Abstract
The transsulfuration pathway converts homocysteine to cysteine and represents the metabolic link between antioxidant and methylation metabolism. The first and committing step in this pathway is catalyzed by cystathionine β-synthase (CBS), which is subject to complex regulation, including allosteric activation by the methyl donor, S-adenosylmethionine (AdoMet). In this study, we demonstrate that methionine restriction leads to a >10-fold decrease in CBS protein levels, and pulse proteolysis studies reveal that binding of AdoMet stabilizes the protein against degradation by ≈12 kcal/mol. These observations predict that under pathological conditions where AdoMet levels are diminished, CBS, and therefore glutathione levels, will be reduced. Indeed, we demonstrate this to be the case in a mouse model for spontaneous steatohepatitis in which the gene for the MAT1A isoenzyme encoding AdoMet synthetase has been disrupted, and in human hepatocellular carcinoma, where MAT1A is silenced. Furthermore, diminished CBS levels are associated with reduced cell viability in hepatoma cells challenged with tert-butyl hydroperoxide. This study uncovers a mechanism by which CBS is allosterically activated by AdoMet under normal conditions but is destabilized under pathological conditions, for redirecting the metabolic flux toward methionine conservation. A mechanistic basis for the coordinate changes in redox and methylation metabolism that are a hallmark of several complex diseases is explained by these observations.
Keywords: glutathione, liver disease
Cellular methylation and antioxidant metabolism are linked by the transsulfuration pathway, which converts the methionine cycle intermediate, homocysteine, to cysteine, the limiting reagent in glutathione synthesis. The balance between conserving methionine via transmethylation under conditions of methionine restriction and committing it to transsulfuration under conditions of plenty is regulated at two key control points, methionine adenosyltransferase (MAT) and cystathionine β-synthase (CBS) (Fig. 1). Aberrations in methylation and redox homeostasis are common to a number of chronic diseases including pathologies of the liver. In alcoholic liver disease and in hepatocellular carcinoma an increase in markers of oxidative stress is observed (1, 2). Furthermore, there is a switch in the expression of MAT genes from MAT1A to MAT2A in liver cancer, which correlates with lower S-adenosylmethionine (AdoMet) levels (3).
Fig. 1.
Methionine metabolism in mammals. CBS, MS, CGL, PPG, and MAT refer to cystathionine β-synthase, methionine synthase, γ-cystathionase, propargylglycine, and methionine adenosyltransferase, respectively. The inhibitors used to block specific enzymes are indicated. AdoHcy, S-adenosylhomocysteine.
Under normal conditions, coordinate regulation of methylation and antioxidant metabolism is achieved by the allosteric activation of CBS by AdoMet (Fig. 1). AdoMet is a V-type allosteric effector that increases CBS activity 2- to 3-fold (4, 5). Under conditions of plenty, methionine is directed toward cysteine synthesis via the transsulfuration pathway for use in glutathione and other cellular functions or directed toward catabolism. Cysteine is the limiting reagent in glutathione synthesis and in liver; ≈50% of the cysteine in glutathione is derived from methionine via the transsulfuration pathway (6–8). Conversely, it is expected that when methionine levels are low, flux through the transsulfuration pathway is down-regulated to conserve the amino acid backbone in the methionine cycle (9).
In this study, we discovered that under conditions of methionine restriction, CBS protein levels are diminished >10-fold. This led us to uncover a mechanism by which a reduction in the methyl donor, AdoMet, leads to destabilization of CBS and, in turn, to a reduction in antioxidant capacity via glutathione. We show that under pathological conditions with reduced AdoMet levels, namely human hepatocellular carcinoma and a mouse model for chronic steatohepatitis, CBS levels are diminished. This decrease in CBS levels correlates with reduced glutathione that is, in turn, associated with increased vulnerability to oxidative stress. Posttranslational regulation of CBS stability by AdoMet provides a mechanism for achieving coordinate changes in cellular methylation and antioxidant status that is observed in a number of disease states.
Results and Discussion
Methionine Restriction Reduces CBS Protein Levels.
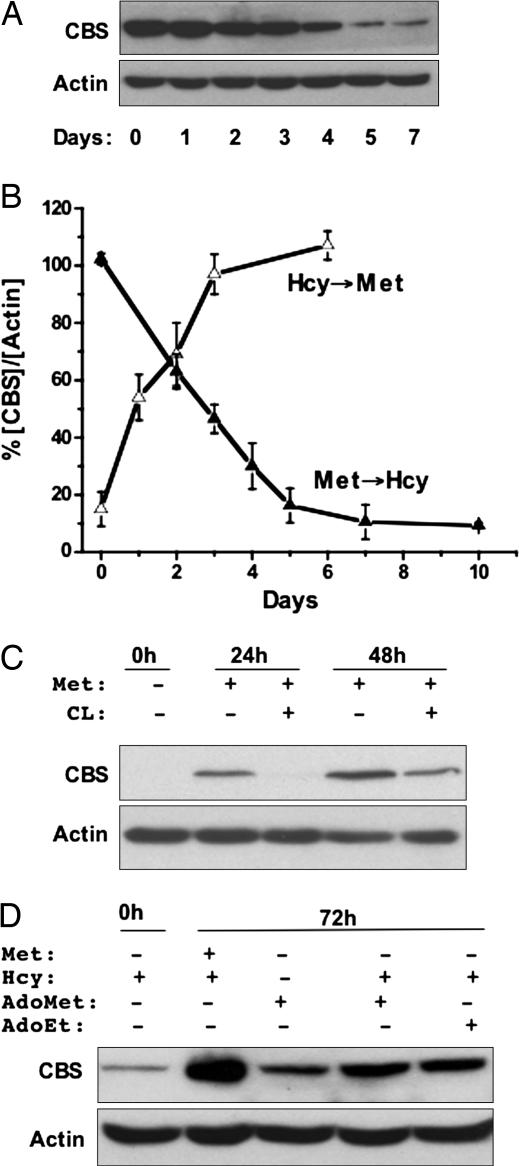
Many carcinomas demonstrate a methionine-dependent phenotype and are unable to grow upon substitution of methionine in the growth medium by homocysteine and folic acid, substrates of the methionine synthase reaction (10). While examining whether a link, if any, exists between methionine dependence of many carcinomas and changes in antioxidant capacity, we discovered that replacement of methionine (Met+) by homocysteine (Met−Hcy+) in folate-replete medium resulted in a marked time-dependent decrease in CBS levels in the human hepatoma cell lines, SkHep-1 (Fig. 2A). This effect was reversible, and addition of methionine to cells grown in Met−Hcy+ medium resulted in restoration of CBS to levels observed in Met+ medium, albeit with faster kinetics (Fig. 2B). CBS levels in cells grown in Met+Hcy+ were identical to those in Met+ medium (data not shown), indicating that the absence of methionine rather than the presence of homocysteine was responsible for the observed change in CBS levels.
Fig. 2.
Regulation of CBS by methionine status. (A) SkHep-1 cells were grown for 7 days in MEM containing 100 μM methionine and switched to Met−Hcy+ medium containing 200 μM dl-homocysteine. CBS and actin (an equal loading control) were detected in cell extracts by Western analysis as described under Methods and are representative of four independent experiments. A similar response was observed in the human hepatoma cell line HepG2 and the transformed monkey kidney cell line COS-1 (not shown). (B) Reversible regulation of CBS by methionine. Change in CBS concentration normalized to the density of actin in each lane was plotted as a function of time in cells switched from Met+ to Met−Hcy+ medium or vice versa. Before switching to Met+ medium, SKHep-1 cells were cultured for 7 days in Met−Hcy+ medium. Data are the average of four independent experiments. (C) AdoMet signals changes in methionine status. SkHep-1 cells were grown in Met−Hcy+ medium for 7 days, CBS expression was induced by transferring cells to Met+ medium with or without 20 mM cycloleucine (CL), and CBS levels were assessed by Western analysis 0, 24, and 48 h later. Data are representative of at least three experiments performed in duplicate. (D) AdoMet or its structural analog, AdoEt, can induce CBS under conditions of methionine restriction. SkHep-1 cells grown for 7 days in Met−Hcy+ medium were transferred to the same medium supplemented with either 100 μM methionine or 500 μM AdoMet or 500 μM AdoEt. CBS levels were assessed by Western blot analysis 72 h later. Data are representative of two experiments performed in duplicate.
AdoMet Signals Methionine Restriction.
To distinguish between the possibilities that methionine itself versus a downstream metabolite signals a change in methionine status, we used inhibitors of pathway enzymes (Fig. 1) in combination with rescue strategies (Fig. 2C). Inhibition of MAT by cycloleucine resulted in lower CBS levels versus untreated controls when cells were transferred from Met−Hcy+ to Met+ medium. The other inhibitors shown in Fig. 1 failed to inhibit restoration of CBS levels in response to methionine provision to cells grown in Met−Hcy+ medium (data not shown). These results suggest that AdoMet rather than methionine signals a change in cellular methionine availability and regulates CBS levels. Consistent with this hypothesis, the effect of methionine restriction on CBS levels was attenuated by provision of AdoMet (Fig. 2D). Intracellular AdoMet concentrations were found to decrease >14-fold from 5.4 ± 0.4 nmol·mg−1 protein (n = 8) in cells grown in Met+ medium to 0.37 ± 0.06 nmol·mg−1 protein (n = 7) in Met−Hcy+ medium. The rapid induction of CBS levels by substitution of Met−Hcy+ with Met+ medium (Fig. 2) was accompanied by an overshoot in AdoMet concentrations after 24 h [19.1 ± 2.6 nmol·mg−1 of protein (n = 4)] and returned to steady-state levels at 48 h.
CBS Is Repressed in MAT1A Knockout Mice with High Methionine and Low AdoMet.
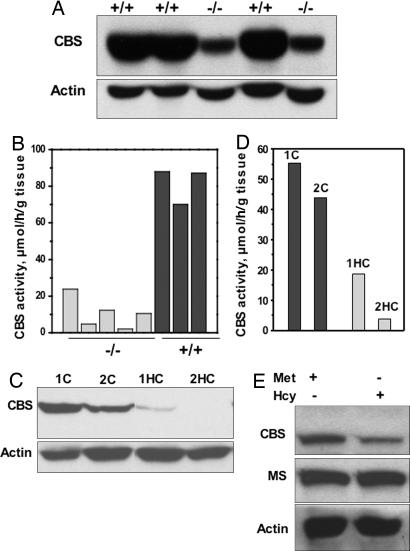
To more clearly interrogate the role of AdoMet in regulating CBS levels, we used the transgenic murine model in which the gene for MAT1A (encoding the MATI/III isoenzyme) is disrupted (11). This knockout induces expression of the MATII isoenzyme in liver with a consequent lowering of hepatic AdoMet and glutathione levels by 4- and 2.5-fold, respectively, and results in an ≈8-fold increase in plasma methionine levels (11). Chronic hepatic insufficiency of AdoMet in the MAT1A knockout mice predisposes them to spontaneous steatohepatitis and hepatocellular carcinoma (11, 12). The MAT1A knockout mice serve as an ideal model to test the effect of reduced AdoMet in a background of high methionine on CBS expression. Western analysis revealed that CBS levels were an excellent predictor of genotype and that steady-state CBS levels and activity were diminished 87 ± 5% (n = 3) and 78 ± 4% (n = 5), respectively (Fig. 3A and B), whereas Northern analysis revealed no change in CBS mRNA levels (data not shown).
Fig. 3.
CBS expression in liver disease states. (A) Hepatic CBS levels were determined by Western blot analysis in wild type (+/+) and MAT1A knockout (−/−) mice. Protein concentration was determined by using the Bradford assay, and equal loading of samples in each lane was ensured by monitoring actin levels. (B) CBS-specific activity was determined by the ninhydrin assay (8) in wild-type (black bars) and MAT1A knockout (gray bars) mouse livers. (C) Hepatic CBS levels were determined by Western blot analysis in control (1C and 2C) and cancerous (1HC and 2HC) biopsied liver tissue from two patients with hepatocellular carcinoma. (D) CBS-specific activity was determined by the ninhydrin assay in normal (black bars) and cancerous (gray bars) liver tissue. Samples 1C and 1HC and 2C and 2HC are derived from normal and hepatocellular cancer tissue from the same patient, respectively. (E) Effect of methionine availability on CBS expression in fibroblasts (F4875) homozygous for the D444N mutation in CBS. The patient fibroblast cell line was grown for 7 days in either Met+ or Met−Hcy+ medium. Levels of CBS, methionine synthase, and actin were monitored by Western analysis. Data are representative of three independent experiments performed in duplicate.
CBS Is Reduced in Human Liver Cancer.
Hepatocellular carcinomas are characterized by a switch in expression of the MAT isoenzyme from MATI/III associated with high AdoMet and a differentiated phenotype, to MATII associated with lower AdoMet and a dedifferentiated phenotype (3). Hepatic cirrhosis is the most common precursor to development of hepatocellular carcinoma and is characterized by increased oxidative stress and decreased glutathione (2). Based on our observations, we hypothesized that down-regulation of CBS may be correlated with the spontaneous oxidative stress and liver injury associated with this pathology. Indeed, a substantial decrease in CBS levels and activity were observed when histologically established noncancerous versus cancerous tissues in the same patient obtained from liver biopsy samples were compared (Fig. 3 C and D). Glutathione levels were decreased from 5.7 to 3.2 μmol·(g of tissue)−1 in control versus malignant tissues in one liver biopsy sample and from 4.1 to 0.7 μmol·(g of tissue)−1 in another.
Stability of CBS Is Decreased Under Conditions of Methionine Restriction.
In principle, a decrease in CBS levels in response to diminished cellular methylation capacity could be effected at multiple levels and various modes of gene regulation by AdoMet are known. In bacteria, AdoMet binds to RNA riboswitches and regulates genes involved in sulfur metabolism (13). In Arabidopsis thaliana, stability of the cystathionine γ-synthase mRNA is negatively regulated by AdoMet (14), and in HepG2 cells, methionine withdrawal results in an AdoMet-dependent increase in MATII expression by stabilization of its mRNA (15). We excluded an effect of AdoMet on transcription (by Northern analysis and reporter assays using the −1b CBS promoter, which is most commonly used in liver) and translation (by in vitro transcription/translation and polysome analysis) (data not shown). In contrast, pulse–chase studies revealed a marked decrease in the t1/2 of CBS from 49 ± 4 h in cells grown in Met+ to 18 ± 2 h in cells grown in Met−Hcy+ medium (data not shown).§ The relatively long half-life of CBS measured in these experiments is consistent with the slow kinetics of its disappearance upon methionine restriction (Fig. 2).
Binding of AdoMet Stabilizes CBS.
AdoMet is an allosteric effector of CBS and binds to the regulatory domain (16), which contains a tandem repeat of “CBS domains,” a secondary structure motif that binds various adenine nucleotides and is believed to play a role in energy sensing (17). AdoMet binding increases CBS activity 2- to 3-fold (5). To distinguish between a structural versus a chemical role (i.e., via methyl transfer) for AdoMet in modulating stability of CBS, the ability of the structural analog, S-adenosyl-l-ethionine (AdoEt), to induce CBS in Met−Hcy+-grown cells was tested (Fig. 2D). AdoEt activates CBS in vitro (4) and was as effective as AdoMet in inducing CBS levels in vivo, suggesting that the stabilizing effect of AdoMet on CBS in cells is associated with binding rather than methyl transfer to CBS or to some other target. To further test whether AdoMet binding enhances CBS stability in vivo, we used the pathogenic D444N variant of CBS, which is unresponsive to the allosteric effects of AdoMet (18, 19). It exists predominantly in an activated conformation in the absence of AdoMet, mimicking that achieved by wild-type enzyme in the presence of AdoMet (20). Our hypothesis that AdoMet binding stabilizes CBS leads to the prediction that the D444N mutant would be less responsive to methionine restriction. Indeed, in a cell line homozygous for the D444N allele, only a 2-fold decrease in CBS levels was observed in cells grown in Met−Hcy+ versus Met+ medium (Fig. 3E) whereas CBS levels decreased >10-fold in normal fibroblast cells grown in Met−Hcy+ versus Met+ medium (data not shown). Methionine synthase levels did not change and served as an equal loading control.
The above results provide strong evidence that AdoMet not only activates but also enhances stability of CBS. Next, we used pulse proteolysis (21) to quantify the effect of AdoMet on CBS stability (Fig. 4A–C). In the presence of AdoMet, the Cm value (i.e., the concentration of urea required to unfold half the CBS) increased from 2.9 ± 0.1 M to 4.6 ± 0.02 M. The global stability of CBS, ΔGunf°, increased from 20.6 kcal·mol−1 in the absence of AdoMet to 33 kcal·mol−1 in its presence. Hence, binding of AdoMet provides significant stabilization to CBS.
Fig. 4.
AdoMet stabilizes CBS. (A and B) Proteolytic digestion of CBS in the absence (A) or presence (B) of 300 μM AdoMet by pulse proteolysis as described in Methods. The first lane in each gel represents molecular-weight markers. (C) Dependence of the fraction of folded protein (ffold) on the denaturant concentration in the absence (open squares) and presence (filled squares) of 300 μM AdoMet. Data are averages of four independent experiments.
CBS Down-Regulation Affects Viability Under Oxidative Stress Conditions.
The effect of CBS down-regulation on transsulfuration flux was measured in SKHep-1 cells as a time-dependent increase in cystathionine levels in cells treated with propargylglycine, an inhibitor of γ-cystathionase. The flux was found to be 220 ± 22 and 118 ± 17 μmol of cystathionine·(liter of cells)−1·h−1 in cells grown in Met+ versus Met−Hcy+ media, respectively. The 2-fold decrease in flux through CBS was associated with the >10-fold difference in protein levels in cells grown in Met+ versus Met−Hcy+ media. This apparent discrepancy in the magnitude of CBS levels and flux through it suggests that CBS is not fully rate limiting in converting methionine to cystathionine. This is consistent with the 2.5-fold decrease in glutathione levels in MAT1A knockout mice (11) in which CBS levels were found to be decreased >5-fold. To assess the physiological effect of CBS regulation on antioxidant capacity, the effect of tert-butyl hydroperoxide on cells containing high and low CBS levels was determined (Fig. 5). Cells grown in Met+ medium were significantly more resistant to the oxidative challenge than were cells grown in Met−Hcy+ medium that had low CBS levels. This difference in viability was partially negated by provision of N-acetylcysteine, a cysteine precursor that can bypass intracellular cysteine deficiency imposed by reduced transsulfuration flux.
Fig. 5.
The effect of diminished CBS on cell viability at increasing concentrations of tert-butyl hydroperoxide in cells grown in the Met+ (filled circles), Met−Hcy+ (filled triangles), or Met−Hcy+ plus N-acetylcysteine (NAC) (open triangles) medium. Viability is represented as a percentage of viable cells in untreated controls, and each data point is the average of three to six independent determinations.
Interestingly, AdoMet has been suggested as a therapeutic agent for chemoprevention and treatment of hepatic cancer (22) and is already being used in clinical trials for the treatment of cirrhosis and alcoholic liver disease, chronic conditions characterized by decreased glutathione and increased oxidative stress. Indeed, it has been reported that long-term treatment of alcoholic liver cirrhosis patients with AdoMet may improve survival or delay liver transplantation (23). Our results suggest that in addition to correcting the methylation status, AdoMet supplementation would enhance transsulfuration capacity by stabilizing CBS, and that an alternative strategy, i.e., administration of N-acetylcysteine, could bypass the block by acting downstream of it. It is interesting to note that oral supplementation with different cysteine-rich whey protein formulas is already used to increase glutathione levels in patients with cystic fibrosis (24) or HIV infection (25) and that the therapy appears to be well tolerated.
In summary, our studies suggest a mechanistic basis for the coordinate changes in methylation and antioxidant capacity that is observed in many diseases. Stabilization of CBS, which catalyzes the committing step in the bridging transsulfuration pathway, by AdoMet, a signal of cellular methylation status, represents a strategy whereby an allosteric effector enhances catalytic activity while also increasing the t1/2 of the protein. The relevance of this mode of regulation to metabolic changes observed in transformed liver cells, in a mouse model susceptible to liver injury and in human liver cancer, suggests therapeutic strategies that may be effective in diminishing the enhanced oxidant burden associated with reduced transsulfuration flux in these conditions.
Methods
MAT1A Knockout Mice and Human Liver Samples.
MAT1A knockout mice were maintained at the University of Southern California, and livers were obtained as described in ref. 11. Cancerous liver tissue was obtained from two patients undergoing surgical resection for primary hepatocellular carcinoma. Noncancerous liver tissue included in the resected specimen was used as the control sample. Written informed consent was obtained from each patient. These tissues were immediately frozen in liquid nitrogen for subsequent measurement of glutathione and proteins as described below. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the University of Southern California School of Medicine’s human research review committee.
Cell Culture and Western Blot Analysis.
Transformed cells (SkHep-1, COS-1, and HepG2) were grown in Eagle’s minimum essential medium containing methionine (Met+) or lacking methionine but containing 200 μM dl-homocysteine (Met−Hcy+) supplemented with 10% dialyzed FBS and an antibiotic–antimycotic mixture (Invitrogen) of 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B. Both media contained cystine and folic acid at concentrations of 100 μM and 2.3 μM, respectively. Primary fibroblast cultures were grown in the same media but lacking amphotericin B. Cultures were maintained at 37°C in a 5% CO2 atmosphere. Inhibitors or other supplements were added to the media at the concentrations indicated in the figure legends. Cells were harvested by trypsinization, washed twice with ice-cold PBS, and, after centrifugation, resuspended in an equal volume of lysis buffer [0.1 M sodium phosphate (pH 7.4), containing 0.1% Triton X-100, 10 μl/ml protease inhibitor mixture (Sigma), 25 μg/ml tosyllysine chloromethyl ketone, 25 μg/ml phenylmethylsulfonyl fluoride, 27 μg/ml aprotinin, and 10 μg/ml leupeptin], placed on ice for 20 min, and stored at −80°C until further use. The lysate was centrifuged at 4°C for 20 min at ≈14,000 × g, and the protein concentration in the supernatant was determined by the Bradford assay (Bio-Rad). After electrophoresis on a 10% polyacrylamide gel and transfer to a PVDF membrane, methionine synthase, CBS, and actin levels were determined by using the respective primary and secondary antibodies and detected by using the chemiluminescent horseradish peroxidase system (Sigma) as described in ref. 8. The intensities of protein and nucleic acid bands were quantified by densitometry with quantity one software (Bio-Rad). For Western analysis, the CBS signal in each lane was first normalized to actin in the same lane before comparison with control values.
CBS Activity, Metabolite Levels, and Flux Measurements.
CBS activity was measured by using the ninhydrin assay as described in ref. 8. For flux measurements, cells were incubated in Met+ or Met−Hcy+ medium for 7 days with the final medium change being performed 1 day before initiation of the experiment. One hour before the experiment, cells were transferred to fresh Met+ medium so that flux measurements were performed at the same concentration of methionine to reflect a difference in CBS level and function, rather than methionine availability. Western blots were performed to establish that no change in CBS levels occurred during the experimental time frame. Flux measurements were conducted as described in ref. 7. Briefly, experiments were initiated by addition of propargylglycine (2.5 mM final concentration), and time points were collected after 0-, 3-, and 7-h incubation periods. Flux was expressed as the amount of cystathionine, μmol·h−1·(liter of cells)−1. Metabolite levels (glutathione and AdoMet) were determined as described in ref. 26. All values reported in this study represent the mean ± SEM.
In Vivo Pulse–Chase Analysis.
SkHep-1 cells were grown for 3 days in 60-mm dishes in either Met+ or Met−Hcy+ medium. After a 1-h incubation in leucine-deficient medium, cells were pulsed for 10 h with 200 μCi of l-[4,5-3H]leucine (Amersham Pharmacia; specific activity 162 Ci/mmol; 1 Ci = 37 GBq) in 1 ml of leucine- and methionine-deficient medium that was supplemented with either 100 μM l-methionine or 200 μM dl-homocysteine, respectively. After 10 h, cells were washed with 2 × 2 ml of PBS to remove unincorporated radioactivity and placed in chase medium containing 1 mM leucine and either methionine or homocysteine. A batch of cells from each condition was collected after 0, 12, 24, 36, and 48 h of incubation in chase medium and washed twice with ice-cold PBS, harvested with a cell scraper in the presence of 600 μl of lysis buffer, and frozen at −80°C until further analysis. For CBS immunoprecipitation, the lysates were thawed on ice and centrifuged at ≈14,000 × g for 15 min at 4°C. Lysates containing equal amounts of total protein (as judged by Bradford; Bio-Rad) were adjusted to a 600-μl volume each and then incubated by using 20 μl of agarose-linked anti-chicken IgY antibody (Aves Labs). After preclearing, the supernatants were incubated with 2.5 ng of purified anti-CBS antibody (developed in chicken by using purified recombinant human CBS) for 2 h at 4°C with vigorous shaking, followed by a 2-h incubation with 20 μl of agarose-linked anti-chicken IgY antibody. The beads were collected by centrifugation and washed according to the manufacturer’s recommendations by using the Immunoprecipitation kit (Sigma). To elute the protein, the beads were incubated with 50 μl of Laemmli sample buffer for 15 min at 95–100°C and spun down, and the clear supernatant was collected, divided into two portions, and separated on two SDS gels. One gel was incubated for 30 min in a fixing solution containing 10% (vol/vol) acetic acid and 30% methanol (vol/vol), then impregnated with the EN3HANCE solution (PerkinElmer) for 30 min and dried at 70°C. The dried gel was exposed to BioMax film (Kodak) at −80°C for 1–2 months. The autoradiography signal was normalized to the amount of CBS in each sample, as judged by densitometric analysis of the Western blot performed with the second gel. The initial experiments established that the radioactivity associated with CBS represented >90% of the total signal observed in the autoradiogram. Therefore, as a time-saving strategy, additional samples were analyzed for tritium content directly by using a liquid scintillation counter, and the dpm were normalized for CBS levels in each sample as judged by Western blot analysis.
In Vitro Pulse Proteolysis.
Pulse proteolysis of CBS was performed as described in ref. 21. Briefly, thermolysin (0.20 mg/ml from Bacillus thermoproteolyticus rokko; Sigma) was used to digest purified recombinant human CBS (0.50 mg/ml) at 25°C that had been equilibrated overnight at 4°C in 20 mM Tris·HCl buffer (pH 8.3) containing 10 mM CaCl2, 50 mM NaCl, and urea (0–7 M). To monitor CBS stability in the presence of AdoMet, the protein was first preincubated with 300 μM AdoMet for 30 min on ice and then exposed to a urea solution containing 300 μM AdoMet. The reaction was initiated by the addition of a 25× stock solution of thermolysin in 1.25 M NaCl and 0.25 M CaCl2. After 1 min, the reaction was quenched by the addition of 50 mM EDTA (pH 8.0) to a final concentration of 12.5 mM. Samples were mixed with Laemmli buffer, boiled for 5 min, separated on a 10% SDS/PAGE gel, and stained with Coomassie blue solution. The gels were scanned and CBS band intensities were quantified by using quantity one image analysis software. The global stability of a protein, ΔGunf°, was calculated by multiplying the Cm by the m-value (the denaturant dependence of ΔGunf°). The m-value of CBS was estimated as described in ref. 21 by multiplying the number of residues by −0.013 and found to be −7.2 kcal/(mol·M).
Cell Viability Studies.
SkHep-1 cells were grown for 7 days in six-well plates in Met+ or Met−Hcy+ medium. Immediately before the experiment, the medium was replaced with a fresh 2-ml aliquot of Met+ medium, to which freshly prepared tert-butyl hydroperoxide in ethanol was added. Controls received the equivalent volume of vehicle only. N-acetylcysteine was added to a final concentration of 200 μM 3 h before peroxide addition to test its effect on viability of Met−Hcy+-grown cells. After a 10-h incubation with peroxide, the viability of cells was assessed by using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide dye method (27) and expressed as a percentage of untreated controls.
Acknowledgments
We thank Dr. Henk Blom (University Medical Center, Nijmegen, The Netherlands) for providing the fibroblast cell line (F4875) and Dan Schellhorn (University of Nebraska) for purifying the CBS used in the pulse proteolysis study. This work was supported by grants from the National Institutes of Health to R.B. (DK64959 and HL58984) and S.C.L. (DK51719, AT1576, AA13847, and AA12677), and a predoctoral fellowship to A.P. from the Heartland Affiliate of the American Heart Association.
Abbreviation
- AdoMet
S-adenosylmethionine.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
The doubling time of SK-Hep1 cells in Met+ medium is ≈3 days and could lead to an underestimation of the t1/2 of CBS under these conditions. In contrast, growth of SK-Hep1 cells stalls in Met−Hcy+ medium (without a visible loss in cell density) as reported previously for other cancer cell lines (28).
References
- 1.Jungst C., Cheng B., Gehrke R., Schmitz V., Nischalke H. D., Ramakers J., Schramel P., Schirmacher P., Sauerbruch T., Caselmann W. H. Hepatology. 2004;39:1663–1672. doi: 10.1002/hep.20241. [DOI] [PubMed] [Google Scholar]
- 2.McKillop I. H., Schrum L. W. Alcohol. 2005;35:195–203. doi: 10.1016/j.alcohol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Cai J., Mao Z., Hwang J. J., Lu S. C. Cancer Res. 1998;58:1444–1450. [PubMed] [Google Scholar]
- 4.Finkelstein J. D., Kyle W. E., Martin J. J., Pick A.-M. Biochem. Biophys. Res. Commun. 1975;66:81–87. doi: 10.1016/s0006-291x(75)80297-x. [DOI] [PubMed] [Google Scholar]
- 5.Taoka S., Widjaja L., Banerjee R. Biochemistry. 1999;38:13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- 6.Beatty P. W., Reed D. J. Arch. Biochem. Biophys. 1980;204:80–87. doi: 10.1016/0003-9861(80)90009-0. [DOI] [PubMed] [Google Scholar]
- 7.Mosharov E., Cranford M. R., Banerjee R. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 8.Vitvitsky V., Dayal S., Stabler S., Zhou Y., Wang H., Lentz S. R., Banerjee R. Am. J. Physiol. 2004;287:R39–R46. doi: 10.1152/ajpregu.00036.2004. [DOI] [PubMed] [Google Scholar]
- 9.Martinov M. V., Vitvitsky V. M., Mosharov E. V., Banerjee R., Ataullakhanov F. I. J. Theor. Biol. 2000;204:521–532. doi: 10.1006/jtbi.2000.2035. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman R. M. Biochim. Biophys. Acta. 1984;738:49–87. doi: 10.1016/0304-419x(84)90019-2. [DOI] [PubMed] [Google Scholar]
- 11.Lu S. C., Alvarez L., Huang Z. Z., Chen L., An W., Corrales F. J., Avila M. A., Kanel G., Mato J. M. Proc. Natl. Acad. Sci. USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Chantar M. L., Corrales F. J., Martinez-Cruz L. A., Garcia-Trevijano E. R., Huang Z. Z., Chen L., Kanel G., Avila M. A., Mato J. M., Lu S. C. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 13.Winkler W. C., Nahvi A., Sudarsan N., Barrick J. E., Breaker R. R. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 14.Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S. Proc. Natl. Acad. Sci. USA. 2003;100:10225–10230. doi: 10.1073/pnas.1831512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Chantar M. L., Latasa M. U., Varela-Rey M., Lu S. C., Garcia-Trevijano E. R., Mato J. M., Avila M. A. J. Biol. Chem. 2003;278:19885–19890. doi: 10.1074/jbc.M211554200. [DOI] [PubMed] [Google Scholar]
- 16.Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. J. Clin. Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A. Trends Biochem. Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 18.Kluitjmans L. A. J., Boers G. H. J., Stevens E. M. B., Renie W. O., Kraus J. P., Trijbels F. J. M., van den Heuvel L. P. W. J., Blom H. J. J. Clin. Invest. 1996;98:285–289. doi: 10.1172/JCI118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evande R., Boers G. H. J., Blom H. J., Banerjee R. Biochemistry. 2002;41:11832–11837. doi: 10.1021/bi026248d. [DOI] [PubMed] [Google Scholar]
- 20.Sen S., Yu J., Yamanishi M., Schellhorn D., Banerjee R. Biochemistry. 2005;43:14210–14216. doi: 10.1021/bi051046d. [DOI] [PubMed] [Google Scholar]
- 21.Park C., Marqusee S. Nat. Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 22.Lu S. C., Mato J. M. Alcohol. 2005;35:227–234. doi: 10.1016/j.alcohol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Mato J. M., Camara J., Fernandez de Paz J., Caballeria L., Coll S., Caballero A., Garcia-Buey L., Beltran J., Benita V., Caballeria J., et al. J. Hepatol. 1999;30:1081–1089. doi: 10.1016/s0168-8278(99)80263-3. [DOI] [PubMed] [Google Scholar]
- 24.Grey V., Mohammed S. R., Smountas A. A., Bahlool R., Lands L. C. J. Cyst. Fibros. 2003;2:195–198. doi: 10.1016/S1569-1993(03)00097-3. [DOI] [PubMed] [Google Scholar]
- 25.Micke P., Beeh K. M., Schlaak J. F., Buhl R. Eur. J. Clin. Invest. 2001;31:171–178. doi: 10.1046/j.1365-2362.2001.00781.x. [DOI] [PubMed] [Google Scholar]
- 26.Prudova A., Martinov M. V., Vitvitsky V., Ataullakhanov F., Banerjee R. Biochim. Biophys. Acta. 2005;1741:331–338. doi: 10.1016/j.bbadis.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Manthorpe M., Fagnani R., Skaper S. D., Varon S. Brain Res. 1986;390:191–198. doi: 10.1016/s0006-8993(86)80227-x. [DOI] [PubMed] [Google Scholar]
- 28.Mecham J. O., Rowitch D., Wallace C. D., Stern P. H., Hoffman R. M. Biochem. Biophys. Res. Commun. 1983;117:429–434. doi: 10.1016/0006-291x(83)91218-4. [DOI] [PubMed] [Google Scholar]