Abstract
Much evidence from studies in humans and animals supports the hypothesis that alcohol addiction is a complex disease with both hereditary and environmental influences. Molecular determinants of excessive alcohol consumption are difficult to study in humans. However, several rodent models show a high or low degree of alcohol preference, which provides a unique opportunity to approach the molecular complexities underlying the genetic predisposition to drink alcohol. Microarray analyses of brain gene expression in three selected lines, and six isogenic strains of mice known to differ markedly in voluntary alcohol consumption provided >4.5 million data points for a meta-analysis. A total of 107 arrays were obtained and arranged into six experimental data sets, allowing the identification of 3,800 unique genes significantly and consistently changed between all models of high or low amounts of alcohol consumption. Several functional groups, including mitogen-activated protein kinase signaling and transcription regulation pathways, were found to be significantly overrepresented and may play an important role in establishing a high level of voluntary alcohol drinking in these mouse models. Data from the general meta-analysis was further filtered by a congenic strain microarray set, from which cis-regulated candidate genes for an alcohol preference quantitative trait locus on chromosome 9 were identified: Arhgef12, Carm1, Cryab, Cox5a, Dlat, Fxyd6, Limd1, Nicn1, Nmnat3, Pknox2, Rbp1, Sc5d, Scn4b, Tcf12, Vps11, and Zfp291 and four ESTs. The present study demonstrates the use of (i) a microarray meta-analysis to analyze a behavioral phenotype (in this case, alcohol preference) and (ii) a congenic strain for identification of cis regulation.
Keywords: alcoholism, gene expression, microarray
An understanding of the molecular mechanisms underlying the genetic propensity toward excessive alcohol consumption is crucial for the development of new treatments for alcoholism. Animal models for alcohol-related traits provide an important opportunity to explore mechanisms responsible for different aspects of the uniquely human disease (1). In particular, selected lines and inbred strains of mice with a divergence in voluntary alcohol drinking represent valuable tools to dissect the genetic components of alcoholism (2, 6, 8, 9, §§). However, each model has different advantages. Inbred strains are homogeneous at all alleles, and genetic (allelic) differences between strains lead to observed phenotypic differences. Several inbred strains have been well characterized for numerous alcohol-related traits, but any two strains will differ for numerous other phenotypes (including gene expression), making identification of the specific genes for a given trait difficult. Selected lines exploit homozygosity for large-effect and some small-effect allelic variants that contribute to the divergent preference for drinking. Although the resulting genome remains relatively polymorphic, as selection continues, fixation of genes unrelated to the selected phenotype occurs because of drift.
We propose to identify candidate genes that contribute to the genetic propensity to drink by combining whole-brain gene expression databases of genetic mouse models with differences in voluntary alcohol consumption. Most importantly, our approach should allow identification of small- or moderate-effect genes. Three different sets of oppositely selected lines bred for high and low amounts of alcohol drinking, five inbred strains known to differ in voluntary alcohol consumption, and a hybrid strain recently shown to have the greatest degree of voluntary alcohol consumption of any known mouse genotype (4) were used (Table 1). Global measurement of brain gene expression in each of the mouse models allowed us to explore which transcripts are consistently changed in different genetic models of high and low amounts of alcohol intake. It is important to note that the mice used for array analysis were not exposed to alcohol; thus, we are defining the transcriptional signatures of genetic predisposition to high and low levels of alcohol consumption. Expression analysis of an additional mouse line congenic for a section of chromosome 9, which is known to contain genes that regulate alcohol consumption (3), allowed us to define candidate quantitative trait genes (QTGs) for this quantitative trait locus (QTL).
Table 1.
Mouse models and microarrays used
| Data set | Study | Platform | Strain/line | Sex | N |
|---|---|---|---|---|---|
| 1 | STS (B6.D2) | Affymetrix | High STS | F/M | 10 |
| Low STS | F/M | 10 | |||
| 2 | HAP/LAP1 | Affymetrix | HAP1 | M | 6 |
| LAP1 | M | 6 | |||
| 3 | HAP/LAP2 | Affymetrix | HAP2 | M | 6 |
| LAP2 | M | 6 | |||
| 4 | Inbred strains | Affymetrix | B6 | M | 6 |
| BalbC | M | 6 | |||
| D2 | M | 6 | |||
| LP/NJ | M | 6 | |||
| 5 | Inbred strains | Custom cDNA | B6 | F | 5 |
| FVB/NJ | F | 5 | |||
| FVB.B6 hybrid | F | 5 | |||
| 6 | B6.D2 congenic 9 | Custom cDNA | B6 | F/M | 12 |
| B6.D2 Congenic 9 | F/M | 12 |
Three selected lines (data sets 1–3) and the inbred strains (data sets 4 and 5) were used to form four groups for the meta-analysis. The congenic line (data set 6) was used as a filter to select genes from the meta-analysis. See Materials and Methods for details. STS, short-term selection; F, female; M, male. Data set 1 was contributed by R.J.H., J.C.C., and J.K.B. Data sets 2 and 3 were contributed by B.T. Data set 4 was contributed by R.J.H. and J.K.B. Data sets 5 and 6 were contributed by S.E.B.
Analysis across data sets for common changes in expression provides more statistical power to detect small and reliable changes than analysis of any one or two of the data sets. Meta-analyses of diverse gene expression data sets were recently used to successfully uncover genes related to carcinogenic phenotypes (10), but a similar approach has not been used for a neurobehavioral trait (for a review of meta-analyses of microarray data, see ref. 11). In this study, we employ a meta-analysis of microarray data from different genetic models of high and low levels of alcohol consumption to define functional pathways and individual genes that may determine a predisposition for a high degree of alcohol intake.
Results and Discussion
Use of Cohen's d as an Effect-Size Measure Allows Comparison of Microarray Experiments from Diverse Sources.
Five experimental microarray (Affymetrix and custom cDNA) data sets of mice genetically divergent in voluntary alcohol consumption were analyzed for differential gene expression with the Cohen's d statistic (12). The initial meta-analysis comprised 13 individual data sets from three groups of selected lines and one group of six isogenic strains (Table 1). (A data set consisting of C57BL/6J (B6).DBA/2J (D2) congenic 9 animals is used later as a filter for the initial meta-analysis.) Two advantages of the Cohen's d approach are important. First, data from different platforms and laboratories can be combined without the use of normalization. Second, small differences that are consistent in direction of change can be detected, even though these changes might not be detected in any single data set.
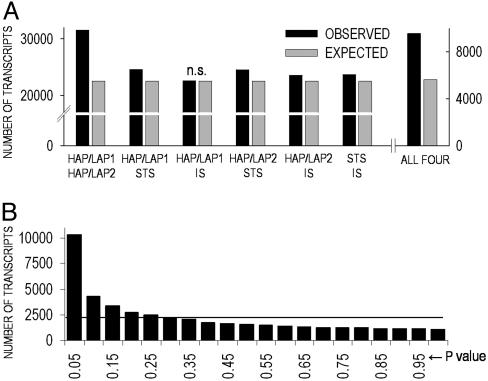
After calculating Cohen's d for each data set (Fig. 1A), a pairwise comparison of common changes was applied to test the compatibility of the four data sets. The number of transcripts regulated in the same direction between each pair of data sets is significantly greater than the number ascribed to chance for five of six pairs. When all four data sets are compared, the number of transcripts regulated in the same direction is nearly 2-fold over the chance level (P < 0.00001, χ2). The lower similarity seen between selected lines and inbred strains, compared with the pairs of selected lines, likely reflects differences in genetic background, an enrichment of alleles linked to alcohol preference in selected lines, and a relative randomness of such alleles in inbred strains.
Fig. 1.
Compatibility of data sets for meta-analysis. (A) Number of transcripts regulated in the same direction between any two (left y axis) or all four (right y axis) data sets (P < 0.0001, χ2). n.s., not significant. (B) Frequency distribution of z test P values (x axis) is shown. The solid line represents theoretical chance distribution. IS, Inbred strain.
An overlap in regulation between mouse models sharing the same alcohol phenotype is readily detectable despite the use of data sets from different animal models and platforms. The distribution of z test P values for the meta-analysis of the four data sets is skewed to the left, indicating a high number of significantly coexpressed genes detected by the meta-analysis (Fig. 1B) and suggesting a low incidence of false positives consistent with the measured Q values. Thus, the use of Cohen's d in our meta-analysis allows for the detection of transcripts significantly divergent between genetically distinct mouse models displaying opposite levels of alcohol consumption.
Meta-Analysis Identifies Candidate Genes for High and Low Amounts of Alcohol Consumption.
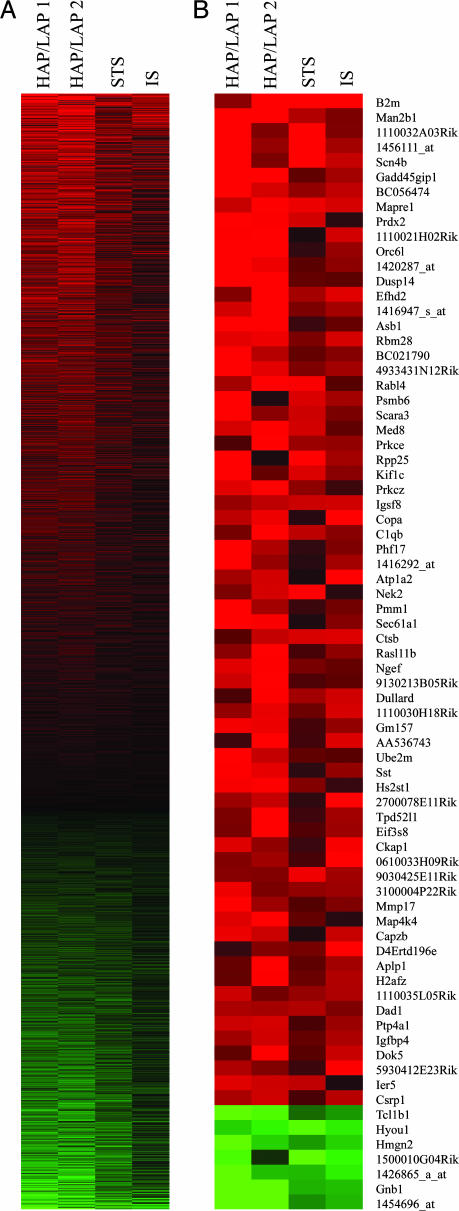
Use of Cohen's d values between the mouse strains and lines displaying high and low levels of alcohol drinking across data sets resulted in the identification of 5,182 significant (∣d∣ < 0.5 and Q < 0.05) differentially expressed transcripts, representing 3,800 unique genes (data for all transcripts is shown in Table 2, which is published as supporting information on the PNAS web site). The highly significant gene expression changes (Q < 0.01) across the four data sets are noticeably consistent when seen in pseudocolor raster display (Fig. 2A). Red and green indicate transcripts overexpressed and underexpressed in animals displaying high degrees of alcohol consumption, respectively. In general, more transcripts appear to be up-regulated in models with high levels of consumption relative to models with low levels of consumption.
Fig. 2.
Visual representation of microarray data. Columns represent microarray databases used in the meta-analysis, and rows represent transcripts. The filtered criterion was Q < 0.01, and genes were sorted by effect size. (A) Transcripts (2,697) are listed from positive to negative Cohen's d values (effect size). Red indicates a positive effect size and higher expression in preferring mice, and green indicates a negative effect size and lower expression in preferring mice. Black indicates an effect size near zero. (B) The 75 unique transcripts with the highest absolute average effect size (Q < 0.05, ∣d∣ > 1.94). STS, short-term selection; IS, Inbred strains.
The sheer number of significant differences suggests that numerous pathways in the brain may be distinct between mouse models with different levels of alcohol consumption. Indeed, the 75 genes with the largest effect size (∣d∣) and lowest Q values (Fig. 2B) fall into broad categories of cellular homeostasis and neuronal function. Several genes of interest expressed higher in preferring models include: B2m, Man2b1, Scn4b, Mapre1, Prkce, and Sst, which function in immunity/cellular defense, glycosylation, ion channel activity, microtubule binding/dynamics, intracellular signaling cascades, and neuronal signaling, respectively. β2-microglobulin may play a role in neuronal plasticity because B2m knockout mice exhibit abnormal synaptic connections as well as altered patterns of long-term potentiation and long-term depression (13). Scn4b is an auxiliary β-subunit of voltage-gated sodium channels, which can alter channel function and interaction between the ion channel and other proteins (14). In cerebellar Purkinje cells, Scn4b may be required for high-frequency firing of neurons containing voltage-gated sodium channels (15). A role for Prkce (PKC-ε) in ethanol consumption is supported by the observation that Prkce knockout mice consume less alcohol compared with controls in a two-bottle choice drinking paradigm, and introduction of conditional Prkce expression in these mice was sufficient to restore alcohol drinking levels (16). Gnb1, Hmgn2, and Hyou function in GTPase activity/signal transduction, DNA binding/packaging, and the cellular response to stress, respectively, and are significantly down-regulated in mice preferring alcohol. The overall results of the meta-analysis show the complexity of the alcohol drinking phenotype as indicated by the surprisingly large number of consistent, small increases or decreases in gene expression between the mouse models.
In Silico Promoter and Functional Analyses Uncover Potential Significant Regulation and Pathways, Respectively.
Nearly 10% of the genes in the mammalian genome were detected as divergent between the alcohol preference lines and strains of mice. One possible explanation is linkage disequilibrium, the tendency of closely linked genes to be coexpressed. Another explanation is that some of the risk-conferring QTG are transcriptional regulators. Although current mouse in silico promoter analysis tools are not yet comprehensive on a genomic scale, the oPOSSUM database (17) is a valuable tool for identifying overrepresented transcription factor (TF) binding sites (TFBS) in promoter regions near a gene's transcription start site. oPOSSUM analysis is very conservative and has strict requirements; simulation studies have suggested that it detects few false positives (17).
TFBS for Zfp143 (Staf, selenorysteine RNA gene transcription activating factor) were identified as overrepresented in the up-regulated genes (for high amounts of alcohol drinking). The Staf TFBS was present in the upstream (2,000 bp) promoter regions of 64 genes. Importantly, Zfp143 expression was also found to be significantly up-regulated across the models with high drinking levels. Similarly, the TFBS for the fork-head box TF Foxa2 (HNF-3β, hepatocyte nuclear factor 3β) was detected as overrepresented across the down-regulated gene group and present in 146 genes, and Foxa2 was found to be significantly down-regulated in the meta-analysis. In both cases, the overrepresented TF has the same pattern of expression as its target genes. It is easy to imagine, as evidenced by the number of gene targets for Zfp143 and Foxa2, how small changes in the levels of a TF could cause profound differences in the brain transcriptome and may account for the large number of gene expression changes (3,800 genes) detected in this meta-analysis. Although neither Zfp143 or Foxa2 were identified as cis-regulated and are not primary candidate genes for alcohol preference, an understanding of how changes in the expression level and/or activity of transcriptional regulators affects the predisposition toward alcohol consumption phenotypes warrants further investigation.
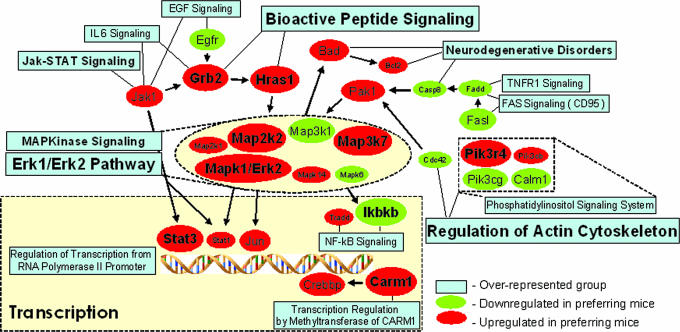
To better understand possible global interactions of the divergent genes, overrepresentation analyses for function were completed. Such analyses apply statistical methods to estimate whether some biological function/pathway is represented in a given data set more than expected by chance. Using the webgestalt search engine, which queries different databases, including KEGG, BioCarta, and GO (Gene Ontology) (18–20), functional group analysis revealed that kinase and signaling pathways were overrepresented in genes divergent between alcohol-preferring and nonpreferring genotypes. The result supports previous suggestions that the mitogen-activated protein kinase, NF-κB, and IL-6 signaling pathways (and CREBBP) are sensitive to alcohol (21). Known functional interactions (summarized in Fig. 3) show that gene transcription pathways predominant in the overrepresented categories and apoptosis/antiapoptosis, neurodegeneration, and neuroplasticity pathways were also functionally overrepresented (for all results, see Fig. 5 and Table 3, which are published as supporting information on the PNAS web site). In silico functional analyses are powerful tools for hypothesis generation, especially for complex phenotypes, such as predisposition toward alcohol drinking. It should now be feasible, for example, to test the effect of candidate genes in Fig. 3 for their impact on alcohol consumption.
Fig. 3.
Complexity of functional group interaction. Genes present in at least three overrepresented functional groups/pathways from BioCarta, KEGG, and Gene Ontology are shown. Larger font sizes represent either smaller P values for functional groups/pathways (from overrepresentation analysis) or larger effect size for individual genes (P < 0.001, Q < 0.01, and ∣d∣ > 0.5 for all genes). Lines connect gene symbols with relevant functional groups. Arrows indicate pathway connections. For full gene names, see Table 2.
Several Genes Identified in the Meta-Analysis Are Located Within QTLs for Human Alcoholism Susceptibility.
A two-pronged approach was taken to use known QTLs in both human and rodent research. First, the list of ≈3,800 significant genes was annotated using source (22) for mouse and human chromosomal location. Two separate lists were created by using Feighner, DSM-III-R, and ICD-10 criteria (23): one list for genes found within the boundaries of mouse QTLs for preference drinking (3) and another for genes within the boundaries of alcoholism susceptibility QTLs in humans. The lists were used as filters to identify overlap of significant genes from the meta-analysis with QTLs between the two species (Table 4, which is published as supporting information on the PNAS web site). Thirty-six genes met criteria as candidate genes with 11 (Gstm1, Gstm2, Gstm5, Il1f5, Il1f6, Il1f8, Il1rn, S100a1, S100a10, and S100a13) belonging to only three gene families: GSTM (glutatione S-transferase activity), S100 class calcium-binding, and cytokine/proinflammatory activity (IL-1) (Table 5, which is published as supporting information on the PNAS web site). GSTM1 alleles have been shown to be associated with increased alcoholism and alcoholic liver disease (24), and recent results from mice with mutant chemokine receptors suggest a role of inflammatory pathways in alcohol consumption (25). QTL analyses for alcohol preference in rats (26, 27) (not listed in Table 5) show two syntenic QTLs with mouse, and the chromosome 2 QTL is syntenic between all three species, including humans. GST8-8, the implicated QTG within the QTL on rat chromosome 8 (28), is syntenic on mouse chromosome 9. However, Gsta4, the mouse alias for GST8-8, was not significantly divergent in this meta-analysis. Interestingly, several other significantly regulated glutatione S-transferase genes found on mouse chromosome 1 and human chromosome 3 were in syntenic QTL regions for both species (again, see Table 5).
It is important to note that, although there is considerable synteny between the genomes of rodents and man, the degree of evolutionary overlap between polymorphisms or alleles of genes is probably much lower. Thus, translation of data from rodent models to humans is more likely to apply on a pathway level rather than as exact mutations in specific genes. The species differences in glutatione S-transferase may, in fact, be such an example.
Use of a B6.D2 Congenic 9 Data Set Filter Identifies cis-Regulated Genes Occupying a QTL for Strong Alcohol Preference in Mice.
QTL mapping and construction of congenic mice based on such maps provides another genetic approach to defining changes in gene expression linked to propensity for high levels of alcohol consumption. Several mouse QTL experiments show that chromosome 9 contains genes that contribute to the genetic propensity toward increased alcohol intake. This QTL was found to be the largest of several identified in B6- and D2-derived populations (3). Therefore, B6 congenic mice containing a region that captured the D2 alleles between 9 and 58 cM on chromosome 9 were analyzed for gene expression and compared against the B6 background strain. The differentially expressed genes located on chromosome 9 are by default cis-regulated and thus are QTG candidates.
To identify cis-acting candidate genes for the chromosome 9 alcohol preference QTL, the current meta-analysis results were filtered with the congenic data set. Significant genes detected by the meta-analysis that passed the congenic filter were mapped on chromosome 9 along with their relative effect size (Fig. 4). These genes include 16 known genes (Arhgef12, Carm1, Cryab, Cox5a, Dlat, Fxyd6, Limd1, Nicn1, Nmnat3, Pknox2, Rbp1, Sc5d, Scn4b, Tcf12, Vps11, and Zfp291) and four expressed sequence tags (2810423O19Rik, 1110032A03Rik, 5730439E10Rik, and 9030425E11Rik). Four of these (Pknox2, Scn4b, 9030425E11Rik, and 1110032A03Rik) are correlated with alcohol preference in a BXD recombinant inbred population (5). Interestingly, some primary candidate genes also have putative transcriptional activity: Carm1, Pknox2, Tcf12, Zfp29, and Limd1. Although their targets are not yet characterized, each may explain some of the consistently divergent gene expression observed in the meta-analysis. The 20 genes identified with the congenic filter are potential QTGs underlying alcohol preference and provide exciting avenues of investigation into the complex behavior of alcohol preference.
Fig. 4.
Congenic 9 expression analysis. Meta-analysis results were filtered by significant cis-regulation on chromosome 9. Gene names and their physical location are indicated on the left, and effect size and direction are shown on the right. Asterisks indicate a correlation between gene expression and the preference for drinking phenotype in a panel of BXD recombinant inbred strains (5) generated with the WebQTL (www.genenetwork.org) Integrative Neuroscience Initiative on Alcoholism Brain mRNA M430 (April 2005 release) PDNN (Positional Dependent Nearest Neighbor) database [referred to as INIA Brain mRNA M430 (Apr05) PDNN by WebQTL] (7).
Conclusions
Predisposition to excessive alcohol consumption is likely a key aspect of human alcoholism, but molecular determinants of this trait are difficult to study in humans. Our work provides a microarray-based meta-analysis of a behavioral phenotype and uses effect-size measures. The Cohen's d metric eliminated the need for standardization across array platforms and experiments, allowing identification of consistently different transcripts between alcohol-preferring and nonpreferring genotypes, even if the transcriptional differences were small with respect to magnitude or significance or both.
The meta-analysis provided 5,182 targets, including 3,800 nonoverlapping genes for further study. The significant differences between the genotypes for divergent amounts of alcohol drinking represent numerous functional groups covering a large range of cellular pathways as well as many genes (≈25%) whose functions are not yet characterized. Thus, some genes whose functions are currently unknown are also likely to be important determinants of alcohol consumption. The 75 genes with the largest effect size (Q < 0.05, ∣d∣ > 1.94) (see Fig. 2) fall into the broad categories of cellular homeostasis and neuronal function. Differences in the ability to maintain or reset homeostasis and adjust neuronal function in brain likely underlie many aspects of alcohol responses. Understanding these differences will be key to a better understanding of alcoholism. For instance, it is possible that expression differences could substantially affect developmental and adaptive brain neurocircuitry relating to alcohol preference.
It should be noted that our approach is designed to detect common differences found across the animal models used but will not detect differences specific for one model or one sex, nor will it detect mutations that affect alcohol preference for which no concomitant change in gene expression results. Unique but potentially important determinants of drinking require detailed analysis of each data set, which has been accomplished for the HAP/LAP (mouse lines demonstrating high/low levels of alcohol preference, respectively; see ref. 6) selected lines (29).
Our meta-analytic approach using an effect-size metric should be applicable to new data sets. Two-bottle choice alcohol drinking is the most widely used model of voluntary consumption, but new models are emerging (30–33). It would be interesting to determine the generality of our results to other measures of alcohol consumption. Follow-up studies for candidate genes identified in previous array experiments have led to the understanding of molecular mechanisms for other behaviors (34, 35) and is likely for our candidate genes as well.
In summary, this meta-analysis shows that distinct mouse models with genetic predisposition for elevated alcohol consumption have very consistent and reproducible differences in brain gene expression, although they have never consumed alcohol. Furthermore, the differences extend to thousands of transcripts forming many functional groups. Combining the meta-analysis with congenic approaches provides specific candidate genes for one QTL for alcohol preference. These genes and most of the key functional groups revealed by the current study indicate our ignorance about the molecular mechanisms driving alcohol consumption and, most importantly, the opportunities to reveal mechanisms through large-scale genomic screening approaches.
Materials and Methods
Animal Husbandry.
Naïve, adult mice (60–100 d old) had 24 h ad libitum access to rodent chow, water, and 12 h:12 h lighting. All animals were housed and treated according to the National Institutes of Health guidelines for the use and care of laboratory animals (36) and approved Institutional Animal Care and Use Committee protocols at each respective institution. Whole-brain total RNA from naïve mice was used for all array analyses. Microarray hybridizations for the HAP/LAP lines, inbred strains, and congenic 9 strains were completed on individual mice. The lines for high and low levels of short-term selection were hybridized with a pool of three samples for each represented N value (n = 10 mice per line). Details for each strain are listed below.
Short-Term Selection Lines.
Reciprocally crossed, individually housed B6×D2 and D2×B6 F2 mice (8–10 weeks of age) were initially given water in a classical two-bottle choice paradigm. One tube was then replaced with a 3% (vol/vol) solution of ethanol in tap water followed by 10% ethanol (4 d each). The selection index for drinking was based on the 10% concentration expressed in grams per kilogram per day for each of the last days immediately before a bottle position change (i.e., the average of days 8 and 10). The 10 highest-scoring and 10 lowest-scoring mice of each gender were used as breeding pairs to initiate the selection lines with high and low amounts of drinking, respectively. Bidirectional selection continued similarly for each of three generations; the highest of the high line and the lowest of the low line were used to breed the next generation of the high and low selection lines, respectively. Naïve mice with high and low levels of drinking from selection generation S3 were used in the array experiment.
HAP/LAP.
Two replicate lines of HAP and LAP mice were originally generated from HS/Ibg mice (2) based on bidirectional selection outcome of two-bottle choice 10% alcohol solution vs. water consumption (6, §§). Male HAP1/LAP1 and HAP2/LAP2 mice (70–90 d old) were used from generations 26 and 19, respectively, for Affymetrix array analysis.
Inbred Strains.
Male BALB/cJ, B6, D2, and LP/NJ inbred mice were raised at the Veterans Affairs Medical Center in Portland, OR. LP/NJ mice were tested for two-bottle choice alcohol drinking and found to drink significantly less alcohol than B6 mice (D.A.F., unpublished data). Female B6, FVB/NJ, and FVB×B6 F1 mice were raised at the University of Texas. F1 mice were recently found to drink significantly more alcohol than the progenitor strains when tested in an accelerating concentration two-bottle choice paradigm (4).
Congenic Strain.
A D2 QTL region for alcohol preference from 9 to 58 cM on chromosome 9 was introgressed onto a B6 genomic background through ≥12 generations of breeder selection by Massachusetts Institute of Technology microsatellite marker genotyping and backcrossing to B6. The B6.D2Ch9 mice drink significantly less than B6 control mice (difference ≈25%, T.J.P., unpublished data). mRNA was isolated from jointly housed and age-matched (70–100 days) congenic and B6 mice.
Oligonucleotide Microarrays.
Samples containing at least 10 μg of total RNA were hybridized after manufacturer's protocol (Affymetrix, Santa Clara, CA). Details for HAP/LAP mice have been published in ref. 29. RNA from STS mice and mice from inbred strains BALB/cJ, B6, D2, and LP/NJ arrays was hybridized to Affymetrix 430 A and B chips and analyzed at the Oregon Health and Science University Gene Microarray Shared Resource Facility (for more details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Custom cDNA Microarrays.
PCR products of clones from several cDNA libraries (see Supporting Materials and Methods for details) were printed in the University of Texas array core facility on poly-l-lysine-coated (Sigma) microscope slides (Erie Scientific, Portsmouth, NH) using a custom-built robotic arrayer as described in ref. 37. Microarray hybridizations were performed with the Array 350 microarray labeling kit according to the manufacturer's protocol (Genisphere, Hatfield, PA).
Meta-Analysis.
Five data sets listed 1–5 in Table 1 were used for meta-analysis. Alcohol-preferring and nonpreferring mice were first compared by using a parametric statistical test. Student's t test for independent samples was used to compare high and low selected lines (studies 1–3). Both sets of inbred strains (studies 4 and 5) were analyzed independently by an F test using single contrasts (see Supporting Materials and Methods for additional details). The resulting t and F values for each transcript were then used to estimate effect size by calculating a Cohen's d statistic, which is the difference between the groups expressed in pooled within-group SD units (12). The following formulas were applied: d = 2t/ = 2 , where df represents the degrees of freedom. The direction of change was coded in the resulting Cohen's d values so, that 〈positive〉 values indicated an up-regulation and 〈negative〉 values indicated a down-regulation of transcripts in alcohol-preferring genotypes compared with nonpreferring genotypes.
Data from the two inbred strain studies (studies 4 and 5 in Table 1) were collapsed by averaging Cohen's d values for each transcript (for rationale, see Supporting Materials and Methods). To minimize any outlier effects, extremely deviant effect sizes were adjusted to a maximum absolute value of d = 4. Finally, the Cohen's d values generated for the three pairs of selected lines and the combined set of inbred strains were averaged, and a z test was used to test the significance of deviation of the mean effect size from zero. qvalue software was used to estimate the false discovery rate (Q) for the meta-analysis results (38, 39), which estimates the proportion of all declared significant results that are expected to be false positives.
Congenic 9 Data Filter.
We used microarray data obtained from the B6.D2Ch9 congenic strain to filter transcripts detected by meta-analysis as regulated significantly between ethanol-preferring and nonpreferring genotypes and located within the introgressed region containing known ethanol preference QTLs on mouse chromosome 9. Specifically, chromosome 9 transcripts significantly regulated between the congenic and control lines (P < 0.05, t test) were identified and matched for direction of change with data from the meta-analysis. Only genes detected by both approaches were selected as putative candidates for cis-regulation of alcohol preference.
TFBS Overrepresentation Analysis.
The oPOSSUM database (www.cisreg.ca/cgi-bin/oPOSSUM/opossum) (17) was used to analyze TFBS in genes that were significantly (Q < 0.05 and ∣d∣ ≥ 0.5) up- or down-regulated in alcohol-preferring vs. nonpreferring mice. The two lists had 2,388 and 1,580 genes with 1,011 and 546 recognized as orthologs in oPOSSUM for up- vs. down-regulation respectively. TFBS overrepresentation was determined based on one-tailed Fisher exact probabilities and the ranking of z scores. After the initial analysis, the genes for the TFs themselves were individually checked to see whether (i) they were present on the arrays and (ii) transcription differences were consistent with the promoter TFBS analysis.
Functional Overrepresentation Analysis.
Transcripts passing statistical thresholds (Q < 0.05, ∣d∣ ≥ 0.5) were functionally annotated with webgestalt (web-based gene set analysis toolkit, http://genereg.ornl.gov.webgestalt) (19). A hypergeometric test was used to detect functional groups overrepresented in a selected gene set (meta-regulated genes) compared with a reference set (all genes in meta-analysis). See Supporting Materials and Methods for additional details.
Supplementary Material
Acknowledgments
This study was a collaborative effort by investigators affiliated with the Integrative Neuroscience Initiative on Alcoholism Consortium and was supported by National Institute on Alcohol Abuse and Alcoholism Grants AA013182 and AA013404 (to S.E.B.); AA013475, AA006243, and AA010760 (to J.K.B.); AA013520 (to Y.A.B.); AA013519 (to J.C.C.); AA013483 (to N.J.G.); AA013518 (to R.A.H.); AA013484 and AA11034 (to R.J.H.); AA013517 (to G.F.K.); and AA013489 (to B.T.); and by five Merit Review Programs from the Department of Veterans Affairs (to J.K.B., J.C.C., D.A.F., R.J.H., and T.J.P.).
Abbreviations
- B6
C57BL/6J
- D2
DBA/2J
- QTG
quantitative trait gene
- QTL
quantitative trait locus
- TF
transcription factor
- TFBS
TF binding site(s).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
§§Behm, A. L. L., Li, T. K. & Grahame, N. (2003) Alcohol. Clin. Exp. Res. 27, 49A, abstr.
References
- 1.Crabbe J. C. Am. J. Med. Genet. 2002;114:969–974. doi: 10.1002/ajmg.b.10984. [DOI] [PubMed] [Google Scholar]
- 2.McClearn G., Wilson J., Meredith W. The Use of Isogenic and Heterogenic Mouse Stocks in Behavioral Research. New York: Appleton–Century–Crofts; 1970. [Google Scholar]
- 3.Belknap J. K., Atkins A. L. Mamm. Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- 4.Blednov Y. A., Metten P., Finn D. A., Rhodes J. S., Bergeson S. E., Harris R. A., Crabbe J. C. Alcohol. Clin. Exp. Res. 2005;29:1949–1958. doi: 10.1097/01.alc.0000187605.91468.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips T. J., Crabbe J. C., Metten P., Belknap J. K. Alcohol. Clin. Exp. Res. 1994;18:931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- 6.Grahame N. J., Li T. K., Lumeng L. Behav. Genet. 1999;29:47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Williams R. W., Manly K. F. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- 8.Phillips T. J., Terdal E. S., Crabbe J. C. Behav. Genet. 1990;20:473–480. doi: 10.1007/BF01067713. [DOI] [PubMed] [Google Scholar]
- 9.Phillips T. J., Belknap J. K., Hitzemann R. J., Buck K. J., Cunningham C. L., Crabbe J. C. Genes Brain Behav. 2002;1:14–26. doi: 10.1046/j.1601-1848.2001.00011.x. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. Proc. Natl. Acad. Sci. USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreau Y., Aerts S., De Moor B., De Strooper B., Dabrowski M. Trends Genet. 2003;19:570–577. doi: 10.1016/j.tig.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal D. Parametric Measures of Effect Size. New York: Russell Sage Found; 1994. [Google Scholar]
- 13.Huh G. S., Boulanger L. M., Du H., Riquelme P. A., Brotz T. M., Shatz C. J. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu F. H., Westenbroek R. E., Silos-Santiago I., McCormick K. A., Lawson D., Ge P., Ferriera H., Lilly J., DiStefano P. S., Catterall W. A., et al. J. Neurosci. 2003;23:7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grieco T. M., Malhotra J. D., Chen C., Isom L. L., Raman I. M. Neuron. 2005;45:233–244. doi: 10.1016/j.neuron.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Hodge C. W., Mehmert K. K., Kelley S. P., McMahon T., Haywood A., Olive M. F., Wang D., Sanchez-Perez A. M., Messing R. O. Nat. Neurosci. 1999;2:997–1002. doi: 10.1038/14795. [DOI] [PubMed] [Google Scholar]
- 17.Ho Sui S. J., Mortimer J. R., Arenillas D. J., Brumm J., Walsh C. J., Kennedy B. P., Wasserman W. W. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris M. A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B., Kirov S., Snoddy J. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong H. J., Hong S. H., Park R. K., An N. H., Kim H. M. Life Sci. 2005;77:2179–2192. doi: 10.1016/j.lfs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Diehn M., Sherlock G., Binkley G., Jin H., Matese J. C., Hernandez-Boussard T., Rees C. A., Cherry J. M., Botstein D., Brown P. O., Alizadeh A. A. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foroud T., Edenberg H. J., Goate A., Rice J., Flury L., Koller D. L., Bierut L. J., Conneally P. M., Nurnberger J. I., Bucholz K. K., et al. Alcohol. Clin. Exp. Res. 2000;24:933–945. [PubMed] [Google Scholar]
- 24.Engracia V., Leite M. M., Pagotto R. C., Zucoloto S., Barbosa C. A., Mestriner M. A. Am. J. Med. Genet. 2003;123:257–260. doi: 10.1002/ajmg.a.20364. [DOI] [PubMed] [Google Scholar]
- 25.Blednov Y. A., Bergeson S. E., Walker D., Ferreira V. M., Kuziel W. A., Harris R. A. Behav. Brain Res. 2005;165:110–125. doi: 10.1016/j.bbr.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carr L. G., Habegger K., Spence J., Ritchotte A., Liu L., Lumeng L., Li T. K., Foroud T. Alcohol. Clin. Exp. Res. 2003;27:1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- 27.Bice P., Foroud T., Bo R., Castelluccio P., Lumeng L., Li T. K., Carr L. G. Mamm. Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- 28.Liang T., Habegger K., Spence J. P., Foroud T., Ellison J. A., Lumeng L., Li T. K., Carr L. G. Alcohol. Clin. Exp. Res. 2004;28:1622–1628. doi: 10.1097/01.alc.0000145686.79141.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saba L., Bhave S., Lapadat R., Hoffman P. L., Belknap J. K., Tabakoff B. Mamm. Genome. 2006 doi: 10.1007/s00335-005-0190-0. in press. [DOI] [PubMed] [Google Scholar]
- 30.Finn D. A., Belknap J. K., Cronise K., Yoneyama N., Murillo A., Crabbe J. C. Psychopharmacology. 2005;178:471–480. doi: 10.1007/s00213-004-2039-8. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes J. S., Best K., Belknap J. K., Finn D. A., Crabbe J. C. Physiol. Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 32.O'Dell L. E., Roberts A. J., Smith R. T., Koob G. F. Alcohol. Clin. Exp. Res. 2004;28:1676–1682. doi: 10.1097/01.alc.0000145781.11923.4e. [DOI] [PubMed] [Google Scholar]
- 33.Roberts A. J., Heyser C. J., Cole M., Griffin P., Koob G. F. Neuropsychopharmacology. 2000;22:581–594. doi: 10.1016/S0893-133X(99)00167-0. [DOI] [PubMed] [Google Scholar]
- 34.McClung C. A., Nestler E. J. Nat. Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 35.Yao W. D., Gainetdinov R. R., Arbuckle M. I., Sotnikova T. D., Cyr M., Beaulieu J. M., Torres G. E., Grant S. G., Caron M. G. Neuron. 2004;41:625–638. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- 36.National Research Council. Guide for the Care and Use of Laboratory Animals. Washington, DC: Nat. Res. Council; 1996. [Google Scholar]
- 37.DeRisi J. L., Iyer V. R., Brown P. O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 38.Storey J. D., Tibshirani R. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storey J. D., Tibshirani R. Methods Mol. Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.