Abstract
γ-Glutamyltranspeptidase (GGT) is a heterodimic enzyme that is generated from the precursor protein through posttranslational processing and catalyzes the hydrolysis of γ-glutamyl bonds in γ-glutamyl compounds such as glutathione and/or the transfer of the γ-glutamyl group to other amino acids and peptides. We have determined the crystal structure of GGT from Escherichia coli K-12 at 1.95 Å resolution. GGT has a stacked αββα fold comprising the large and small subunits, similar to the folds seen in members of the N-terminal nucleophile hydrolase superfamily. The active site Thr-391, the N-terminal residue of the small subunit, is located in the groove, from which the pocket for γ-glutamyl moiety binding follows. We have further determined the structure of the γ-glutamyl-enzyme intermediate trapped by flash cooling the GGT crystal soaked in glutathione solution and the structure of GGT in complex with l-glutamate. These structures revealed how the γ-glutamyl moiety and l-glutamate are recognized by the enzyme. A water molecule was seen on the carbonyl carbon of the γ-glutamyl-Thr-391 Oγ bond in the intermediate that is to be hydrolyzed. Notably the residues essential for GGT activity (Arg-114, Asp-433, Ser-462, and Ser-463 in E. coli GGT) shown by site-directed mutagenesis of human GGT are all involved in the binding of the γ-glutamyl moiety. The structure of E. coli GGT presented here, together with sequence alignment of GGTs, may be applicable to interpret the biochemical and genetic data of other GGTs.
Keywords: acyl-enzyme intermediate, Ntn-hydrolase family, product–enzyme complex, x-ray crystallography
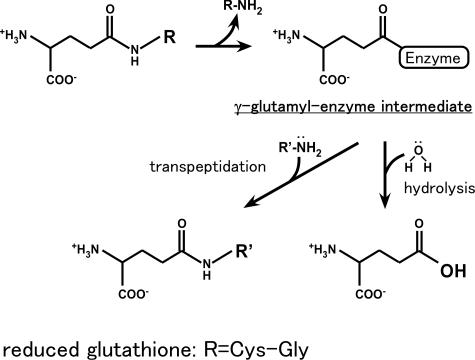
The enzyme γ-glutamyltranspeptidase (GGT; EC 2.3.2.2) catalyzes the cleavage of the γ-glutamyl linkage of γ-glutamyl compounds, e.g., glutathione (GSH), and the transfer of their γ-glutamyl group to other amino acids and peptides (1). The reaction catalyzed by GGT has been thought to proceed via a γ-glutamyl-enzyme intermediate followed by nucleophilic substitution by water, amino acids, or peptides (Scheme 1). GGT is evolutionarily conserved and found in organisms ranging from bacteria to mammals; the enzyme has a variety of physiological roles. Glutathione/xenobiotic conjugates are cleaved by GGT at the γ-glutamyl linkage, and these products are metabolized via the mercapturic acid pathway leading to the excretion of mercapturic acids into the bile and urine (2). GGT also cleaves exogenous glutathione for use as a cysteine and nitrogen source in Escherichia coli, yeast, and mammalian cells (3–5). In mammals, GGT is involved in the conversion of leukotriene C4 to D4, although GGT is not the only enzyme that catalyzes this reaction (6, 7). GGT is a key enzyme in glutathione metabolism, and genetic diseases of GGT deficiency, including glutathionemia and glutathionuria, are associated with mental retardation (8, 9).
Scheme 1.
Reactions catalyzed by GGT.
Clinically, GGT is used in a blood test and, because GGT in serum is mainly derived from liver, high levels of GGT in the blood are indicative of hepatic or biliary tract-associated diseases. Additionally, GGT is a virulence factor associated with the colonization of the gastric mucosa by Helicobacter pylori, the pathogen responsible for gastritis and ulcer and gastric cancer (10, 11).
GGT from E. coli K-12 shares a similar primary structure and enzymatic characteristics with mammalian GGTs (12, 13), suggesting that the residues of human and rat GGTs involved in the catalysis are conserved in E. coli GGT. There are, however, two notable differences between mammalian and E. coli enzymes. First, the precursor protein of E. coli GGT has a signal peptide at its N-terminal region (13) directing export to the periplasmic space (14), but mammalian GGTs have anchor domains in their N-terminal regions leading to membrane association (15). Additionally, mammalian GGTs are heterologously glycosylated, whereas E. coli GGT is nonglycosylated (2). The ability to highly purify large quantities of soluble enzyme makes E. coli GGT an ideal enzyme to study the general reaction mechanisms of this class of enzyme. Using this system, we established that autocatalytic posttranslational processing occurs in the precursor protein, in which Thr-391 acts as the nucleophile, to produce the large (L) and small (S) subunits (16). We also identified that the Oγ atom of N-terminal Thr-391 of the S subunit acts as the nucleophile in its enzymatic reaction (17). However, the molecular mechanisms controlling the enzymatic reaction and autocatalytic processing remain unclear because the 3D structure of GGT is unknown.
We report here the crystal structure of E. coli GGT refined at 1.95 Å resolution, revealing that GGT exhibits a folding pattern similar to the N-terminal nucleophile hydrolase family members (18) and the detailed environment of the active site. Furthermore, we prepared crystals of the γ-glutamyl-enzyme intermediate and monitored the time course of the enzymatic reaction by x-ray crystallography, providing the structural basis how the substrate is recognized and processed. Given the high degree of conservation, the structures presented here likely represent the enzyme characteristics of GGTs from other sources, including mammalian GGTs.
Results and Discussion
Overall Structure.
The crystal structure of SeMet-GGT was refined at 1.95 Å resolution to Rwork and Rfree factors of 0.207 and 0.231, respectively. The mature GGT molecule is a heterodimer comprising the L subunit (residues 25–390) and S subunit (residues 391–580) (residues 1–24 are signal peptide) (13). The asymmetric unit contains two GGT molecules (A and B). Residues 25–28 and 388–390 in A molecule and 25–29 and 388–390 in B molecule were disordered. The structures of the A and B molecules are superimposable with a rms deviation of 0.22 Å for the main-chain atoms.
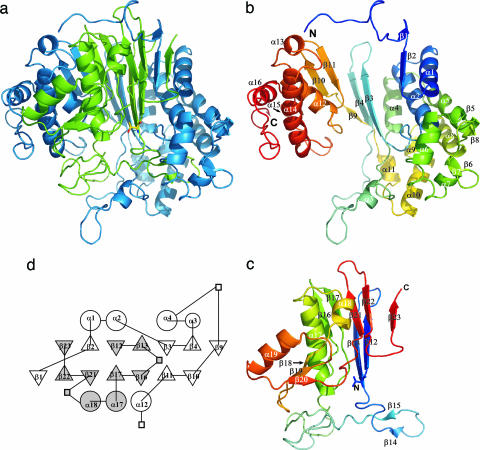
The structure of GGT is shown in Fig. 1. One side of the S subunit is surrounded by the L subunit. The GGT heterodimer has a stacked αββα-core; the β-strands of the L and S subunits form the two central β-sheets, which are sandwiched by α-helices. The topology of GGT is similar to those observed for Ntn-hydrolase superfamily members such as aspartylglucosaminidase (22) and penicillin acylase (23). The β-strands comprising the two central β-sheets and nearby α-helices are topologically conserved among the members of this superfamily; the main-chain atoms of the 66 residues comprising the core of E. coli GGT are superimposable on those of human aspartylglucosaminidase with a rms deviation of 1.3 Å. However, the relative orientation of the sheets, α-helices, and the secondary structure elements distant from the core are considerably variable.
Fig. 1.
Structure of E. coli GGT. (a) Ribbon drawing of the GGT heterodimer. The L subunit is colored blue, and the S subunit is colored green. (b) Ribbon drawing of the L subunit. (c) Ribbon drawing of the S subunit. α-Helices and β-strands are labeled. In each of the L and S subunits, the N terminus is blue and the C terminus is red, with intermediate colors following the distance in the sequence from the N terminus. The N-terminal residue of the S subunit (Thr-391) is shown with a stick model. (d) A topology diagram of E. coli GGT. Circle, triangle, and square indicate α-helix, β-strand, and insertion not conserved among Ntn-hydrolases, respectively. The secondary structures were defined with dssp (19). The figures were prepared with pymol (20) and tops (21).
Interestingly, the C terminus of the L subunit and the N terminus of the S subunit are quite distant from one another with Ser-387 C and Thr-391 N being 36 Å apart. The peptide bond between Gln-390 and Thr-391 is cleaved autocatalytically by posttranslational processing (16). Although exact positions of the three C-terminal residues (388–390) are unknown because of disorder of these residues, it is certain that conformational change has occurred upon processing, most probably in the C-terminal segment of the L subunit. This interpretation is based on the fact that the C-terminal segment of the L subunit (Ser-375-Ser-388) adopts mostly an extended conformation, whereas the N-terminal segment of the S subunit forms a β-strand that comprises one of the central β-sheets. A continuous groove is seen on the molecular surface of the S subunit from Ser-375 to the N terminus (Thr-391) (Fig. 2a), reminiscent of a path for the peptide of the precursor protein. The conformational change in the Ser-375-Ser-388 segment appears necessary for the proper formation of the substrate-binding pocket.
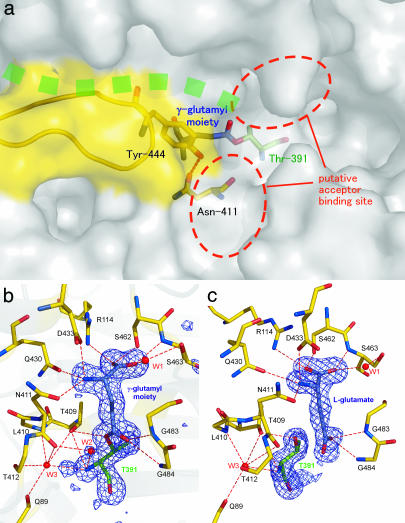
Fig. 2.
The structure of the substrate binding pocket of E. coli GGT. (a) Surface drawing of substrate binding pocket. The stick model of the γ-glutamyl moiety, nucleophile (Thr-391), and residues forming the wall (Asn-411 and Tyr-444) are shown in blue, green, and yellow, respectively. Green dots represent the groove in which the peptide of the precursor protein is assumed to be present. The hydrogen bond between Asn-411 Oδ and Tyr-444 Oη is shown as a dashed line. The ribbon model shown in yellow represents residues Pro-438–Gly-449, which are absent in B. subtilis GGT. (b) The (Fo − Fc) omit map contoured at the 3σ level for GGT-γG. The omit map was generated by omitting the γ-glutamyl moiety, Thr-391, and a water molecule (labeled W2) from the model. Ball-and-stick models of γ-glutamyl–enzyme complex are overlaid on the map. The residues involved in substrate binding and enzyme reaction are shown in the model. For the clarity, the side chains of Gln-89, Leu-410, and Thr-412 are omitted from the model. Water molecules involved in substrate binding and the catalytic reaction are labeled (W1∼W3). The hydrogen bonds are shown as dashed lines. (c) The (Fo − Fc) omit map for GGT-Glu prepared as for GGT-γG. The view direction is rotated by 40° around the vertical axis relative to b. The figures were prepared with pymol (20).
We identified one Ca2+ ion chelated by Asp-569 O, Ser-572 O, Asp-575 Oδ1, and two or three water molecules in the crystals grown in the presence of Ca2+. No electron density was found at this site in the crystals grown in the absence of Ca2+, but the conformations of the Ca2+-binding residues changed little. The Ca2+-binding site is far distant from the enzyme active site, suggesting that Ca2+ binding is not involved in enzyme catalysis.
Substrate-Binding Pocket.
The substrate-binding pocket must be the cavity in which established active residue of Thr-391 resides. Thr-391 is located at the bottom of the deep groove, and the binding pocket extends from Thr-391 into the enzyme (Fig. 2a). The (Fo − Fc) map for SeMet-GGT had a broad electron density in this pocket (data not shown), suggesting that this pocket is partially occupied by small molecules during protein preparation. To gain insight into the mechanisms of substrate recognition and enzyme reaction, the structures of GGT crystals soaked in the solutions containing GSH (γ-glutamyl-enzyme intermediate of GGT, GGT-γG) and l-glutamate (GGT-Glu) also were determined (Table 1).
Table 1.
Data collection and refinement statistics
| SeMet-GGT (remote) | SeMeT-GGT (edge) | SeMeT-GGT (peak) | GGT-γG | GGT-Glu | |
|---|---|---|---|---|---|
| Data collection | |||||
| Unit cell a, b, and c, Å | 78.7, 126.9, 128.8 | 78.8, 126.7, 128.9 | 77.7, 126.5, 129.2 | ||
| Wavelength, Å | 0.98214 | 0.97931 | 0.97902 | 1.0000 | 1.0000/0.9000 |
| Resolution range, Å | 50–1.95 | 50–1.95 | 50–1.95 | 50–1.80 | 50–1.68 |
| No. of measurements | 1,152,158 | 1,112,378 | 895,433 | 961,007 | 829,979 |
| No. of unique reflections* | 94,722 | 181,389 | 180,401 | 120,057 | 144,777 |
| Redundancy*† | 12.2 (12.0) | 6.1 (6.0) | 5.0 (4.9) | 8.0 (8.0) | 5.7 (3.2) |
| Completeness,*† % | 100.0 (100.0) | 100.0 (99.9) | 99.4 (99.4) | 100.0 (100.0) | 99.8 (99.7) |
| Rsym,*†‡ % | 5.9 (36.9) | 6.2 (35.1) | 7.0 (36.5) | 6.3 (34.8) | 11.2 (33.6) |
| Refinement | |||||
| Resolution range, Å | 50–1.95 | 50–1.80 | 50–1.70 | ||
| No. of water/glycerol molecules | 605/0 | 896/4 | 790/1 | ||
| No. of Ca2+ ions | 0 | 0 | 2 | ||
| No. of ligand atoms | 0 | 18 | 20 | ||
| Rwork/Rfree,§ % | 20.7/23.1 | 18.0/20.0 | 19.4/21.2 | ||
| rms deviations from ideal values | |||||
| Bond length, Å | 0.005 | 0.005 | 0.007 | ||
| Bond angles, ° | 1.3 | 1.3 | 2.2 | ||
| Ramachandran plot | |||||
| Most favored regions, % | 90.1 | 90.7 | 90.5 | ||
| Additional allowed regions, % | 9.4 | 8.8 | 9.0 | ||
| Disallowed regions,¶ % | 0.5 | 0.5 | 0.5 |
*The intensities of Bijvoet pairs were treated independently for the edge and the peak data of SeMet-GGT.
†The value in parentheses are for the highest resolution shell.
‡Rsym = Σhki Σi|Ii(hkl) − 〈I(hkl)〉|/Σhkl ΣiIi(hkl).
§R = Σ|Fobs(hkl) − Fcalc(hkl)|/Σ|Fobs(hkl)|. Rfree is the R value calculated for 5% of the data set not included in the refinement.
¶Asn-411 and Trp-518 in the two crystallographically independent molecules.
Fig. 2b shows the (Fo − Fc) map near the substrate-binding pocket for the crystal flash cooled after soaking for 10 s in a GSH-containing solution. The γ-glutamyl moiety bound within the pocket is clearly visible in the electron density map. Remarkably, there was a covalent bond between the carbonyl group of the γ-glutamyl moiety and the Oγ atom of Thr-391. This result provides a clear illustration of the γ-glutamyl-enzyme intermediate formed when Thr-391 Oγ attacks the carbonyl carbon at the γ-position of GSH. The (Fo − Fc) map for GGT-Glu also showed a clear density representing l-glutamate within the binding pocket (Fig. 2c), and the electron density between l-glutamate and Thr-391 was clearly resolved. The binding of l-glutamate to GGT is similar to that of γ-glutamyl moiety seen in GGT-γG, but the torsion angle of the Cα-Cβ bond differs. Despite the presence or absence of a covalent bond between Thr-391 of GGT and γ-glutamyl moiety or l-glutamate, respectively, the conformation of the enzyme-binding pocket was unchanged.
When bound within the enzymatic pocket, the α-carboxyl and α-amino groups of the γ-glutamyl moiety are at the bottom of the pocket, and this moiety is held in this position by many hydrogen bonds and salt bridges. The carboxyl group is bonded with Arg-114 Nη, Ser-462 Oγ, Ser-463 N, and Ser-463 Oγ via W1 (Fig. 2b) and the α-amino group with Asn-411 Oδ, Gln-430 Oε, and Asp-433 Oδ. The γ-glutamyl carbonyl oxygen is hydrogen-bonded with two main-chain amino groups of Gly-483 and Gly-484. Except for Arg-114, all residues involved in γ-glutamyl binding are of the S subunit. The large side chain of Tyr-444 forms a wall that shields the pocket from solvent (Fig. 2a). The hydrogen bond between Tyr-444 Oη and Asn-411 Oδ may contribute to firm wall formation. Indeed, no conformational change in the side chain of Tyr-444 was detected upon substrate/product binding. The length and width of the pocket appear perfectly sized to accept the γ-glutamyl moiety, with the active site being solvent accessible at the γ-glutamyl linkage. Thus, when GSH is bound to E. coli GGT, its cysteinyl-glycine moiety would be solvent-exposed.
Enzyme Reaction.
The reaction catalyzed by GGT consists of two steps: (i) the active Oγ atom of Thr-391 attacks the carbonyl carbon atom of the γ-glutamyl compound to form the γ-glutamyl-enzyme intermediate, and (ii) the γ-glutamyl moiety is transferred to another substrate or the γ-glutamyl-enzyme bond is hydrolyzed to reform the resting enzyme. The present structure of GGT crystal soaked in 5 mM GSH solution for 10 s (Fig. 2b) has established that this GGT is active in the crystalline state to form the γ-glutamyl-enzyme intermediate. This result also indicates that the second step of the reaction, the hydrolysis of the intermediate, is much slower than the first reaction step.
The second step of reaction was pursued by varying the soaking time of the crystal in the GSH-containing solution and by examining each structure. The active-site structure in the crystal soaked for 1 min (GGT-γG-1min) resembled that seen in GGT-Glu but differed markedly from that seen in GGT-γG soaked for 10 s (Fig. 4, which is published as supporting information on the PNAS web site). The active-site structure seen in GGT-γG-1min appeared identical to those seen in the crystals soaked for 2.5 min and overnight (data not shown). Thus, hydrolysis of the γ-glutamyl-enzyme intermediate occurs in a minute in the crystalline state under these conditions. We have not observed an electron density consistent with enzyme-bound GSH; the first step of the reaction appears too fast to monitor by crystallography.
It should be noted that in the structure of γ-glutamyl-enzyme intermediate, the electron-density ascribable to a water molecule (W2) was observed on the carbonyl group that connects the γ-glutamyl moiety and the enzyme (Fig. 2b). W2 is hydrogen-bonded to Thr-391 N and Asn-411 O. It is most likely that this water molecule acts as the nucleophile that attacks the carbonyl carbon of the intermediate in the second step of the enzymatic reaction. Additionally, in the intermediate, Thr-391 Oγ is hydrogen-bonded to Thr-409 Oγ, and the carbonyl oxygen atom is hydrogen-bonded to the nitrogen atoms of Gly-483 and Gly-484. In the transition state from the intermediate to the resting state, the carbonyl carbon likely adopts a tetrahedral arrangement upon W2 attacking the carbonyl carbon atom of the intermediate. The structure presented here appears analogous to that seen in the acyl intermediates of serine proteases (24), where the site surrounded by the carbonyl oxygen and the nitrogen atoms of Gly-483 and Gly-484 is the oxyanion hole.
In the first and second steps of the reaction catalyzed by GGT, the candidate that accepts a proton from Thr-391 Oγ or W2 is the amino group of Thr-391. The amino group is further hydrogen bonded to Thr-409 Oγ and W3; these hydrogen bonds may contribute to the stabilization of the positive charge on the amino group of Thr-391.
Implication to Other GGTs.
The sequence alignment of several GGTs from typical organisms is shown in Fig. 3. The residues essential for GGT activity are completely conserved across bacterial and mammalian proteins, including the active residue Thr-391, Thr-409, which forms hydrogen-bonding with Thr-391 Oγ, and Gly-483 and Gly-484, which form the oxyanion hole. Additionally, the residues involved in substrate binding are also highly conserved. Extensive site-directed mutagenesis studies for human GGT showed that mutation of R107N, R107H, D423A, D423E, S451A, and S452A significantly reduced enzymatic activity (27–29). Arg-107, Asp-423, Ser-451, and Ser-452 in human GGT correspond to Arg-114, Asp-433, Ser-462, and Ser-463, respectively, in E. coli GGT and are strictly conserved in all GGTs. All of these residues are involved in the binding of the α-amino and α-carboxyl groups of the γ-glutamyl moiety. These results suggest that the mechanism of substrate binding is conserved in bacterial and mammalian GGTs, and, curiously, that slight deviations in substrate binding dramatically affect enzymatic activity. Based on the mutation of E108Q in human GGT, Glu-108 was postulated to be involved in acceptor binding for the transfer of the γ-glutamyl group (27). The corresponding residue in E. coli GGT, Glu-115, is located inside the enzyme and bonded to Arg-114 Nη1 via a water molecule and Gln-430 Nε. Therefore, it is unlikely that this glutamyl residue is involved in acceptor binding. Instead, the glutamyl residue may contribute to fixing the side chains of those residues at the correct positions because Arg-114 and Gln-430 are hydrogen-bonded to the α-carboxyl group and α-amino group of the γ-glutamyl moiety, respectively.
Fig. 3.
Multiple sequence alignment of GGT of several representative organisms. The sequence numbering corresponds to E. coli GGT. The secondary structure of E. coli GGT is shown above the sequence. Identical residues are shaded, and similar residues are boxed. The residues of catalytic nucleophile, stabilizing the nucleophile, substrate binding, and comprising the oxyanion hole are shown with the letters N, S, B, and O, respectively. The residues that form the substrate-binding pocket wall (Asn-411 and Tyr-444 in E. coli GGT) are shown as triangles, and the segment from Pro-438 to Gly-449 in E. coli is shown in the gray bar. The residues that significantly reduced enzymatic activity by site-directed mutagenesis for human GGT are underlined. Sequences shown are for GGTs of E. coli (ECOLI), B. subtilis (BACSU), human, and rat. The figure was prepared with clustalw (25) and espript (26).
The segment from Pro-438 to Gly-449 in E. coli GGT, which corresponds to the loop extending toward the active site, is variable in other GGTs. As described earlier, the side chain of Tyr-444, located at the middle of the loop in E. coli GGT, is hydrogen-bonded with Asn-411 Oδ to form the wall of the substrate-binding pocket. This loop must be absent in Bacillus subtilis GGT, and, consequently, it is likely that its pocket is more open than the pockets of other GGTs. Tyr-444 is substituted to Phe in mammalian GGTs, suggesting the walls of the pockets of these GGTs are more flexible than that of E. coli GGT. These structural features may be consistent with the observation that E. coli GGT is several hundred- and 33-fold less active than mammalian and B. subtilis GGTs, respectively (28–31).
Materials and Methods
Expression and Purification.
The 5′ overhang of plasmid pHW71 (32) after EcoRI digestion (AATT) was deleted with mung bean nuclease to adjust the distance between the Shine–Dalgarno sequence and the initiation codon of the ggt gene to obtain plasmid pSH1291. E. coli strain MY1 (F− metA Δggt-2 zig::Tn10) was transformed with pSH1291 to obtain strain SH1603, and this strain was grown at 37°C to an A600 of 0.5 in 8 liters of M9 glucose medium supplemented with 100 μg/ml of ampicillin and 25 μg/ml of SeMet with reciprocal shaking. GGT expression was induced by adding isopropyl-1-thio-β-d-galactopyranoside to the medium (final 0.5 mM) with an additional 12 h of shaking incubation. The periplasmic fraction was prepared, and the enzyme was purified as described in ref. 14 and 33. GGT eluted from a PBE94 column (GE Healthcare, Piscataway, NJ) was concentrated by the addition of ammonium sulfate to 80% saturation. This precipitate was dissolved in a small amount of 20 mM Tris·HCl (pH 8) and applied to a Cellulofine GC-700m column (1 × 113 cm; Seikagaku Kogyo, Tokyo) equilibrated with the same buffer for further purification. Fractions containing GGT were precipitated by the addition of ammonium sulfate to 80% saturation and stored at 4°C until use. The specific activity of SeMet GGT was the same as that of native GGT.
Crystallization and Data Collection.
The ammonium sulfate precipitate of SeMet GGT was dissolved in 50 mM Hepes buffer at pH 7.0. The SeMet GGT solution was desalted by repeated concentration and dilution with the buffer by using Vivaspin filter (Sartorius, Goettingen, Germany) and then concentrated to 6 mg/ml for crystallization. Although crystallization of GGT from E. coli was reported in ref. 34, we surveyed a wide range of crystallization conditions for SeMet GGT by the hanging-drop vapor diffusion method with the PEG/Ion Screen kit (Hampton Research, Aliso Viejo, CA) and JB Screen kit (Jena Bioscience, Jena, Germany). Promising crystals were produced at 4°C in two crystal forms (space groups P21 and P212121) when either JB Screen No. 2/B6 or No. 3/B5 was used as the reservoir solution.¶ In addition to condition optimization, we also applied microseeding; microcrystals were transferred to the preequilibrated drops. Most SeMet GGT crystals used in the present study were produced by equilibrating a 3-μl drop containing 2 μl of protein solution and 1 μl of reservoir solution (12.5–15% PEG 4000/0.2 M CaCl2/0.1 M Tris·HCl, pH 8.0–8.5 or 12.5–15% PEG 4000/0.2 M MgSO4/5% glycerol) against a 0.2-ml reservoir solution. Crystals grew to a typical size of ≈0.4 × 0.1 × 0.1 mm3 in a few weeks.
The orthorhombic form (P212121) of crystal was used in the present analysis. Crystals were soaked in cryoprotectant solution and flash cooled with a nitrogen gas stream at 100 K, where the cryoprotectant solution was prepared by the adding PEG 4000 and glycerol to the reservoir solution to final concentrations of 22.5–25% and 15%, respectively. Most crystals of samarium derivative and substrate/product bound forms were prepared by the soaking method, where each soaking solution contains the cryoprotectant and the heavy-atom reagent or substrate/product. Diffraction data were collected at beamline BL41XU at SPring-8 and the Area Detector Systems (Poway, CA) Quantum 315 detector. MAD data were collected for SeMet-GGT. Because the asymmetric unit of this crystal has 32 Se sites, the SAD data were collected at λ = 1.20 Å for the SeMet-GGT-Sm crystal to obtain initial phase angles for locating the Se sites. The data were processed and scaled by using the hkl2000 suite (35). Details of the data collection and the preparation of the derivative crystals are given in Table 1 (see also Table 2, which is published as supporting information on the PNAS web site).
Structure Determination.
We first located two Sm sites from the Bijvoet difference Patterson map with SeMet-GGT-Sm. These sites were used to calculate the initial phase angles on the basis of which Bijvoet difference Fourier map with SeMet-GGT data located five Se sites. After three more rounds of refinement of the atomic parameters of the Se sites and inspection of the difference Fourier map, a total of 20 Se sites were located (figure of merit = 0.556). The phase angles derived from these sites were refined further by density truncation and solvent modification to produce an interpretable map at 2.1 Å resolution.
Automated tracing of the polypeptide chain by using arp/warp (36) placed 1,038 of 1,112 residues (two GGT molecules in an asymmetric unit) in 20 fragments of chains. These fragments were combined manually by the symmetry and/or translation operations to build one whole GGT model. Using this model as the search model, the other crystallographically independent molecule was located by molecular replacement with molrep in the ccp4 suite (37). Several cycles of model refinement and water picking with cns (38) followed by manual revision of the model with xtalview/xfit (39) were performed by using SeMet-GGT data to 1.95-Å resolution. The stereochemical check of the model was made with procheck (40). The refinement of GGT-γG and GGT-Glu began with the model of GGT-SeMet. Iterative cycles of model refinement and water picking were performed by using lafire (41) and cns. Finally, ligands were incorporated manually with xfit, and further refinement was carried out with cns. The stereochemistry of all models was checked by using procheck. Refinement statistics are presented in Table 1.
Supplementary Material
Acknowledgments
We thank Drs. Masahide Kawamoto and Nobutaka Shimizu (Japan Synchrotron Radiation Research Institute, Hyogo, Japan) and Shintaro Kitaoka (Osaka University) for their aid with data collection by using the synchrotron radiation at SPring-8 (proposal nos. 2004A0796 and 2005B0400) and Drs. Kiwako Sakabe and Noriyoski Sakabe for encouragement. This work was supported, in part, by Grants-in-Aid for Scientific Research 17053014 (to K.F.) and 15580061 (to H.S.) and by a grant from the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K.F.).
Abbreviations
- GGT
γ-glutamyltranspeptidase
- GGT-γG
γ-glutamyl-enzyme intermediate of GGT
- GGT-Glu
GGT in complex with l-glutamate
- GSH
glutathione
- L
large
- S
small.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2DBU for the native GGT, 2DBW for GGT-γG, 2DBX for GGT-Glu, and 2DG5 for GGT-γG-1min).
This paper was submitted directly (Track II) to the PNAS office.
The monoclinic (P21) crystal had the cell dimension of a = 65.4 Å, b = 128.2 Å, c = 75.6 Å, and β = 95.2° with two GGT molecules in the asymmetric unit. This crystal had high mosaicity, and we did not try to improve the crystal quality.
References
- 1.Tate S. S., Meister A. Mol. Cell. Biochem. 1981;39:357–368. doi: 10.1007/BF00232585. [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi N., Ikeda Y. Adv. Enzymol. Relat. Areas Mol. Biol. 1998;72:239–278. doi: 10.1002/9780470123188.ch7. [DOI] [PubMed] [Google Scholar]
- 3.Hanigan M. H., Ricketts W. A. Biochemistry. 1993;32:6302–6306. doi: 10.1021/bi00075a026. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H., Hashimoto W., Kumagai H. J. Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehdi K., Penninckx M. J. Microbiology. 1997;143:1885–1889. doi: 10.1099/00221287-143-6-1885. [DOI] [PubMed] [Google Scholar]
- 6.Anderson M. E., Allison R. D., Meister A. Proc. Natl. Acad. Sci. USA. 1982;79:1088–1091. doi: 10.1073/pnas.79.4.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter B. Z., Wiseman A. L., Orkiszewski R., Ballard K. D., Ou C. N., Lieberman M. W. J. Biol. Chem. 1997;272:12305–12310. doi: 10.1074/jbc.272.19.12305. [DOI] [PubMed] [Google Scholar]
- 8.Schulman J. D., Goodman S. I., Mace J. W., Patrick A. D., Tietze F., Butler E. J. Biochem. Biophys. Res. Commun. 1975;65:68–74. doi: 10.1016/s0006-291x(75)80062-3. [DOI] [PubMed] [Google Scholar]
- 9.Wright E. C., Stern J., Ersser R., Patrick A. D. J. Inherit. Metab. Dis. 1979;2:3–7. doi: 10.1007/BF01805554. [DOI] [PubMed] [Google Scholar]
- 10.Marshall B. J., Warren J. R. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 11.Chevalier C., Thiberge J. M., Ferrero R. L., Labigne A. Mol. Microbiol. 1999;31:1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H., Kumagai H., Tochikura T. J. Bacteriol. 1986;168:1325–1331. doi: 10.1128/jb.168.3.1325-1331.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki H., Kumagai H., Echigo T., Tochikura T. J. Bacteriol. 1989;171:5169–5172. doi: 10.1128/jb.171.9.5169-5172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki H., Kumagai H., Tochikura T. J. Bacteriol. 1986;168:1332–1335. doi: 10.1128/jb.168.3.1332-1335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughey R. P., Curthoys N. J. Biol. Chem. 1976;251:7863–7870. [PubMed] [Google Scholar]
- 16.Suzuki H., Kumagai H. J. Biol. Chem. 2002;277:43536–43543. doi: 10.1074/jbc.M207680200. [DOI] [PubMed] [Google Scholar]
- 17.Inoue M., Hiratake J., Suzuki H., Kumagai H., Sakata K. Biochemistry. 2000;39:7764–7771. doi: 10.1021/bi000220p. [DOI] [PubMed] [Google Scholar]
- 18.Brannigan J. A., Dodson G., Duggleby H. J., Moody P. C. E., Smith J. L., Tomchick D. R., Murzin A. G. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W., Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 20.DeLano W. L. The PyMol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 21.Michalopoulos I., Torrance G. M., Gilbert D. R., Westhead D. R. Nucleic Acids Res. 2004;32:D251–D254. doi: 10.1093/nar/gkh060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oinonen C., Tikkanen R., Rouvinen J., Peltonen L. Nat. Struct. Biol. 1995;2:1102–1108. doi: 10.1038/nsb1295-1102. [DOI] [PubMed] [Google Scholar]
- 23.Duggleby H. J., Tolley S. P., Hill C. P., Dodson E. J., Dodson G., Moody P. C. E. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 24.Voet D., Voet J. G. Biochemistry. Vol. 1. New York: Wiley; 2004. pp. 515–528. [Google Scholar]
- 25.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouet P., Courcelle E., Stuart D. I., Metoz F. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda Y., Fujii J., Taniguchi N. J. Biol. Chem. 1993;268:3980–3985. [PubMed] [Google Scholar]
- 28.Ikeda Y., Fujii J., Anderson M. E., Taniguchi N., Meister A. J. Biol. Chem. 1995;270:22223–22228. doi: 10.1074/jbc.270.38.22223. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda Y., Fujii J., Taniguchi N., Meister A. J. Biol. Chem. 1995;270:12471–12475. [PubMed] [Google Scholar]
- 30.Ikeda Y., Fujii J., Taniguchi N. J. Biochem. (Tokyo) 1996;119:1166–1170. doi: 10.1093/oxfordjournals.jbchem.a021363. [DOI] [PubMed] [Google Scholar]
- 31.Minami H., Suzuki H., Kumagai H. Enzyme Microb. Technol. 2003;32:431–438. [Google Scholar]
- 32.Hashimoto W., Suzuki H., Yamamoto K., Kumagai H. J. Biochem. (Tokyo) 1995;118:75–80. doi: 10.1093/oxfordjournals.jbchem.a124894. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H., Kumagai H., Echigo T., Tochikura T. Biochem. Biophys. Res. Commun. 1988;150:33–38. doi: 10.1016/0006-291x(88)90482-2. [DOI] [PubMed] [Google Scholar]
- 34.Kumagai H., Nohara S., Suzuki H., Hashimoto W., Yamamoto K., Sakai H., Sakabe K., Fukuyama K., Sakabe N. J. Mol. Biol. 1993;234:1259–1262. doi: 10.1006/jmbi.1993.1677. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Perrakis A., Morris R., Lamzin V. S. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 38.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.McRee D. E. J. Mol. Graphics. 1992;10:44–46. [Google Scholar]
- 40.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 41.Yao M., Zhou Y., Tanaka I. Acta Crystallogr. D. 2006;62:189–196. doi: 10.1107/S0907444905038965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.