Abstract
The Rad6–Rad18 ubiquitin-conjugating enzyme complex promotes replication through DNA lesions by means of at least three different pathways: the DNA polymerase (Pol) η- and ζ-dependent translesion DNA synthesis (TLS) and a Rad5–Mms2–Ubc13-dependent pathway. In DNA-damaged yeast cells proliferating cell nuclear antigen (PCNA) becomes monoubiquitylated at the K164 residue, and genetic studies in yeast have indicated a requirement for this modification in TLS mediated by Polη and Polζ. To be able to decipher the role of PCNA monoubiquitylation in the TLS process, we have reconstituted this PCNA modification in vitro from purified yeast proteins. We show that, in addition to the requirement for Rad6–Rad18, the reaction depends on the loading of the PCNA homotrimeric ring onto the DNA by replication factor C and that all three PCNA monomers become efficiently ubiquitylated. The availability of PCNA monoubiquitylated on all of its three monomers has enabled us to examine the effects of this PCNA modification on DNA synthesis by Pols δ, η, ζ, and Rev1. Contrary to the prevailing ideas that presume a role for PCNA ubiquitylation in the disruption of Polδ’s binding to PCNA or in the enhancement of the binding affinity of the TLS Pols for PCNA, we find that PCNA ubiquitylation does not affect any of these processes. These observations lead us to suggest a role for PCNA monoubiquitylation in disrupting the PCNA binding of a protein(s) that otherwise is inhibitory to the binding of PCNA by TLS Pols.
Keywords: proliferating cell nuclear antigen ubiquitylation, Rad6–Rad18 ubiquitin-conjugating enzyme, Rev1, DNA polymerase η, DNA polymerase ζ
DNA lesions in the template strand block the progression of replication fork. Genetic and biochemical studies in the yeast Saccharomyces cerevisiae have indicated a preeminent role of Rad6–Rad18 ubiquitin (Ub)-conjugating enzyme complex (1, 2) in promoting replication through DNA lesions (3). Rad6–Rad18-mediated Ub conjugation is indispensable for lesion bypass, which occurs via at least three independent pathways: DNA polymerase (Pol) η- and ζ-mediated translesion DNA synthesis (TLS) and a Rad5–Mms2–Ubc13-dependent pathway whose mechanism of action is not known (4).
The RAD30 gene of yeast encodes Polη, which is exceptional among eukaryotic TLS Pols in its proficient and relatively error-free ability to replicate through UV-induced cyclobutane pyrimidine dimers (5–8). Consequently, inactivation of Polη in yeast and humans confers enhanced UV mutagenesis (9–13) and in humans results in the cancer-prone syndrome, the variant form of xeroderma pigmentosum (14, 15). Polζ, comprising the Rev3 catalytic and Rev7 accessory subunits (16), promotes TLS by extending from the nucleotide inserted opposite DNA lesions by another DNA Pol (17, 18). Rev1, which, like Polη, is a member of the Y family of Pols, differs from the other Pols of this family in its high degree of specificity for inserting a C opposite template G (19, 20). Although Rev1 is strongly inhibited from inserting nucleotides opposite lesions that form at the template bases A, T, and C, it proficiently incorporates a C opposite N2-adducted guanines that obstruct synthesis by replicative Pols (21). The recently determined ternary crystal structure of Rev1 has revealed an elegant mechanism by which Rev1 can efficiently perform nucleotide incorporation opposite such lesions (22).
Genetic and biochemical studies in yeast and humans have indicated a pivotal role of proliferating cell nuclear antigen (PCNA) in Rad6–Rad18-dependent lesion bypass processes. Polη from yeast (23) and Pols η, ι, and κ from humans (24–26) have been shown to interact physically and functionally with PCNA, and mutations in the PCNA binding motif of yeast Polη render this Pol nonfunctional in TLS in vivo (23). The evidence that TLS Pols interact physically and functionally with PCNA and that PCNA is also required for the other Rad6–Rad18-dependent lesion bypass processes (27) has indicated that the various TLS Pols and the other lesion bypass proteins gain access to the replication fork stalled at the lesion site via their binding to PCNA.
In yeast cells treated with DNA-damaging agents, PCNA becomes monoubiquitylated at the K164 residue in a Rad6–Rad18-dependent manner, and subsequently this residue becomes polyubiquitylated via a K63-linked chain in a Rad5–Mms2–Ubc13-dependent manner (28). Genetic studies in yeast have suggested the requirement of PCNA monoubiquitylation for Polη- and Rev1/Polζ-dependent TLS and of PCNA polyubiquitylation for the Rad5-dependent lesion bypass process (28–30), which repairs the discontinuities in the newly synthesized DNA strand opposite DNA lesions by a mechanism that is not understood.
The requirement of PCNA monoubiquitylation for Polη- and Rev1/Polζ-dependent TLS processes has raised the strong possibility that the access of TLS Pols to PCNA in the stalled replication fork is governed by this modification. There are several distinct possibilities by which PCNA ubiquitylation could modulate the TLS process: (i) it could destabilize the binding of the replicative Pol, Polδ, from the PCNA, and that enables the TLS Pols to trade places with Polδ on PCNA; (ii) it could promote the binding of TLS Pols to PCNA without adversely affecting the binding of Polδ to PCNA. Thus, Polδ could remain bound to one of the PCNA monomers, and TLS Pols could gain access to one of the other PCNA monomers after its ubiquitylation. In this case TLS Pols would bind to ubiquitylated PCNA with a much higher affinity than to nonubiquitylated PCNA; (iii) alternatively, this modification could disrupt the PCNA binding of some protein(s) in whose presence the TLS Pols are unable to gain access to PCNA. For example, one could envision the possibility that Polδ bound to one of the PCNA monomers is tightly held onto PCNA, not only through the binding of that PCNA monomer via its various subunits but also through its tight association with the other proteins that are bound to the other two PCNA monomers. In that case the TLS Pols might be able to gain access to PCNA only when one of the PCNA monomers has been freed of the bound protein(s). Finally, (iv) a combination of some or all of the above-mentioned possibilities could be involved.
To be able to define the role of PCNA ubiquitylation in the TLS process we reconstituted the PCNA monoubiquitylation reaction using the purified protein components from yeast. Here we describe the requirements for this reaction, and we determine the effects of PCNA monoubiquitylation on the DNA synthetic activities of Pols δ, η, ζ, and Rev1. Our observations allow us to choose from among the different possibilities enumerated above, and they provide strong evidence against a role of ubiquitylation in the activation of PCNA binding by the TLS Pols.
Results
PCNA Encircling DNA, but Not Free PCNA, Is Ubiquitylated by Rad6–Rad18.
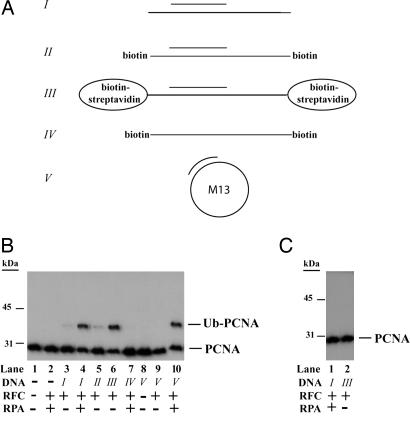
In preliminary experiments with fractionated yeast extracts we found that, in addition to the requirement of purified Rad6–Rad18, Uba1, and Ub, a replication factor C (RFC)-containing protein fraction and DNA were also required for the in vitro ubiquitylation of PCNA. The requirement of an RFC-containing protein fraction as well as of DNA for the in vitro ubiquitylation of PCNA led us to hypothesize that PCNA has to be loaded onto DNA by RFC to become an adequate substrate for Rad6–Rad18-dependent ubiquitylation. To test this idea we examined the effect of single-stranded, partial-heteroduplex, and circular DNAs, and of purified RFC and replication protein A (RPA), on PCNA ubiquitylation. The DNAs used in our assays are represented schematically in Fig. 1A. As expected, in the absence of DNA, Rad6–Rad18 was not able to ubiquitylate PCNA (Fig. 1B, lane 1), and the addition of purified RFC and RPA did not result in PCNA ubiquitylation either (Fig. 1B, lane 2). In the presence of RFC and a 75/31-nt heteroduplex DNA, however, Rad6–Rad18 was able to monoubiquitylate PCNA (Fig. 1B, lane 3), and we speculated that the very low efficiency of ubiquitylation of PCNA was due to the fact that, after the loading PCNA onto DNA by RFC, the PCNA ring does not remain stably bound on this DNA and slides off at the ends. To examine for this possibility we also added RPA to the reaction, which can bind to the single-stranded regions at each end of this DNA, thereby preventing the sliding off of PCNA from DNA. As shown in Fig. 1B, lane 4, in the presence of RFC, RPA, and the 75/31-nt partial-heteroduplex DNA, Rad6–Rad18 was able to efficiently monoubiquitylate PCNA.
Fig. 1.
Monoubiquitylation of PCNA by Rad6–Rad18. (A) Schematic representation of DNAs used in the PCNA ubiquitylation reactions. I, 75/31-nt partial-heteroduplex DNA containing a single-stranded region at each end; II, in addition to I, the template had biotin bound at each end; III, in addition to I, template had biotin–streptavidin bound at each end; IV, single-stranded 75-nt oligomer containing biotin bound at each end; V, ssM13 singly primed with a 38-nt oligonucleotide. (B) PCNA monoubiquitylation by Rad6–Rad18 in the presence or absence of DNA, RFC, and RPA. The complete reaction mixture contained 10 ng of PCNA, 0.5 μg of Rad6–Rad18, 100 ng of Uba1, 2.5 μg of Ub, 15 ng of RFC, 50 ng of RPA along with 0.5 pmol of oligomeric DNAs (I–IV) or 300 ng of RPA along with 0.1 pmol of M13 DNA (V). DNA, RFC, RPA, and combinations of these factors were omitted from the reaction mix as indicated below the blot. Monoubiquitylation of PCNA was followed by Western blot using anti-PCNA antibody. (C) Mutant PCNA K164R is not ubiquitylated. Mutant PCNA K164R was subjected to ubiquitylation reaction under the conditions described for wild-type PCNA in Fig. 2B, lanes 4 and 6.
To provide further support for the importance of DNA encircling by PCNA for its ubiquitylation, we modified the 75/31-nt heteroduplex DNA in a way that its 75-nt oligomer had biotin moieties bound at each end (Fig. 1A, II). Previously we have shown that binding streptavidin to the biotin on such a DNA can serve as a barrier, which then prevents the sliding off of PCNA from the ends of DNA (25). Indeed, when we preincubated this biotin-containing DNA with streptavidin before adding it together with RFC to the reaction, we observed efficient PCNA ubiquitylation (Fig. 1B, lane 6), whereas without adding streptavidin PCNA ubiquitylation remained inefficient (Fig. 1B, lane 5). Using ssDNA we observed no PCNA ubiquitylation (Fig. 1, lane 7), whereas adding circular singly primed DNA along with RFC and RPA resulted in efficient PCNA ubiquitylation (Fig. 1B, lane 10). As a control we determined that the PCNA K164R mutant protein was not ubiquitylated (Fig. 1C). This result confirmed that, similar to the in vivo situation, in our in vitro reconstituted system Ub is conjugated to the K164 residue of PCNA. From these experiments we conclude that the Rad6–Rad18 enzyme complex does not ubiquitylate free PCNA and that Ub conjugation to PCNA occurs only if PCNA encircles DNA.
All Three Monomers of Homotrimeric PCNA Can Be Monoubiquitylated at the Same Time.
Because it is quite possible that all of the three monomers of the homotrimeric PCNA ring can each interact, respectively, with the replicative Pol, the TLS Pol, and with some other replication proteins at the same time, it is important to determine whether Rad6–Rad18 is able to ubiquitylate all three subunits of PCNA at the same time. This issue was also important for another, very practical reason: because we had planned to use purified ubiquitylated PCNA in the replication assays in vitro, it would have been difficult to interpret the effects of PCNA ubiquitylation on the DNA synthetic activity of various Pols if the PCNA had a mixture of monoubiquitylated and unmodified subunits.
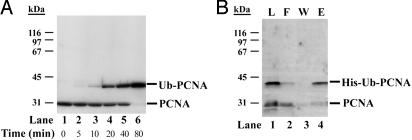
To test whether all three monomers of PCNA can be monoubiquitylated by the Rad6–Rad18 enzyme at the same time we carried out PCNA ubiquitylation reactions for increasing time under predetermined optimal reaction conditions. As shown in Fig. 2A, monoubiquitylation of monomers of PCNA increased with the incubation time, and it progressed to the point where almost all of the PCNA monomers were monoubiquitylated. This observation indicates that Rad6–Rad18 is able to monoubiquitylate all three subunits of PCNA at the same time.
Fig. 2.
Purification of PCNA bearing monoubiquitin at the K164 residue of all its three monomers. (A) All three monomers of PCNA can be monoubiquitylated at the same time. Standard ubiquitylation reactions of PCNA (10 ng) were carried out in the presence of 50 ng of RFC at 37°C for increasing time as indicated below the blot. PCNA and its monoubiquitylated form were visualized by anti-PCNA antibody. (B) Purification of monoubiquitylated PCNA. PCNA ubiquitylation reaction described in A was scaled up for 500 ng of PCNA, but His–Ub was used instead of Ub. After an 80-min incubation, the sample containing DNA-bound His–Ub–PCNA was treated by DNaseI and S1 nuclease followed by the purification of the free His–Ub–PCNA on Ni2+ beads. Aliquots of each sample before loading on the beads (L, lane 1), the flow-through (F, lane 2), the wash (W, lane 3), and the eluted proteins (E, lane 4) were analyzed on a 10% denaturing polyacrylamide gel followed by Western blot using anti-PCNA antibody. The positions of unmodified PCNA and PCNA conjugated with His–Ub are indicated on the right.
To facilitate the purification of PCNA monoubiquitylated on all its three subunits we used His–Ub instead of Ub and also scaled up the reaction shown in Fig. 2A, lane 6. After ubiquitylation of PCNA encircling DNA, the DNA was removed by nuclease treatment, and the free His–Ub–PCNA conjugate was purified on Ni2+ beads, which selectively bound only the His-ubiquitylated PCNA (Fig. 2B, lane 4), whereas nonubiquitylated PCNA remained in the flow-through (Fig. 2B, lane 2). As the Western blot using anti-PCNA antibody revealed, our final His–Ub–PCNA protein sample contained almost exclusively the PCNA in which all three subunits were monoubiquitylated. Fig. 2B, lane 4, shows this purified His–Ub–PCNA, and in the experiments discussed below Ub–PCNA refers to this purified sample.
Effect of Monoubiquitylated PCNA on the DNA Synthetic Activity of Polδ, Polη, Rev1, and Polζ.
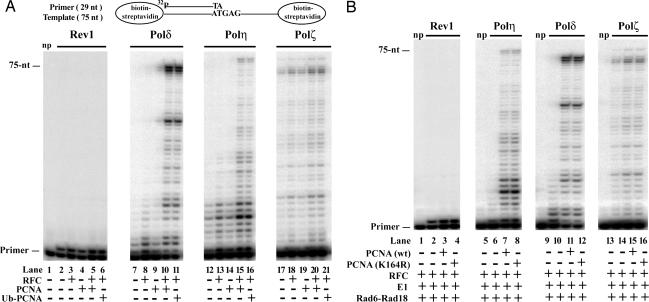
PCNA stimulates the DNA synthetic activity of Polδ by increasing its processivity (31, 32), and we have previously shown a stimulatory effect of PCNA on DNA synthesis by yeast Polη (23). To examine the possibility that monoubiquitylation alters PCNA binding of various DNA Pols, we examined the effects of monoubiquitylated PCNA on the DNA synthetic activity of Polδ, Polη, Rev1, and Polζ. For these experiments we constructed a DNA substrate in which the 75-nt template was bound to biotin at both ends, and after its annealing to the 29-nt 5-32P-labeled oligonucleotide primer both of these biotins were coupled to streptavidin. The first nucleotide on the template after the primer–template junction is a G residue (Fig. 3A).
Fig. 3.
Effects of PCNA and monoubiquitylated PCNA on the DNA synthetic activity of replicative and translesion synthesis DNA Pols. (A) Purified PCNA and Ub–PCNA are equally able to stimulate DNA synthesis by Polδ and Polη, but neither of them has a significant effect on the activity of Rev1 or Polζ. DNA synthesis reactions were carried out with Rev1, Polδ, Polη, and Polζ (2 nM each) on biotin–streptavidin bearing 75/29-nt 32P-labeled-primer–template DNA (10 nM) (schematically shown at the top) in the presence or absence of RFC (5 nM), PCNA (10 nM), or Ub–PCNA (10 nM) as indicated on the bottom. After incubation at 30°C for 10 min, reaction products were separated on 10% polyacrylamide gel containing 8 M urea and analyzed by PhosphoImager. In lane 1 no Pols were added (np). (B) Monoubiquitylation of PCNA has no effect on the DNA synthetic activity of Pols. Ubiquitylation reactions were carried out in the absence or presence of wild-type PCNA or K164R mutant PCNA (10 nM each), 75/29-nt biotin–streptavidin-containing 32P-labeled primer–template DNA (10 nM), 0.5 μg of Rad6–Rad18, 100 ng of Uba1, 2.5 μg of Ub, and 15 ng of RFC in 10-μl reactions containing 100 μM ATP. After an 80-min incubation at 30°C, 150 mM NaCl, 10 μM of each of the four dNTPs, and Rev1, Polη, Polδ, or Polζ (2 nM each) were added followed by incubation for 10 min at 30°C. Reactions were quenched and run on 10% polyacrylamide gel. np, no DNA Pol added.
First we compared the effect of purified Ub–PCNA and PCNA on the DNA synthetic activity of Polδ, Polη, Rev1, and Polζ. As shown in Fig. 3A, PCNA alone did not stimulate any of these Pols (Fig. 3A, lanes 4, 9, 14, and 19). As expected, a robust stimulation of the DNA synthetic activity of Polδ as well as of Polη occurred when we added PCNA together with RFC (Fig. 3A, lanes 10 and 15). In contrast, under the same reaction conditions PCNA along with RFC had no significant effect on the DNA synthetic activity of Rev1 or Polζ (Fig. 3A, lanes 5 and 20). Importantly, Ub–PCNA in the presence of RFC stimulated the DNA synthetic activity of Polδ and Polη to the same degree as unmodified PCNA (Fig. 3A, compare lanes 11 and 16 with lanes 10 and 15) and did not affect synthesis by Rev1 or Polζ (Fig. 3A, lanes 6 and 21).
To further examine the effect of PCNA ubiquitylation on DNA synthesis by various Pols, next we altered our experimental approach. Instead of testing for the effect of purified Ub–PCNA, which was added together with the Pol to the DNA substrate, we carried out the ubiquitylation reaction of PCNA loaded right on the radioactively labeled DNA substrate before adding a DNA Pol and the four dNTPs (Fig. 3B). First we incubated the reaction mixture containing no PCNA (Fig. 3B, lanes 2, 6, 10, and 14), wild-type PCNA (Fig. 3B, lanes 3, 7, 11, and 15), or mutant PCNA K164R (Fig. 3B, lanes 4, 8, 12, and 16) along with RFC, DNA substrate, Rad6–Rad18, E1, Ub, and ATP for 80 min. Under these conditions efficient ubiquitylation of PCNA occurs, whereas no modification of PCNA K164R can be detected (see Figs. 1C and 2A). Next we added all four dNTPs along with Rev1 (Fig. 3B, lanes 1–4), Polη (Fig. 3B, lanes 5–8), Polδ (Fig. 3B, lanes 9–12), or Polζ (Fig. 3B, lanes 13–16) followed by further incubation for 10 min. As expected, the PCNA-containing samples stimulated DNA synthesis by Polδ and Polη but not by Rev1 or Polζ. Again, and most importantly, we observed no difference in the effect of wild-type PCNA or mutant K164R PCNA on the DNA synthetic activity of various Pols. Thus, monoubiquitylation at the K164 residue of PCNA procures no significant effect on the DNA synthetic activity of yeast Polδ, Polη, Rev1, or Polζ.
PCNA Ubiquitylation Does Not Stimulate the Lesion Bypass Activity of Rev1 or Polη.
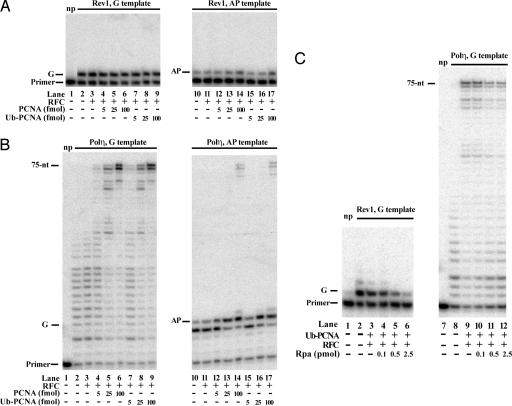
In a recently published study (33) ubiquitylated PCNA was reported to be more effective than unmodified PCNA in promoting synthesis through a DNA lesion by Rev1 and Polη. In particular, nucleotide incorporation opposite an abasic site by Rev1 was more efficient with Ub–PCNA than with PCNA, and Ub–PCNA was also more effective than PCNA in promoting TLS by Polη across an abasic site. In our studies, however, we have found no evidence for such a stimulatory effect of Ub–PCNA on the lesion bypass ability of Rev1 or Polη. As shown in Fig. 4A, the ability of Rev1 to incorporate a nucleotide opposite an undamaged G template or opposite an abasic site was the same with Ub–PCNA as with PCNA; furthermore, neither form of PCNA was stimulatory to synthesis by Rev1 on either of these DNA templates. And for Polη (Fig. 4B) we observed the same level of enhancement of DNA synthesis with Ub–PCNA as with PCNA, irrespective of whether the DNA contained an abasic site or not.
Fig. 4.
PCNA ubiquitylation does not affect the efficiency of nucleotide insertion opposite an abasic site by Rev1 or Polη. (A) Comparison of increasing amounts of PCNA or Ub–PCNA on the DNA synthetic activity of Rev1. DNA synthesis reactions were carried out with Rev1 (4 nM) on biotin–streptavidin bearing 75/31-nt 32P-labeled-primer–template DNA (10 nM) containing either a G (lanes 1–9) or an abasic site (AP) (lanes 10–17) at standing start positions after the primer–template junction in the presence or absence of RFC (5 nM) and increasing amounts of PCNA (lanes 4–6 and 12–14) or Ub–PCNA (lanes 7–9 and 15–17) (5–100 fmol) as indicated. After incubation at 30°C for 10 min, reaction products were separated on polyacrylamide gel containing 8 M urea. np, no Rev1 protein was added. (B) Comparison of the effect of increasing amounts of PCNA or Ub–PCNA on the DNA synthetic activity of Polη. Polη (4 nM) was incubated with biotin–streptavidin bearing 75/27-nt 32P-labeled-primer–template DNA (10 nM) containing either a G (lanes 1–9) or an abasic site (AP) (lanes 10–17) at running start positions as indicated, in the presence or absence of RFC (5 nM) and increasing amounts of PCNA (lanes 4–6 and 12–14) or Ub–PCNA (lanes 7–9 and 15–17) (5–100 fmol). (C) Inhibitory effect of RPA on DNA synthesis by Rev1 and Polη. Pol reactions were carried out as described in A and B. Where indicated, 25 fmol of Ub–PCNA and increasing amounts of RPA (0.1–2.5 pmol) were added to the reactions.
How might we then account for the discrepancy between our results and those published earlier (33)? Although we cannot be certain of the underlying reasons, we note that the experimental protocols used for obtaining Ub–PCNA differ in an important way in the two studies. In our experiments, to ensure that there was no contaminating DNA remaining bound to Ub–PCNA, we carried out DNaseI and S1 nuclease digestions to get rid of the DNA encircled by Ub–PCNA before loading it onto Ni2+ beads. However, because no such nuclease digestions were reported to have been carried out in the other study (33), one cannot exclude the possibility of some circular ssDNA remaining bound to Ub–PCNA throughout the purification, and that could account for the differences seen in the two studies.
We find that RPA is inhibitory to synthesis by Rev1 and Polη, particularly when short DNA templates are used, as was the case in both these studies. As we show in Fig. 4C, the addition of RPA together with Ub–PCNA and RFC inhibits synthesis by Rev1 as well as Polη, and for this reason we did not add any RPA to the experiments presented in Figs. 3 and 4 A and B. Because RPA was added in the other study (33), the stimulatory effect ascribed to Ub–PCNA could then have arisen from the lessening of the inhibitory effect of RPA resulting from the presence of circular ssDNA to which RPA would bind. However, because of the absence of any circular ssDNA with the nonubiquitylated PCNA, the inhibitory effect of RPA on DNA synthesis would be greater for unmodified PCNA than for Ub–PCNA. The stimulatory effect of Ub–PCNA on DNA synthesis by both Rev1 and Polη, then, could have arisen from the lessening of RPA inhibition in the Ub–PCNA sample because of the presence of ssDNA.
Discussion
Here we show that all three monomers of PCNA can be efficiently monoubiquitylated at their K164 residue. However, the reaction requires that PCNA be loaded on the DNA by RFC. These observations indicate that the Rad6–Rad18 enzyme complex is unable to bind PCNA in the absence of DNA and that it can conjugate Ub at the K164 residue only when PCNA has been loaded on the DNA. Because Rad18 is a DNA binding protein, we presume that Ub conjugation of PCNA by Rad6–Rad18 can occur only when this enzyme complex is bound to DNA via its Rad18 DNA binding subunit (1, 2).
PCNA ubiquitylation could affect TLS in a number of ways. One of the prevailing ideas presumes that the TLS Pol trades places with the replicative Pol on PCNA. For this idea to be valid we would expect ubiquitylation to be disruptive to the binding of the replicative Pol to PCNA. However, we find that PCNA-dependent stimulation of the DNA synthetic activity of Polδ is not impaired in the presence of ubiquitylated PCNA. The absence of any significant effect of PCNA ubiquitylation on Polδ activity indicates that the binding of Polδ to PCNA is not jeopardized. We infer from this observation that TLS does not involve the removal of Polδ from PCNA and its replacement by a TLS Pol.
The other alternative possibility is that PCNA monoubiquitylation greatly stimulates the binding affinity of TLS Pols for PCNA whereas the binding of a replicative Pol is not affected. In that case Polδ would remain bound to one of the PCNA monomers, and the TLS Pol would gain access to one of the other PCNA monomers via its increased binding affinity for PCNA. Although we have shown previously that yeast Polη (23) as well as human Pols η, ι, and κ (24–26) can all physically bind PCNA in the absence of ubiquitylation and that their DNA synthetic activities are stimulated as a consequence, in vivo studies with human Polη have been inferred to suggest that PCNA monoubiquitylation is indispensable for its physical interaction with PCNA (34). Thus, it has been suggested that the physical interaction between unmodified PCNA and Polη is too weak to be manifested in the in vivo conditions, and for the interaction to occur in vivo PCNA needs to be ubiquitylated. Our results, however, provide no support for this suggestion, because we observe no stimulatory effect of PCNA ubiquitylation on the DNA synthetic activity of Polη. We conclude from our observations that the role of PCNA ubiquitylation does not lie in the stimulation of the physical binding of Polη to PCNA; rather, it affects the TLS process at another step.
Although we consider it paramount that the TLS Pols be able to physically bind PCNA to gain entry into the replication fork, despite our persistent efforts over the years we have been unable to gather evidence for the physical interactions of yeast Rev1 or Polζ with PCNA. These experiments have been carried out under a variety of conditions, using DNAs where PCNA is efficiently loaded by RFC and conditions under which we find consistent evidence of physical binding and of stimulation of the DNA synthetic activities of yeast Polη and human Pols η, ι, and κ (23–26). Also, we have been unable to obtain any evidence of PCNA stimulation of Rev1 or Polζ activity on undamaged or a variety of damaged DNAs, e.g., DNA containing an abasic site for Rev1, and DNA containing a TT dimer, a (6–4) TT photoproduct, or an abasic site for Polζ (L.H., N. Acharya, L.P., and S.P., unpublished observations).
The data we present here also yield no evidence for the stimulation of the synthetic activity of Rev1 or Polζ by PCNA. Moreover, and interestingly, PCNA monoubiquitylation also procures no stimulation of the synthetic activity of these Pols. The inability of Rev1 or Polζ to bind PCNA (data not shown) and to be stimulated by it could reflect the need for additional protein factors or for a protein modification such as phosphorylation. The requirement of the Cdc7 kinase for Rev1/Polζ-dependent TLS could be indicative of such a requirement (35).
Our observations that neither Polζ or Rev1 are stimulated by unmodified PCNA and that ubiquitylation of PCNA has no stimulatory effect on Polη activity or on Rev1 activity are at variance with a recently published study (33). Although we cannot be certain of the reasons for the differences observed between these two studies, we note that the stimulatory effects that have been reported for the unmodified vs. modified PCNAs are relatively small (33) and might well be ascribed to the different experimental protocols used in the two studies (see Results).
In view of our findings that PCNA monoubiquitylation has no significant effect on the activity of Polδ or of Polη, Rev1, or Polζ, how can we account for the requirement of this modification for the TLS process? Our results leave open the possibility for a role of this PCNA modification in the removal of a protein(s) that binds PCNA and that is inhibitory to the PCNA binding of TLS Pols. We elaborate on this idea below.
To achieve the high rate of synthesis during DNA replication, Polδ, in addition to binding a PCNA monomer at multiple sites through its different subunits, could bind to the other PCNA monomers via its binding to other proteins, which, in turn, are bound to these other PCNA monomers. We envision that when the replication fork stalls at a lesion site and PCNA becomes monoubiquitylated this modification hinders the PCNA binding of such a protein(s). Consequently, the PCNA monomer is freed of the bound protein(s) and thus becomes available for the binding of a TLS Pol. In summary, we suggest a role for ubiquitylation in promoting the disassembly of a PCNA-bound protein(s), which normally act to improve the processivity of Polδ but whose continued presence on PCNA is inhibitory to the binding of TLS Pols.
Materials and Methods
Proteins and Antibodies.
Anti-RFC2 and anti-PCNA antibodies were purchased from Santa Cruz Biotechnology. His–Ub and Ub were purchased from Boston Biochem (Cambridge, MA) and from Sigma, respectively. Yeast Polη, Polζ, Rev1, Polδ, Rad6–Rad18, RFC, and RPA were purified as described previously (2, 5, 19, 36–39). The POL30 gene encoding PCNA was cloned into plasmid, pPM1088, which contains the GST gene under the control of the galactose-inducible phosphoglycerate promoter. The GST tag can be proteolytically cleaved from the GST fusion protein produced from pPM1088. Single-point mutation K164R was generated in the wild-type POL30 gene by PCR with the QuikChange site-directed mutagenesis kit from Stratagene. The plasmids used for overexpressing the wild-type and K164R mutant PCNAs were designated pPCNA1.33 and pPCNA1.40, respectively. The wild-type and mutant GST–PCNA proteins were expressed in the yeast strain BJ5464 and bound to a glutathione-Sepharose 4B column as described for GST-Polη in ref. 5. GST–PCNA protein bound to 100 μl of glutathione-Sepharose 4B was incubated overnight at 4°C with 4 units of PreScission protease, which cleaves the GST–PCNA fusion protein 7 aa amino-terminal from the first methionine of PCNA, in a buffer containing 40 mM Tris·HCl (pH 7.5), 100 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, and 10% glycerol. The cleaved protein was concentrated by using a Microcon 30 (Millipore, Billerica, MA), and purified PCNA was aliquoted and frozen at −70°C.
DNA Substrates.
Linear DNA molecules shown in Fig. 1A were generated by annealing a 75-nt oligomer, 5′-AGC TAC CAT GCC TGC CTC AAG AAT TCC CAT TAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′ (Fig. 1A, I), or the same but containing one biotin molecule at each end (Fig. 1A, II), to a 31-nt oligomer primer, 5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA T-3′. Substrate DNA shown in Fig. 1A, III, was generated by preincubation of the above biotin-containing DNA 75/31-nt partial heteroduplex (2.5 pmol) with streptavidin (5 μg) in a 25-μl sample. DNA shown in Fig. 1A, IV, contained only the 75-nt single-stranded biotinylated oligomer. The circular DNA shown in Fig. 1A, V, was a circular single-stranded M13 derivative (M13mp7L2) DNA primed with a 38-nt oligomer primer, 5′-GGG TTT TCC CAG TCA CGA CGT TGT AAA ACG ACG GCC AG-3′. DNAs used as substrate for DNA Pol assays shown in Figs. 3 and 4 were generated by annealing of the 75-nt oligonucleotide template containing biotin at each end and a G or an abasic site (AP) at position 45 from the 3′ end to either the 29-nt 5′ 32P-labeled oligomer primer, 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TA-3′, the 27-nt primer, 5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG-3′, or the above 31-nt oligomer primer. Before DNA Pol reaction these partial-heteroduplex DNAs were preincubated with streptavidin.
Ubiquitylation Reactions.
A standard in vitro ubiquitylation reaction of PCNA was carried out in 10 μl of P0 buffer (40 mM Tris·HCl, pH 7.5/8 mM MgCl2/100 μg/ml BSA/10% glycerol/100 μM ATP) in the presence of 10 ng of PCNA, 0.5 μg of Rad6–Rad18, 100 ng of Uba1, 2.5 μg of Ub, 15 ng of RFC, and 0.5 pmol of 75/31-nt partial-heteroduplex DNA containing biotin–streptavidin at each end of the 75-nt oligomer at 30°C for 60 min. Samples containing unmodified and monoubiquitylated PCNA were separated on 10% denaturing polyacrylamide gel and visualized by Western blot by using anti-PCNA antibody. The following modifications were included in some of the experiments: reactions were carried out in the absence or presence of combinations of RFC (15 ng), RPA (50 ng or 300 ng), and various linear (0.5 pmol) and circular (0.1 pmol) DNAs (Fig. 1B); mutant K164R PCNA was used instead of wild-type PCNA (Fig. 1C); reactions were incubated at 37°C for increasing time (0–80 min) in the presence of 50 ng of RFC (Fig. 2A); His–Ub was used instead of Ub in the presence of 50 ng of RFC for 80 min at 37°C in a 500-μl scaled-up reaction (Fig. 2B).
For obtaining purified His–Ub–PCNA protein, first the DNA from the 500-μl His–Ub–PCNA-containing reaction was digested by DNaseI and S1 nuclease enzymes (Roche) followed by incubation of the sample with 20 μl of Ni2+ bead (Qiagen) at 4°C for 30 min. After washing with buffer B (40 mM Tris·HCl/150 mM NaCl/0.01% Nonidet P-40/10% glycerol) supplemented with 10 mM imidazole, His–Ub–PCNA was eluted from the bead with 250 mM imidazole in buffer B. Finally, imidazole was removed by repeated washing with buffer B by using a Microcon 30, and purified His–Ub–PCNA was aliquoted and frozen at −70°C.
DNA Pol Assays.
A standard DNA Pol reaction was carried out in P0 buffer (40 mM Tris·HCl, pH 7.5/8 mM MgCl2/1 mM DTT/100 μg/ml BSA/10% glycerol/100 μM ATP) supplemented with 150 mM NaCl and 10 μM of each dGTP, dATP, dTTP, and dCTP. As indicated in the figure legends, Rev1, Polδ, Polη, or Polζ (2 nM each) in the presence or absence of RFC (5 nM), PCNA (10 nM), or His–Ub–PCNA (10 nM) were incubated with biotin–streptavidin-containing 75/29-nt primer–template DNA substrate (10 nM). Assays were assembled on ice, incubated at 30°C for 10 min, and stopped by the addition of loading buffer (40 μl) containing EDTA (20 mM), 95% formamide, 0.3% bromophenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea and visualized by using a Molecular Dynamics STORM PhosphoImager and imagequant software.
For DNA Pol reactions shown in Fig. 3B, reaction mixtures (10 μl) containing no PCNA or wild-type PCNA or K164R mutant PCNA (10 nM each) were first preincubated with the 75/29-nt biotin–streptavidin-containing 32P-labeled primer–template DNA (10 nM), RFC (5 nM), Rad6–Rad18 (0.5 μg), Uba1 (100 ng), and Ub (2.5 μg) in P0 buffer for 80 min at 30°C. Next the reaction mixtures were supplemented with 150 mM NaCl and 10 μM of each dGTP, dATP, dTTP, and dCTP followed by addition of Rev1, Polδ, Polη, or Polζ (2 nM each) and further incubation at 30°C for 10 min.
Acknowledgments
We thank Mike O’Donnell (Rockefeller University, New York) for providing the RFC plasmids and to Peter Burgers (Washington University in St. Louis) for Polδ. This work was supported by National Institutes of Health Grant CA107650, a Wellcome Trust International Senior Research Fellowship, Hungarian Science Foundation Grant OTKA T043354, Marie Curie International Reintegration Grant 022345, and Howard Hughes Medical Institute Grant 55005612.
Abbreviations
- TLS
translesion DNA synthesis
- PCNA
proliferating cell nuclear antigen
- Pol
polymerase
- Ub
ubiquitin
- RFC
replication factor C
- RPA
replication protein A.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bailly V., Lamb J., Sung P., Prakash S., Prakash L. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 2.Bailly V., Lauder S., Prakash S., Prakash L. J. Biol. Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 3.Prakash L. Mol. Gen. Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 4.Torres-Ramos C., Prakash S., Prakash L. Mol. Cell. Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson R. E., Prakash S., Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 6.Johnson R. E., Washington M. T., Prakash S., Prakash L. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 7.Washington M. T., Johnson R. E., Prakash S., Prakash L. Proc. Natl. Acad. Sci. USA. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Washington M. T., Prakash L., Prakash S. Proc. Natl. Acad. Sci. USA. 2003;100:12093–12098. doi: 10.1073/pnas.2134223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald J. P., Levine A. S., Woodgate R. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stary A., Kannouche P., Lehmann A. R., Sarasin A. J. Biol. Chem. 2003;278:18767–18775. doi: 10.1074/jbc.M211838200. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y.-C., Maher V. M., Mitchell D. L., McCormick J. J. Mol. Cell. Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters H. L., Seetharam S., Seidman M. M., Kraemer K. H. J. Invest. Dermatol. 1993;101:744–748. doi: 10.1111/1523-1747.ep12371686. [DOI] [PubMed] [Google Scholar]
- 13.Yu S.-L., Johnson R. E., Prakash S., Prakash L. Mol. Cell. Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R. E., Kondratick C. M., Prakash S., Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 15.Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 16.Nelson J. R., Lawrence C. W., Hinkle D. C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 17.Prakash S., Prakash L. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 18.Prakash S., Johnson R. E., Prakash L. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 19.Haracska L., Prakash S., Prakash L. J. Biol. Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 20.Nelson J. R., Lawrence C. W., Hinkle D. C. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 21.Washington M. T., Minko I. G., Johnson R. E., Haracska L., Harris T. M., Lloyd R. S., Prakash S., Prakash L. Mol. Cell. Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L., Kondratick C. M., Unk I., Prakash S., Prakash L. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 24.Haracska L., Johnson R. E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haracska L., Johnson R. E., Unk I., Phillips B. B., Hurwitz J., Prakash L., Prakash S. Proc. Natl. Acad. Sci. USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haracska L., Unk I., Johnson R. E., Phillips B. B., Hurwitz J., Prakash L., Prakash S. Mol. Cell. Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres-Ramos C. A., Yoder B. L., Burgers P. M. J., Prakash S., Prakash L. Proc. Natl. Acad. Sci. USA. 1996;93:9676–9681. doi: 10.1073/pnas.93.18.9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoege C., Pfander B., Moldovan G.-L., Pyrowolakis G., Jentsch S. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 29.Haracska L., Torres-Ramos C. A., Johnson R. E., Prakash S., Prakash L. Mol. Cell. Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelter P., Ulrich H. D. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 31.Prelich G., Kostura M., Marshak D. R., Matthews M. B., Stillman B. Nature. 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 32.Tan C. K., Castillo C., So A. G., Downey K. M. J. Biol. Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 33.Garg P., Burgers P. M. Proc. Natl. Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kannouche P. L., Wing J., Lehmann A. R. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 35.Pessoa-Brandao L., Sclafani R. A. Genetics. 2004;167:1597–1610. doi: 10.1534/genetics.103.021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgers P. M. J., Gerik K. J. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 37.Haracska L., Unk I., Johnson R. E., Johansson E., Burgers P. M. J., Prakash S., Prakash L. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 39.Yao N., Coryell L., Zhang D., Georgescu R. E., Kinelstein J., Coman M. M., Hingorani M. M., O’Donnell M. J. Biol. Chem. 2003;278:50744–50753. doi: 10.1074/jbc.M309206200. [DOI] [PubMed] [Google Scholar]