Many new microbial lineages have recently been discovered in the course of ribosomal RNA-based environmental surveys (1). Although the recognition of such extensive diversity has been exhilarating, it also raises large questions about the nature, activities, and ecological significance of all this newly recognized microbial life. What are the specific biological properties of environmentally abundant but previously unrecognized microbial groups? Are these new microbes significantly different from commonly cultivated laboratory strains? Do ubiquitous, newly recognized microbial species significantly impact their surrounding environment, and if so, how? In this issue of PNAS, the work of Ingalls et al. (2) illustrates one powerful approach for addressing such questions. These authors, led by Ann Pearson of Harvard University, used compound-specific isotope analyses (3) to track the flow of organic and inorganic carbon into the lipids of naturally occurring Archaea in deep ocean waters. Their results extend and confirm the notion that one group of marine Archaea, the Crenarchaea, are autotrophic and derive their carbon from CO2. Ingalls et al. (2) convincingly demonstrate that CO2 is a main carbon source for deep-sea Crenarchaea, indicating one major biogeochemical role for Archaea in the sea.
Archaea have a dramatically different lipid composition compared with other cellular life. Although Bacteria and Eucarya possess mainly ester-linked fatty acids in their lipids, archaeal cell membranes are composed predominantly of ether-linked isoprenoid core lipids (structure I, for example, in Fig. 1B). Some Archaea biosynthesize lipid bilayers composed of 20-carbon isoprenoid units, individually linked to a single glycerol backbone by an ether bond—these form conventional lipid bilayers. Other archaeal lipids include 40-carbon isoprenoids that are ether-linked at either end of their carbon chain to glycerol, thereby spanning the entire membrane in a tetraether monolayer (structure I in Fig. 1B). These unique lipids provide specific biomarkers that aid in tracking Archaea in the environment.
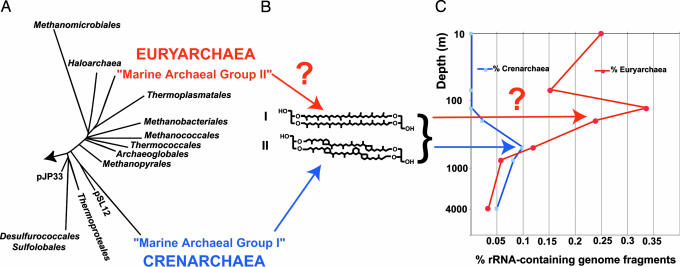
Fig. 1.
Planktonic archaeal phylogeny, lipid structures, and environmental distributions. (A) Phylogenetic relationships of cultivated Archaea and two dominant planktonic archaeal groups (group I Crenarchaea and group II Euryarchaea). (B) Two of the dominant archaeal lipids observed by Ingalls et al. (2). (C) Distributions of planktonic archaea inferred from recovery of rRNA-containing genome fragments reported in a recent study (15). Group II Euryarchaea rRNA-containing genome fragments were more abundant in the upper water column. Question marks indicate the uncertain contribution of marine Euryarchaea to planktonic archaeal lipids.
Evidence for planktonic Archaea was first reported based on a handful of archaeal rRNA clones recovered from deep Pacific Ocean waters (4) and in an independent, quantitative molecular survey of several coastal habitats (5). The coastal study revealed the presence and abundance of two different planktonic marine archaeal groups, one related to deep-water crenarchaea (Marine Archaeal Group I in Fig. 1A), and another new euryarchaeal group, peripherally related to the Thermoplasmatales (Marine Archaeal Group II in Fig. 1A) (5). Not long after planktonic Archaea were discovered, lipid analyses in marine sediments and plankton revealed high levels of crenarchaeal-like, tetraether lipids (6, 7). These cyclohexyl-containing lipids, dubbed crenarchaeol, have subsequently been used to track the presence, quantity, and isotopic composition of archaeal lipids in many different habitats (6–8).
Ingalls et al. (2) analyzed archaeal-associated lipids (e.g., structures I and II in Fig. 1B) using a compound-specific isotope approach (3) to directly ask the question: “What are deep-sea planktonic marine Archaea eating in the deep ocean?” More specifically, the authors sought to determine whether deep-sea Archaea were primarily autotrophic (using CO2 as their sole carbon source) or heterotrophic (using organic material as their carbon source). The foundation for their approach derives from the adage “You are what you eat” (with a little give and take, due to downstream metabolism). More specifically, the diet of an organism can sometimes be inferred by comparing the isotopic signatures of its potential food sources (for instance, the 13C/12C ratio in organic versus inorganic carbon) relative to the isotopic signature found in the organism. Organisms deriving sustenance from isotopically “light” carbon sources (e.g., food with a low 13C/12C ratio) will biosynthesize correspondingly “light,” 13C-depleted tissues or daughter cells.
The study of Ingalls et al. (2) aimed to identify the true carbon sources of deep-sea planktonic Archaea in situ. It was no small task to collect enough biomass for the compound-specific 14C analyses. Working at the Natural Energy Laboratory of Hawaii Authority, Ingalls et al. concentrated plankton cells from up to 200,000 liters of seawater (containing ≈2 × 1013 cells) from each of two depths to perform their study. (They also used some innovative laboratory equipment to achieve their goals, including a “combusted hacksaw” to access the filters!) After archaeal lipids were extracted, the 14C content of different fractions [including compounds I and II (Fig. 1B)] were determined. The archaeal 14C signature was then compared with that of the surrounding organic matter, sea surface-derived organic matter, and deep-water dissolved inorganic carbon. Quantitative estimates of archaeal carbon sources required several explicit assumptions about the inputs that contribute to deep-water archaeal biomass. The model of Ingalls et al. assumes that (i) sinking archaeal biomass from the surface keeps its original isotopic signature, (ii) all deep-water archaeal lipid biomarkers are derived from one and the same pooled carbon source, and (iii) no individual archaeal lipid biomarkers are produced disproportionately by metabolically disparate archaea coexisting in the same population. The other major assumption implicit in the study is that group I Crenarchaea [not group II Euryarchaea or other sources (Fig. 1A)] are the main contributors to the archaeal tetraether lipids. With these assumptions in hand, the authors derived a two-end-member mixing model to estimate the sources of dietary carbon for deep-water archaea. The model calculations suggested that at 670 m, 71% of the archaeal biomass was derived from inorganic carbon, implying that a large fraction of the deep-water Archaea were growing autotrophically. This is an important finding, because the study of Ingalls et al. required no tracer addition or environmental perturbations. Their measurements directly probed the in situ metabolism and autotrophic activities of the deep-water Archaea in their natural habitat. The results of Ingalls et al. then suggest that group I Crenarchaea derive much of their carbon from CO2 fixation and therefore may contribute significantly to the deep-ocean carbon cycle.
The report of Ingalls et al. (2) adds significantly to a growing body of evidence suggesting that group I Crenarchaea are key participants in ocean biogeochemical cycles. Schleper and colleagues (9), for example, recently showed that group I Crenarchaea contain key genes required for chemolithoautotrophic oxidation of ammonia. Subsequently, many of the genetic components required for both chemolithotrophic ammonia oxidation and CO2 fixation have been identified in symbiotic and free-living marine Crenarchaea (10). Other studies using stable isotopic tracers (11) or microautoradiography (12) also point to the prevalence of marine crenarchaeal CO2 fixation. Most significantly, one member of the marine group I crenarchaeal clade was recently isolated in pure culture and shown to grow by using ammonia as its energy source and CO2 as its carbon source (13). Considering the above, along with the new observations of Ingalls et al. (2) and the sheer abundance of planktonic Archaea (14), it seems clear that planktonic Crenarchaea play major roles in both carbon and nitrogen cycling in the ocean.
Of course, the puzzle of archaeal metabolism in the ocean is by no means solved. The mixed autotrophic and heterotrophic signal observed by Ingalls et al. (2) is curious. Does this represent crenarchaeal “mixotrophy” (different metabolic pathways for carbon or energy incorporation that are coexpressed in same microorganisms)? Or could these data reflect differential metabolic expression among different members of the same community? Are the signatures of disparate metabolic and phylogenetic types of Archaea hidden within the compound-specific isotope analyses reported by Ingalls et al. and others (see Fig. 1C)?
At least two very different groups of Archaea appear to contribute very significantly to archaeal biomass in marine plankton (5, 14, 15). Group II planktonic Euryarchaea (Fig. 1A) can be quite abundant at times, especially near the ocean’s surface (15, 16). Evidence for large numbers of group II Euryarchaea in the upper water column has recently been reported near the study site of Ingalls et al. (15). So far, organic geochemists have assumed that Euryarchaea do not contribute significantly to archaeal tetraether biomarkers, because most members of this group contain the shorter, 20-carbon isoprenoids (2, 6). But this assumption may not be correct. Group II Euryarchaea are related to the Thermoplasmatales (Fig. 1A) and share a number of properties in common with them. Furthermore, Thermoplasmatales membrane lipids are made up of up to 80% tetraether (17)—the same sorts of lipids now being ascribed to planktonic Crenarchaea. It seems quite possible, perhaps even likely, that planktonic group II Euryarchaea contain tetraether lipids very similar to those of group I Crenarchaea. Planktonic tetraether lipid signatures in the water column may well originate in part from planktonic Euryarchaea. If so, this complicates quantitative deconvolution of planktonic archaeal isotope signatures (see question mark in Fig. 1C). Such considerations, if true, also cast some doubt on the fundamental validity of recently proposed paleo-oceanographic temperature proxies that rely on the quantitative distributions of these archaeal lipids (18). It seems apparent that considerable work remains to more confidently interpret archaeal lipid signatures in the environment. The phylogenetic relationships and environmental distributions of marine Archaea reflect considerable diversity—this is probably also expressed in both the metabolic characteristics and ecological roles of these ubiquitous members of life’s third domain.
Conflict of interest statement: No conflicts declared.
See companion article on page 6442.
References
- 1.Pace N. R. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Ingalls A. E., Shah S. R., Hansman R. L., Aluwihare L. I., Santos G. M., Druffel E. R. M., Pearson A. Proc. Natl. Acad. Sci. USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes J. M., Freeman K. H., Popp B. N., Hoham C. H. Org. Geochem. 1990;16:1115–1128. doi: 10.1016/0146-6380(90)90147-r. [DOI] [PubMed] [Google Scholar]
- 4.Fuhrman J. A., McCallum K., Davis A. A. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 5.DeLong E. F. Proc. Natl. Acad. Sci. USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoefs M., Schouten S., De Leeuw J. W., King L. L., Wakeham S. G., Damste J. Appl. Environ. Microbiol. 1997;63:3090–3095. doi: 10.1128/aem.63.8.3090-3095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E. F., King L. L., Massana R., Cittone H., Murray A., Schleper C., Wakeham S. G. Appl. Environ. Microbiol. 1998;64:1133–1138. doi: 10.1128/aem.64.3.1133-1138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson A., McNichol A. P., Benitez-Nelson B. C., Hayes J. M., Eglinton T. I. Geochim. Cosmochim. Acta. 2001;65:3123–3137. [Google Scholar]
- 9.Treusch A. H., Leininger S., Kletzin A., Schuster S. C., Klenk H. P., Schleper C. Environ. Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 10.Hallam S. J., Mincer T. J., Schleper C., Preston C. M., Roberts K., Richardson P. M., DeLong E. F. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wuchter C., Schouten S., Boschker H. T., Sinninghe Damste J. S. FEMS Microbiol. Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 12.Herndl G. J., Reinthaler T., Teira E., van Aken H., Veth C., Pernthaler A., Pernthaler J. Appl. Environ. Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konneke M., Bernhard A. E., de la Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 14.Karner M. B., DeLong E. F., Karl D. M. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 15.DeLong E. F., Preston C. M., Mincer T., Rich V., Hallam S. J., Frigaard N. U., Martinez A., Sullivan M. B., Edwards R., Brito B. R., et al. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 16.Pernthaler A., Preston C. M., Pernthaler J., DeLong E. F., Amann R. Appl. Environ. Microbiol. 2002;68:661–667. doi: 10.1128/AEM.68.2.661-667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langworthy T. A., Smith P. F., Mayberry W. R. J. Bacteriol. 1972;112:1193–1200. doi: 10.1128/jb.112.3.1193-1200.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schouten S., Hopmans E. C., Schefuss E., Damsté J. S. S. Earth Planet. Sci. Lett. 2002;204:265–274. [Google Scholar]