Abstract
Environmental cues modulate a variety of intracellular pathways whose signaling is integrated by the molecular mechanism that constitutes the circadian clock. Although the essential gears of the circadian machinery have been elucidated, very little is known about the signaling systems regulating it. Here, we report that signaling mediated by the dopamine D2 receptor (D2R) enhances the transcriptional capacity of the CLOCK:BMAL1 complex. This effect involves the mitogen-activated protein kinase transduction cascade and is associated with a D2R-induced increase in the recruiting and phosphorylation of the transcriptional coactivator cAMP-responsive element-binding protein (CREB) binding protein. Importantly, CLOCK:BMAL1-dependent activation and light-inducibility of mPer1 gene transcription is drastically dampened in retinas of D2R-null mice. Because dopamine is the major catecholamine in the retina, central for the neural adaptation to light, our findings establish a physiological link among photic input, dopamine signaling, and the molecular clock machinery.
Keywords: circadian clock, dopamine receptors, light, retina
Most organisms possess intrinsic time-tracking circadian systems that enable them to anticipate environmental changes and thereby adapt their behavior and physiology to the appropriate time of day (1, 2). The molecular mechanisms underlying the establishment and maintenance of circadian rhythms comprise interconnected transcriptional–translational feedback loops in which specific transcription factors control their own activity once they reach critical levels in the cell (1, 3). Complexed CLOCK and BMAL1 control clock genes Period (Per) and Cryptochrome (Cry), which encode transcriptional inhibitors that counteract CLOCK:BMAL1, thereby generating a negative feedback loop (4–6). Circadian rhythms are autonomous and self-sustained, but they need to be entrained through the perception of light, the major synchronizer of circadian clocks (7–9). The intricate process of synchronization involves neural connections and the activation of multiple transduction pathways (10–12).
The eye is the principal mediator of light input to the central nervous system in mammals (13, 14). The light signal to the circadian clock is integrated by a specific subset of cells, the retinal ganglion cells, localized in the ganglion cell layer (GCL) of the retina (15). Photic information is then conveyed through the retinohypothalamic tract to the suprachiasmatic nucleus (SCN), the central clock structure in mammals (16, 17). Here, the light stimulus induces the mitogen-activated protein kinase (MAPK) pathway (18, 19) and the phosphorylation of cAMP-responsive element (CRE)-binding protein (CREB) (11, 20). These events are thought to contribute to clock phase shifting through the induction of several genes, including c-fos and clock components Per1 and Per2 (12, 21, 22). Although progress has been made in elucidating the molecular components of the light input pathway (7, 8, 12), the identification of the circadian mediators of light signaling in the retina remains elusive.
Dopamine is the major catecholamine in the vertebrate retina and plays a central role in neural adaptation to light (23). Indeed, light stimulates the synthesis, turnover, and release of retinal dopamine, and it has been shown that dopaminergic activity is higher during the day than during the night (24–27). Thereby, dopamine is a likely mediator of light signaling to the retinal circadian clock. Among members of the dopamine receptor family (28, 29), the dopamine D2 receptor (D2R) has been shown to be implicated in light- and dopamine-reset of the circadian phase in the Xenopus eye (30) and to induce xPer2 expression (31). Also, quinpirole, a selective D2R agonist, mimics light in its acute effects on various rhythmic retinal phenomena, suggesting that endogenous retinal dopamine might modulate the circadian phase through the activation of D2R-mediated effects (30).
We have investigated the implication of D2R-mediated signaling in the control of clock gene expression. Our studies reveal a molecular mechanism by which dopamine-activated signaling pathways regulate CLOCK:BMAL1 activity. In addition, clock gene expression and light responsiveness are altered in the retinas of D2R knockout mice. Our findings uncover a role for D2R-mediated signaling in regulating clock gene expression and in controlling physiological pathways leading to light-responsiveness of the circadian clock.
Results
D2R-Mediated Signaling Increases CLOCK:BMAL1 Transactivation Potential.
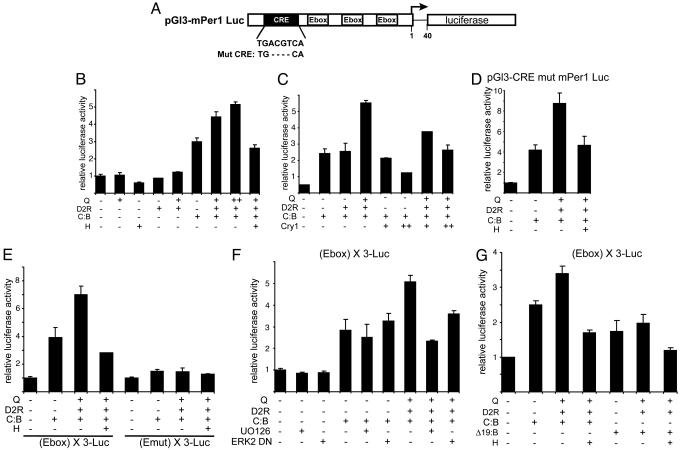
We investigated the role of D2R-dependent signaling in the expression of mPer1, a clock gene whose transcription is acutely induced in response to light in the SCN (21, 22) and to a variety of stimuli in cultured cells (32–34). NG108-15 cells were transiently transfected with a luciferase-based mPer1 reporter (Fig. 1A). As expected, CLOCK:BMAL1 transcriptionally stimulated this promoter. Interestingly, coexpression of D2R caused a considerable, dose-dependent elevation of mPer1 promoter activity, exclusively in the presence of the D2R-specific agonist quinpirole. Importantly, the enhancing effect of D2R coexpression was blocked by pretreatment haloperidol, a D2R-specific antagonist (Fig. 1B). In addition, D2R-mediated stimulation of CLOCK:BMAL1 seems to be cell-specific. Indeed, enhanced CLOCK:BMAL1 transactivation potential was observed only in NG108-15 cells, not in nonneuronal cell lines such as COS-1 or CHO (data not shown).
Fig. 1.
D2R-mediated signaling increases CLOCK:BMAL1 transactivation potential on the mPer1 promoter. (A) Schematic representation of the mPer1 promoter. The upstream sequence of the mPer1 gene was fused to a luciferase reporter. The sequence of the CRE site and its deletion mutant are indicated below the box. (B) Effect of D2R activation on CLOCK:BMAL1-dependent transcription. CLOCK:BMAL1 (C:B, 100 ng) was cotransfected with D2R (100 ng) activated with increasing amounts of quinpirole (Q, 6 h, 1 μM and 10 μM) and with a construct containing the mPer1 promoter (pGL3-mPer1-Luc, 50 ng) in NG108-15 cells. When indicated, cells were pretreated with haloperidol (H, 10 μM) 1 h before quinpirole treatment. The total DNA amount was kept constant by adding empty vector as required. After normalization for transfection efficiency using β-galactosidase activity, reporter gene activities were expressed relative to those of a control transfected only with the empty vectors. All values are the mean ± SD (n = 3). Shown are the representative data of triplicate experiments. (C) Effect of D2R activation on CRY1-mediated repression. Experimental conditions were as in B except that increasing amounts of pcDNA3.1-hemagglutinin (HA)-tagged mCry1 (5 and 10 ng) were used. (D) D2R-dependent induction of CLOCK:BMAL1 was conserved in the absence of a functional CRE. Experimental conditions were as in B except that pGL3-CREmut mPer1 Luc was used as reporter gene construct. (E) The E box element is mediating D2R-induced activation of CLOCK:BMAL1. Experimental conditions were as in B except that reporter constructs containing three copies of the E box consensus sequence [pGL3 promoter (E box) X3 LUC] or its mutated form [pGL3 promoter (Emut) X3 LUC] were used. (F) D2R-induced activation of CLOCK:BMAL1 is mediated by the MAPK pathway. Experimental conditions were as in B except that the mitogen-activated ERK kinase inhibitor UO126 (20 μM) was applied to cells 1 h before quinpirole treatment or that pcDNA3-mERK2 Y185F (ERK2DN, 200 ng) was coexpressed when indicated. (G) D2R-induced signaling has no effect on the CLOCK-Δ19:BMAL1 complex. Experimental conditions were as in E except that the CLOCK-Δ19 expression vector was used when indicated (Δ19:B).
D2R Activation Relieves CRY1-Mediated Repression of the mPer1 Promoter.
CRY proteins act as strong repressors of CLOCK:BMAL1-mediated transcription (35–37). We investigated whether D2R-dependent induction of mPer1 could influence mCRY1-mediated repression of CLOCK:BMAL1. As expected, coexpression of increasing amounts of mCRY1 resulted in dose-dependent transcriptional repression of CLOCK:BMAL1-mediated transcription (Fig. 1C). Interestingly, activation of D2R signaling significantly relieved the CLOCK:BMAL1 complex from mCRY1-mediated repression. This finding suggests that D2R-mediated mPer1 induction is elicited by means of a direct effect of receptor signaling on CLOCK:BMAL1 transcriptional activity.
The E Box Elements, but Not the CRE, Mediate D2R-Induced mPer1 Expression.
The CRE in the mPer1 promoter plays an important role in response to signaling (34). To assess the role of the CRE in D2R-mediated induction of mPer1, we generated an internal deletion that generates a nonfunctional CRE (Fig. 1A). D2R-mediated induction of mPer1 expression was fully conserved even in the absence of a functional CRE (Fig. 1D), strongly suggesting that D2R-dependent stimulation converges to CLOCK:BMAL1.
Having established that the CRE is not involved in D2R-dependent induction of mPer1 and taking into account that D2R-mediated activation relieves CRY-mediated repression (Fig. 1C), we analyzed in further detail the effect on the E boxes. We generated a heterologous reporter with three consensus E box sequences in tandem upstream of the luciferase gene [pGL3 promoter (E box) X3 LUC]. Interestingly, E box elements alone were able to relay D2R-inducible activation. Both CLOCK:BMAL1- and D2R-mediated responses were abolished when the E boxes were mutated (Emut X3-LUC) (Fig. 1E). Our results demonstrate that D2R signaling is specifically modulating CLOCK:BMAL1 activity.
D2R-Mediated Signaling on CLOCK:BMAL1 Involves the MAPK Pathway.
To identify the intracellular pathway responsible for D2R-mediated activation of mPer1 through CLOCK:BMAL1, we treated cells with a panel of protein kinase inhibitors. Only the mitogen-activated extracellular signal-regulated kinase (ERK) kinase (MEK)-specific inhibitor, UO126, caused a complete block of D2R-dependent mPer1 induction (Fig. 1F and data not shown). Importantly, UO126 had no significant effect on CLOCK:BMAL1 function. The involvement of the ERKs of the MAPK pathway was further strengthened by coexpression of a dominant negative form of ERK2 (Fig. 1F) (38). Interestingly, coexpression of a constitutively active mutant of MEK1 mimicked the effect of D2R-mediated signaling on CLOCK:BMAL1 (data not shown). Because activation of D2R signaling results in the induction of the MAPK pathway (39–42), our findings reveal a mechanism by which MAPKs regulate transcription through a clock-controlling complex.
Transcriptionally Inactive CLOCK-Δ19 Is Insensitive to D2R-Mediated Signaling.
Our results show that D2R-induced signaling specifically modulates CLOCK:BMAL1 activity. Next, we wanted to ascertain whether D2R-mediated mPer1 activation required a functionally active CLOCK protein. Thus, we used CLOCK-Δ19, a mutant CLOCK protein that operates as a dominant negative factor (43). In vitro studies show that CLOCK-Δ19:BMAL1 heterodimers, although still capable of binding DNA, have defective transcriptional activity. Importantly, D2R-mediated mPer1 induction was not observed when we replaced WT CLOCK by CLOCK-Δ19 (Fig. 1G). This observation emphasizes the requirement of a functional CLOCK:BMAL1 complex for dopamine to mediate the stimulation of the mPer1 promoter.
Dopamine Increases CRE-Binding Protein (CREB) Binding Protein (CBP) Recruitment to CLOCK:BMAL1.
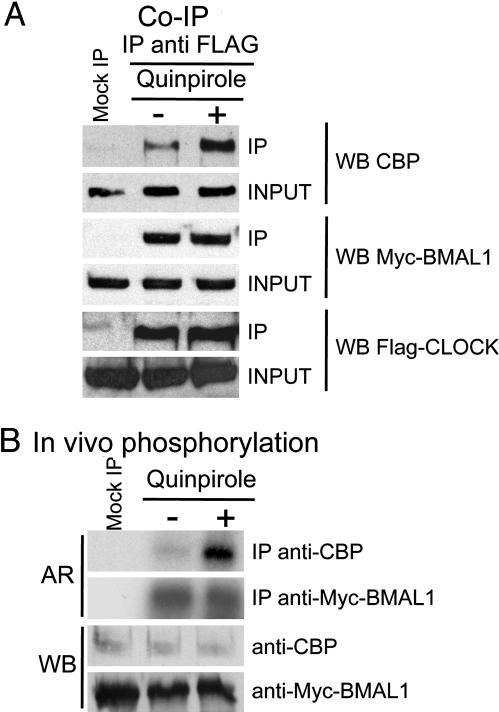
To elucidate the molecular mechanisms by which dopamine elicits mPer1 induction, we analyzed a number of regulatory parameters for CLOCK:BMAL1, including subcellular localization, dimerization, DNA binding, and ability to recruit the coactivator CBP. CBP was shown to be implicated in modulating clock gene expression (44, 45). After treatment with quinpirole of NG108-15 cells, no changes were observed in CLOCK:BMAL1 subcellular localization and E box binding (data not shown). Next, coimmunoprecipitation experiments revealed the presence of BMAL1 and CBP associated with FLAG-CLOCK (Fig. 2A). Quinpirole treatment induced a significant increase in CBP recruitment to CLOCK:BMAL1, whereas CLOCK:BMAL1 interaction was unmodified. Finally, we found that quinpirole induced a remarkable increase in CBP phosphorylation, without affecting that of BMAL1, as shown by in vivo phosphorylation assays (Fig. 2B). Because CBP phosphorylation has been coupled to its coactivator function in response to intracellular signaling (46), our results provide a scenario in which control of clock gene expression in response to dopamine signaling is relayed to CLOCK:BMAL1 by increased CBP recruitment.
Fig. 2.
CBP recruitment to CLOCK:BMAL1. CBP phosphorylation levels increase in response to D2R activation. (A) Myc-mBMAL1 and FLAG-mCLOCK were coexpressed in NG108-15 cells together with D2R in the presence or absence of quinpirole (1 h, 10 μM). Cell extracts were subjected to immunoprecipitation (IP) with anti-Flag M2 antibody or with mouse IgG (Mock IP). Precipitates were immunoblotted with anti-CBP [Western blot CBP (WB CBP)], anti-Myc 9E10 antibody (WB Myc-BMAL1), or anti-Flag M2 (WB Flag-Clock). The whole-cell extracts were also immunoblotted with the same antibodies (Input). (B) In vivo phosphorylation of CBP and mBMAL1 was investigated in NG108-15 cells upon treatment with quinpirole. Shown are the autoradiography (AR) and the Western blot (WB).
Reduction of mPer1 Light-Inducibility in Retinas of D2R-Null Mice.
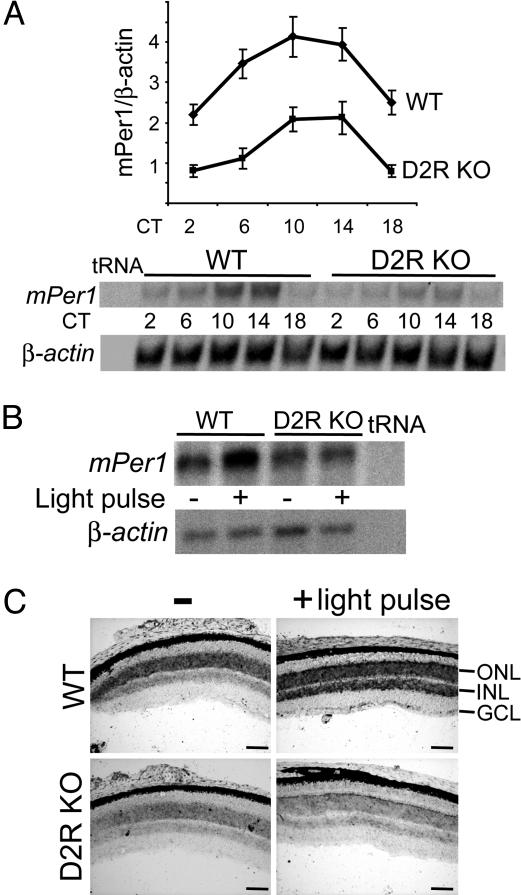
Because dopamine is the major catecholamine of the vertebrate retina and plays a central role in neural adaptation to light (23), we compared mPer1 oscillatory expression and light-inducibility in the eye of WT and D2R-null mice (47). As previously described (22), mPer1 exhibited prominent circadian expression with a peak at circadian time 10 (CT10) in the whole eye (Fig. 3A). In contrast, mPer1 oscillatory levels were markedly lower in the eye of D2R-null mice (Fig. 3A). Next, we determined the effect of a light pulse (30 min) at CT18 on mPer1 induction in the eye of WT and D2R−/− mice. The significant induction in WT mice was strongly reduced in D2R−/− mice (Fig. 3B).
Fig. 3.
Light-inducibility of mPer1 is reduced in the retina of D2R-null [knockout (KO)] mice. (A) RNA analysis of mPer1 expression in the eye of WT and D2R-null mice (D2R KO). (Upper) Quantitation by quantitative RT-PCR of mPer1 expression levels relative to those of β-actin at different circadian times (CT) in the eye of WT and D2R knockout mice. CT0 corresponds to “light on.” (Lower) RPA analysis of mPer1 and β-actin mRNA levels at each CT. In all experiments, a tRNA control was used. (B) Light regulation of mPer1 expression in the eye of WT and D2R knockout mice. RPA analysis of mPer1 and β-actin mRNA levels before and after a 30-min light pulse given at CT18. (C) Retinal expression patterns of mPer1 at CT18 before and after a 30 min light-pulse in WT and D2R-null mice. Retinal sections (10 μm) were examined for mPer1 RNA by in situ hybridization using antisense probes. ONL, Outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (Scale bar, 100 μm.)
Histological analysis of the retinas revealed no anatomical differences between WT and D2R-null mice (data not shown). In situ hybridization experiments showed that mPer1 was strongly expressed in the outer nuclear layer (ONL) and, to a lesser extent, in the inner nuclear layer (INL) and ganglion cell layer. After a 30-min light pulse, mPer1 was induced within the inner nuclear layer and ganglion cell layer in the inner retina of WT mice and greatly diminished in that of D2R−/− mice (Fig. 3C). Expression of mChox10 was used as internal control because it is restricted to the INL and unmodified by the genotype or lighting condition (48) (data not shown).
Lack of mPer1 Light-Inducibility in the Retina of clock/clock Mice.
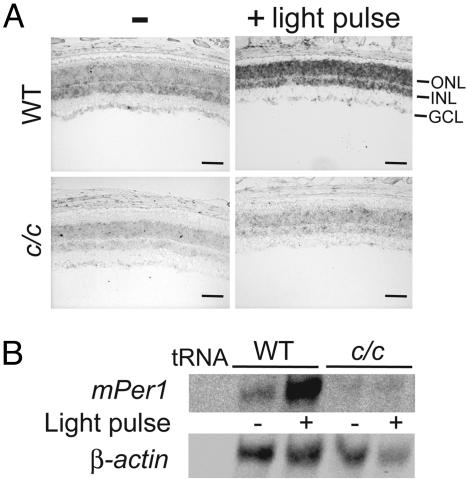
These findings prompted us to test mPer1 light-inducibility in the retina of clock/clock mice. These mice express the transcriptionally inactive mutant protein CLOCK-Δ19 (35, 49). We performed RNase protection assay (RPA) and in situ hybridization analyses before and after a light pulse given at CT18. Histological characterization revealed no anatomical differences in the retinas of the two genotypes (data not shown). Interestingly, the light pulse induced mPer1 expression in WT mice, but not in clock/clock mutants, confirming that CLOCK:BMAL1 relays photic signaling to mPer1 gene activation (Fig. 4). Our findings confirm previous results indicating that CLOCK plays a central role in circadian photoreception in the SCN (50).
Fig. 4.
Lack of mPer1 light-inducibility in the retina of clock/clock mutant mice. (A) Retinal expression patterns of mPer1 at CT18 before and after a 30-min light pulse in WT and clock/clock (c/c) mice. Retinal sections (10 μm) were examined for mPer1 RNA by in situ hybridization using an antisense probe. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (Scale bar, 100 μm.) (B) Light regulation of mPer1 expression in the eye of WT and clock/clock mice. RPA analysis of mPer1 and β-actin before and after a 30-min light pulse given at CT18.
Discussion
Transcriptional activation of Per genes in response to signaling pathways has been coupled to stimulation of CRE-binding protein (20, 34, 51). Here, we demonstrate that transcription mediated by the CLOCK:BMAL1 complex is also subjected to signaling control. Indeed, activation of D2R-dependent signaling results in stimulation of the MAPK transduction cascade, increased CBP recruitment to CLOCK:BMAL1, and a combined enhancement of CBP phosphorylation (Fig. 5). Our findings nicely parallel studies in cell culture showing a role for MAPKs in inducing rhythmic clock gene expression in response to extracellular signals (33).
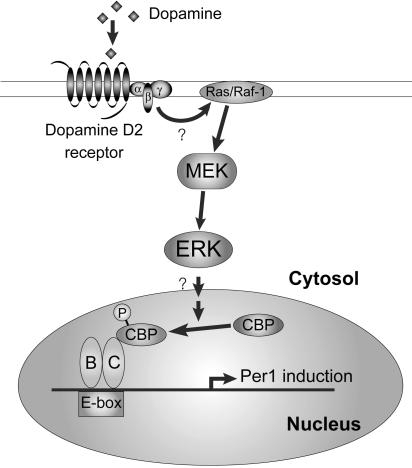
Fig. 5.
Proposed mechanism of D2R-mediated mPer1 induction. Activation of the dopamine D2R signaling cascade results in enhancement of CLOCK:BMAL1-driven transcription of clock genes. This signaling system involves the activation of MAPKs and the increased recruitment and phosphorylation of the transcriptional coactivator CBP to the CLOCK:BMAL1 complex. P, phosphate; B, BMAL1; C, CLOCK.
A relevant conclusion of our study is the elucidation of the intracellular pathway that links D2R-mediated signaling to clock control. The role of CBP is of particular interest. Importantly, ERKs can stimulate CBP coactivator function by targeting its C-terminal region (52) in response to growth factors (53) and phenylephrine (54). In addition, the C-terminal activation domain of CBP interacts with components of the basal transcription machinery such as TFIIB (55) and the RNA polymerase holoenzyme complex, as well as the transcriptional coactivator p/CIP (56, 57). Thus, in our working model (Fig. 5), CBP integrates second messenger signaling to allow the circadian clock to sense diverse stimuli and thereby serves an important physiological function by coordinating clock-controlled gene expression.
The markedly reduced oscillatory level of retinal mPer1 gene expression in D2R-null mice underscores the essential and specific role played by D2R-mediated signaling in circadian expression in the retina (Fig. 3). Our findings constitute an important advance in the understanding of the mechanism of oscillatory clock gene expression at the retinal level. Indeed, in the retina, dopamine operates prominently through D2Rs, which localize to the inner segments of photoreceptor cells, specifically to the inner nuclear layer and to the ganglion cell layer. The inner plexiform layer exhibits small areas of staining (58) that, interestingly, have been reported to express all of the proteins of the clock machinery (49, 59) and to contain a functional oscillator (60). In addition, light has been shown to induce dopamine release in the retina (24, 61, 62). Our results with the D2R-null mice provide evidence about the identity of the receptor and signaling cascade that dopamine utilizes to impact on the clock molecular mechanism.
The CRE in the mPer1 gene is not required to elicit dopamine-mediated transcriptional response (Fig. 1), suggesting that the cAMP pathway may not be essential for mPer1 light-inducibility in the retina. Furthermore, it has been shown that illumination of retinas results in a reduction of cAMP levels (63). This effect is thought to be mediated by the dopamine D4 receptor, which localizes to the photoreceptor layer (64). Although the neural retina of mammals possesses an oscillator that can be directly entrained by light (65), it remains to be established whether light-induced mPer1 expression in the retina participates in the entrainment of the retinal clock, as has been shown for the SCN (66).
Our observations favor a scenario where CLOCK plays an essential role in relaying light responses at the retinal level, a notion supported by the impaired mPer1 photic induction in the retina of clock/clock mice (Fig. 4). Light-inducibility of mPer1 is reduced also in the SCN of Clock mutant mice (50). Finally, the involvement of CLOCK in light response is suggested by previous findings in Drosophila. Indeed, dClock mutant flies are less photosensitive and show altered responses to light when compared with other arrhythmic lines (per0 and tim0) (67). It was also reported that, in addition to a role in generating circadian rhythms, dCLOCK modulates the direct effects of light on locomotion in Drosophila (68). Thus, Clock may play similar conserved roles in photoreception in mammals and flies. Our findings unravel a signaling route that links light to D2Rs in the mouse retina, leading to the physiological control of the circadian clock molecular mechanism.
Materials and Methods
Animals.
All mice were 7- to 12-week-old males housed in individual cages and entrained on a 12-h light/12-h dark cycle for 2 weeks and then placed in constant darkness for 4 days before sampling. Mice were decapitated and dissected under dim red light conditions. DR2-null mice have been described (47).
Materials and Plasmids.
Quinpirole, haloperidol, and anti-FLAG M2 antibody were purchased from Sigma. UO126 was from Calbiochem. Myc-mBMAL1 was detected with an anti-Myc 9E10 antibody (Transduction Laboratories, Lexington, KY). CBP was detected with A-22 rabbit polyclonal antibody (sc-369; Santa Cruz Biotechnology). The expression vector for D2R has been described (69). pSG5-FLAG-tagged mClock, pSC2-Myc-tagged mBmal1, pGL3-mPer1-Luc promoter, and the CRE mutated luciferase reporter constructs have been described (34). To generate the E box Luc reporter, an oligonucleotide containing three E box consensus sequences was cloned into the pGL3 promoter. An equivalent reporter was generated with mutated E boxes (Emut X3-Luc) (70). The Y185F mutation was introduced into the pcDNA3-mERK2 by using the QuikChange mutagenesis system (Stratagene).
In Situ Hybridization.
Tissues were placed in optimal cutting temperature (OCT) compound (Shandon, Pittsburgh) and frozen on dry ice, and 10-μm-thick coronal cryosections were prepared. The riboprobes were generated by using an in vitro transcription kit (Promega). The probe used covers nucleotides 1–336 of the mouse Per1 reading frame (71). In situ hybridization on frozen sections was as described (72).
Quantitative (q) RT-PCR and RPA.
Whole-tissue RNA was extracted by using RNA-Solv (Omega Bio-Tek, Doraville, GA) according to the manufacturer's instructions. Total RNA was then reverse-transcribed into cDNA and subjected to qPCR analysis as described (73). A miniaturized RPA was performed as described (74). A mouse β-actin riboprobe (nucleotides 193–331 of the mouse coding sequence) was used as internal control to monitor the loading of equal RNA amounts. The primer sequences used for the qRT-PCR are available upon request.
Cell Culture, Transient Transfections, and Luciferase Assays.
NG108-15 cells were grown in DMEM (1 g/liter glucose) supplemented with heat-inactivated 10% FCS and antibiotics and cultured at 37°C in 5% CO2. Cells were transfected with FuGENE 6 (Roche Molecular Biochemicals) according to the manufacturer's protocol. Cells growing in 24-well plates were transfected with various combinations of expression plasmids. Each transfection contained 50 ng of a luciferase reporter plasmid and 20 ng of a β-galactosidase internal control reporter plasmid (pcDNA3.1/lacZ; Invitrogen). The total amount of DNA applied per well was adjusted to 500 ng by adding pSG5 vector. Cell extracts were subjected to a luminometry-based luciferase assay (Promega), and luciferase activity was normalized by β-galactosidase activity. All experiments were repeated at least three times.
Coimmunoprecipitations, in Vivo Phosphorylation, and Immunoblotting.
NG108-15 cells grown on 10-cm plates were collected in PBS, and cell extracts were prepared in modified radioimmunoprecipitation buffer (50 mM Tris·HCl, pH 8/170 mM NaCl/5 mM EDTA/0.5% Nonidet P-40/1 mM DTT/50 mM NaF/100 μM sodium orthovanadate/1 μg/ml leupeptin/1 μg/ml aprotinin/1 mM phenylmethylsulfonyl fluoride). Immunoprecipitations were performed with mouse monoclonal anti-FLAG M2 antibody (Sigma) and protein G Sepharose beads. For in vivo phosphorylation, NG108-15 cells were starved overnight in a phosphate-deprived medium. After 4 h of incubation with 250 μCi/ml (1 Ci = 37 GBq) of [32P]orthophosphate (ICN) in the presence or absence of quinpirole (10 μM), cell extracts were prepared as described (75). CBP was immunoprecipitated with A-22 antibody, and anti-Myc 9E10 antibody was used for Myc-mBMAL1 immunoprecipitation.
Acknowledgments
We thank N. Fischer, E. Heitz, and C. Ziegler-Birling for technical assistance and all of the members of the Borrelli and Sassone-Corsi laboratories for help and discussions. We thank J. Takahashi, G. van der Horst, and S. Eblen for reagents. This work was supported by Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Régional Universitaire, Fondation pour la Recherche Médicale, Université Louis Pasteur, Electricité de France, Association pour la Recherche sur le Cancer, and La Ligue contre le Cancer.
Abbreviations
- SCN
suprachiasmatic nucleus
- MAPK
mitogen-activated protein kinase
- CRE
cAMP-responsive element
- CBP
CRE-binding protein binding protein
- CTn
circadian time n
- D2R
dopamine 2 receptor
- ERK
extracellular signal-regulated kinase
- RPA
RNase protection assay.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dunlap J. C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Panda S., Hogenesch J. B., Kay S. A. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 3.Schibler U., Sassone-Corsi P. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 4.Young M. W., Kay S. A. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 5.Cermakian N., Sassone-Corsi P. Nat. Rev. Mol. Cell Biol. 2000;1:59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 6.Reppert S. M., Weaver D. R. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 7.Zordan M. A., Rosato E., Piccin A., Foster R. Semin. Cell Dev. Biol. 2001;12:317–328. doi: 10.1006/scdb.2001.0259. [DOI] [PubMed] [Google Scholar]
- 8.Kavakli I. H., Sancar A. Mol. Interv. 2002;2:484–492. doi: 10.1124/mi.2.8.484. [DOI] [PubMed] [Google Scholar]
- 9.Duffy J. F., Wright K. P., Jr J. Biol. Rhythms. 2005;20:326–338. doi: 10.1177/0748730405277983. [DOI] [PubMed] [Google Scholar]
- 10.Shirakawa T., Moore R. Y. Neurosci. Lett. 1994;178:47–50. doi: 10.1016/0304-3940(94)90286-0. [DOI] [PubMed] [Google Scholar]
- 11.Ding J. M., Faiman L. E., Hurst W. J., Kuriashkina L. R., Gillette M. U. J. Neurosci. 1997;17:667–675. doi: 10.1523/JNEUROSCI.17-02-00667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cermakian N., Sassone-Corsi P. Curr. Opin. Neurobiol. 2002;12:359–365. doi: 10.1016/s0959-4388(02)00347-1. [DOI] [PubMed] [Google Scholar]
- 13.Nelson R. J., Zucker I. Neuroendocrinology. 1981;32:266–271. doi: 10.1159/000123171. [DOI] [PubMed] [Google Scholar]
- 14.Lee H. S., Nelms J. L., Nguyen M., Silver R., Lehman M. N. Nat. Neurosci. 2003;6:111–112. doi: 10.1038/nn1006. [DOI] [PubMed] [Google Scholar]
- 15.Moore R. Y., Speh J. C., Card J. P. J. Comp. Neurol. 1995;352:351–366. doi: 10.1002/cne.903520304. [DOI] [PubMed] [Google Scholar]
- 16.Moore R. Y., Lenn N. J. J. Comp. Neurol. 1972;146:1–14. doi: 10.1002/cne.901460102. [DOI] [PubMed] [Google Scholar]
- 17.Inouye S. T., Kawamura H. Proc. Natl. Acad. Sci. USA. 1979;76:5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obrietan K., Impey S., Storm D. R. Nat. Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- 19.Butcher G. Q., Dziema H., Collamore M., Burgoon P. W., Obrietan K. J. Biol. Chem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- 20.Ginty D. D., Kornhauser J. M., Thompson M. A., Bading H., Mayo K. E., Takahashi J. S., Greenberg M. E. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht U., Sun Z. S., Eichele G., Lee C. C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 22.Shearman L. P., Zylka M. J., Weaver D. R., Kolakowski L. F., Jr, Reppert S. M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 23.Witkovsky P. Doc. Ophthalmol. 2004;108:17–40. doi: 10.1023/b:doop.0000019487.88486.0a. [DOI] [PubMed] [Google Scholar]
- 24.Kramer S. G. Invest. Ophthalmol. 1971;10:438–452. [PubMed] [Google Scholar]
- 25.Iuvone P. M., Galli C. L., Garrison-Gund C. K., Neff N. H. Science. 1978;202:901–902. doi: 10.1126/science.30997. [DOI] [PubMed] [Google Scholar]
- 26.Iuvone P. M. Fed. Proc. 1984;43:2709–2713. [PubMed] [Google Scholar]
- 27.Nir I., Haque R., Iuvone P. M. Brain Res. 2000;870:118–125. doi: 10.1016/s0006-8993(00)02409-4. [DOI] [PubMed] [Google Scholar]
- 28.Missale C., Nash S. R., Robinson S. W., Jaber M., Caron M. G. Physiol. Rev. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 29.Vallone D., Picetti R., Borrelli E. Neurosci. Biobehav. Rev. 2000;24:125–132. doi: 10.1016/s0149-7634(99)00063-9. [DOI] [PubMed] [Google Scholar]
- 30.Cahill G. M., Besharse J. C. J. Neurosci. 1991;11:2959–2971. doi: 10.1523/JNEUROSCI.11-10-02959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steenhard B. M., Besharse J. C. J. Neurosci. 2000;20:8572–8577. doi: 10.1523/JNEUROSCI.20-23-08572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balsalobre A., Marcacci L., Schibler U. Curr. Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 33.Akashi M., Nishida E. Genes Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- 34.Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. Proc. Natl. Acad. Sci. USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., Takahashi J. S. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Horst G. T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A. P., van Leenen D., et al. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 37.Griffin E. A., Jr, Staknis D., Weitz C. J. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 38.Robbins D. J., Zhen E., Owaki H., Vanderbilt C. A., Ebert D., Geppert T. D., Cobb M. H. J. Biol. Chem. 1993;268:5097–5106. [PubMed] [Google Scholar]
- 39.Luo Y., Kokkonen G. C., Wang X., Neve K. A., Roth G. S. J. Neurochem. 1998;71:980–990. doi: 10.1046/j.1471-4159.1998.71030980.x. [DOI] [PubMed] [Google Scholar]
- 40.Welsh G. I., Hall D. A., Warnes A., Strange P. G., Proud C. G. J. Neurochem. 1998;70:2139–2146. doi: 10.1046/j.1471-4159.1998.70052139.x. [DOI] [PubMed] [Google Scholar]
- 41.Iaccarino C., Samad T. A., Mathis C., Kercret H., Picetti R., Borrelli E. Proc. Natl. Acad. Sci. USA. 2002;99:14530–14535. doi: 10.1073/pnas.222319599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Z., Feng J., Fienberg A. A., Greengard P. Proc. Natl. Acad. Sci. USA. 1999;96:11607–11612. doi: 10.1073/pnas.96.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King D. P., Zhao Y., Sangoram A. M., Wilsbacher L. D., Tanaka M., Antoch M. P., Steeves T. D., Vitaterna M. H., Kornhauser J. M., Lowrey P. L., et al. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Etchegaray J. P., Lee C., Wade P. A., Reppert S. M. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 45.Curtis A. M., Seo S. B., Westgate E. J., Rudic R. D., Smyth E. M., Chakravarti D., FitzGerald G. A., McNamara P. J. Biol. Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 46.Chan H. M., La Thangue N. B. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 47.Baik J. H., Picetti R., Saiardi A., Thiriet G., Dierich A., Depaulis A., Le Meur M., Borrelli E. Nature. 1995;377:424–428. doi: 10.1038/377424a0. [DOI] [PubMed] [Google Scholar]
- 48.Liu I. S., Chen J. D., Ploder L., Vidgen D., van der Kooy D., Kalnins V. I., McInnes R. R. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 49.Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 50.Shearman L. P., Weaver D. R. NeuroReport. 1999;10:613–618. doi: 10.1097/00001756-199902250-00031. [DOI] [PubMed] [Google Scholar]
- 51.Obrietan K., Impey S., Smith D., Athos J., Storm D. R. J. Biol. Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 52.Janknecht R., Nordheim A. Biochem. Biophys. Res. Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y. Z., Chrivia J. C., Latchman D. S. J. Biol. Chem. 1998;273:32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 54.Gusterson R., Brar B., Faulkes D., Giordano A., Chrivia J., Latchman D. J. Biol. Chem. 2002;277:2517–2524. doi: 10.1074/jbc.M104626200. [DOI] [PubMed] [Google Scholar]
- 55.Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bachinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 56.Kee B. L., Arias J., Montminy M. R. J. Biol. Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 57.Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen-Legros J., Chanut E., Versaux-Botteri C., Simon A., Trouvin J. H. J. Neurochem. 1996;67:2514–2520. doi: 10.1046/j.1471-4159.1996.67062514.x. [DOI] [PubMed] [Google Scholar]
- 59.Miyamoto Y., Sancar A. Proc. Natl. Acad. Sci. USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tosini G., Fukuhara C. Cell Tissue Res. 2002;309:119–126. doi: 10.1007/s00441-002-0578-z. [DOI] [PubMed] [Google Scholar]
- 61.Doyle S. E., Grace M. S., McIvor W., Menaker M. Vis. Neurosci. 2002;19:593–601. doi: 10.1017/s0952523802195058. [DOI] [PubMed] [Google Scholar]
- 62.Zawilska J. B., Bednarek A., Berezinska M., Nowak J. Z. J. Neurochem. 2003;84:717–724. doi: 10.1046/j.1471-4159.2003.01559.x. [DOI] [PubMed] [Google Scholar]
- 63.Nir I., Harrison J. M., Haque R., Low M. J., Grandy D. K., Rubinstein M., Iuvone P. M. J. Neurosci. 2002;22:2063–2073. doi: 10.1523/JNEUROSCI.22-06-02063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen A. I., Todd R. D., Harmon S., O'Malley K. L. Proc. Natl. Acad. Sci. USA. 1992;89:12093–12097. doi: 10.1073/pnas.89.24.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tosini G., Menaker M. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 66.Akiyama M., Kouzu Y., Takahashi S., Wakamatsu H., Moriya T., Maetani M., Watanabe S., Tei H., Sakaki Y., Shibata S. J. Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allada R., White N. E., So W. V., Hall J. C., Rosbash M. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 68.Kim E. Y., Bae K., Ng F. S., Glossop N. R., Hardin P. E., Edery I. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- 69.Montmayeur J. P., Borrelli E. Proc. Natl. Acad. Sci. USA. 1991;88:3135–3139. doi: 10.1073/pnas.88.8.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin X., Shearman L. P., Weaver D. R., Zylka M. J., de Vries G. J., Reppert S. M. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 71.Cermakian N., Monaco L., Pando M. P., Dierich A., Sassone-Corsi P. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strahle U., Blader P., Adam J., Ingham P. W. Trends Genet. 1994;10:75–76. doi: 10.1016/0168-9525(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 73.Hirayama J., Cardone L., Doi M., Sassone-Corsi P. Proc. Natl. Acad. Sci. USA. 2005;102:10194–10199. doi: 10.1073/pnas.0502610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hirayama J., Kaneko M., Cardone L., Cahill G., Sassone-Corsi P. Methods Enzymol. 2005;393:186–204. doi: 10.1016/S0076-6879(05)93005-X. [DOI] [PubMed] [Google Scholar]
- 75.Kotaja N., Macho B., Sassone-Corsi P. J. Biol. Chem. 2005;280:31739–31745. doi: 10.1074/jbc.M505971200. [DOI] [PubMed] [Google Scholar]