
We thank Ray Dixon (John Innes Centre, Norwich, U.K.) for providing us with the A. vinelandii strain. This work was supported by a grant-in-aid for Scientific Research on Priority Areas from Monkasho (to S.H.) and the Waksman Foundation of Japan (to N.F.).
Abbreviations
- ACP
acyl carrier protein
- ars
alkylresorcinol synthesis
- CHS
chalcone synthase
- LC-APCIMS
liquid chromatography–atmospheric pressure chemical ionization mass spectrometry
- PKS
polyketide synthase
- STS
stilbene synthase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kozubek A., Tyman J. H. P. Chem. Rev. (Washington, D.C.) 1999;99:1–25. doi: 10.1021/cr970464o. [DOI] [PubMed] [Google Scholar]
- 2.Reusch R. N., Sadoff H. L. Nature. 1983;302:268–270. doi: 10.1038/302268a0. [DOI] [PubMed] [Google Scholar]
- 3.Su C.-J., Sadoff H. L. J. Bacteriol. 1981;147:91–96. doi: 10.1128/jb.147.1.91-96.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawlings J. B. Nat. Prod. Rep. 2001;18:190–227. doi: 10.1039/b009329g. [DOI] [PubMed] [Google Scholar]
- 5.Shen B. Top. Curr. Chem. 2000;209:1–51. [Google Scholar]
- 6.Austin M. B., Noel J. P. Nat. Prod. Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 7.Kozubek A., Pietr S., Czerwonka A. J. Bacteriol. 1996;178:4027–4030. doi: 10.1128/jb.178.14.4027-4030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer J.-L., Jez J. M., Bowman M. E., Dixon R. A., Noel J. P. Nat. Struct. Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 9.Lambalot R. H., Gehring A. M., Flugel R. S., Zuber P., La Celle M., Marahiel M. A., Reid R., Khosla C., Walsh C. T. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 10.Sankaranarayanan R., Saxena P., Marathe U. B., Gokhale R. S., Shanmugam V. M., Rukmini R. Nat. Struct. Mol. Biol. 2004;11:894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 11.Saxena P., Yadav G., Mohanty D., Gokhale R. S. J. Biol. Chem. 2003;278:44780–44790. doi: 10.1074/jbc.M306714200. [DOI] [PubMed] [Google Scholar]
- 12.Austin M. B., Bowman M. E., Ferrer J.-L., Schröder J., Noel J. P. Chem. Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Rock C. O., Cronan J. E. Biochim. Biophys. Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 14.Schweizer E., Hofmann J. Microbiol. Mol. Biol. Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su C.-J., Reusch R., Sadoff H. L. J. Bacteriol. 1979;137:1434–1436. doi: 10.1128/jb.137.3.1434-1436.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pal S., Manna A., Paul A. K. Curr. Microbiol. 1997;35:327–330. doi: 10.1007/s002849900263. [DOI] [PubMed] [Google Scholar]
- 17.Segura D., Cruz T., Espín G. Arch. Microbiol. 2003;179:437–443. doi: 10.1007/s00203-003-0553-4. [DOI] [PubMed] [Google Scholar]
- 18.Lin L. P., Sadoff H. L. J. Bacteriol. 1968;95:2336–2343. doi: 10.1128/jb.95.6.2336-2343.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch E. F., Maniatis T. In: Molecular Cloning: A Laboratory Manual. Nolan C, editor. Woodbury, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Bush J. A., Wilson P. W. Nature. 1959;184:381. [Google Scholar]
- 21.Kennedy C., Gamal R., Humphrey R., Ramos J., Brigle K., Dean D. Mol. Gen. Genet. 1986;205:318–325. [Google Scholar]
- 22.Schwink L., Knochel P. Tetrahedron Lett. 1994;35:9007–9010. [Google Scholar]
- 23.Funa N., Ohnishi Y., Ebizuka Y., Horinouchi S. J. Biol. Chem. 2002;277:4628–4635. doi: 10.1074/jbc.M110357200. [DOI] [PubMed] [Google Scholar]
- 24.Blecher M. Methods Enzymol. 1981;72:404–408. doi: 10.1016/s0076-6879(81)72030-5. [DOI] [PubMed] [Google Scholar]
- 25.Page W. J., von Tigerstrom M. J. Bacteriol. 1979;139:1058–1061. doi: 10.1128/jb.139.3.1058-1061.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.