Abstract
The folding environment in the endoplasmic reticulum (ER) depends on multiple abundant chaperones that function together to accommodate a range of substrates. The ways in which substrate engagement shapes either specific chaperone dynamics or general ER attributes in vivo remain unknown. In this study, we have evaluated how changes in substrate flux through the ER influence the diffusion of both the lectin chaperone calreticulin and an inert reporter of ER crowdedness. During acute changes in substrate load, the inert probe revealed no changes in ER organization, despite significant changes in calreticulin dynamics. By contrast, inhibition of the lectin chaperone system caused rapid changes in the ER environment that could be reversed over time by easing new substrate burden. Our findings provide insight into the normal organization and dynamics of an ER chaperone and characterize the capacity of the ER to maintain homeostasis during acute changes in chaperone activity and availability.
Keywords: castanospermine, fluorescence loss in photobleaching, fluorescence recovery after photobleaching, pactamycin, puromycin
The initial folding and maturation of nearly all secretory and membrane proteins in eukaryotic cells occurs in the endoplasmic reticulum (ER). Nascent proteins are translocated as largely unfolded polypeptides into this tremendously crowded environment (1, 2) through the translocon (3, 4). The ER lumen contains numerous abundant enzymes that facilitate the folding and modification of these nascent polypeptides (5, 6). These enzymes include polypeptide-binding proteins [i.e., BiP and calreticulin (Crt)], isomerases (i.e., PDI), proteases (i.e., signal peptidase), and glycosylation enzymes [i.e., oligosaccharyl transferase (OST) and glucosidases].
The primary sequence of each protein defines which subsets of these maturation factors will be required for its biogenesis. Because the maturation machinery of the ER lumen cannot anticipate a priori the polypeptide that will emerge from a translocon, it is critical for the maturation machinery to be highly flexible in adapting to the needs of the substrate. Hence, each maturation factor must be able to access any potential substrate quickly and efficiently to prevent protein misfolding and ER dysfunction. The organizing principles that permit complex maturation machinery to balance efficient substrate access with tremendous flexibility are poorly understood.
Conceptually, two distinct mechanisms can be envisaged. In one strategy, enzymes are effectively immobilized at the site of their intended functions. For example, the OST is positioned with its active site close to the lumenal side of the translocon where nascent chains enter the ER (7). The OST is immobile relative to the translocating nascent chain (8), providing the OST with uniform access to all potential substrates at precisely the point in their maturation that requires OST function. Indeed, given that most maturation enzymes, such as ER chaperones, are at least as abundant as translocons (9), their effective immobilization in a spatially defined manner near the site of translocation is an attractive model. Such a “matrix” would afford each maturation factor the opportunity to access any given nascent chain, and each nascent chain could sample the maturation machinery in a defined manner (10–12).
An alternative strategy would be a modular system in which maturation enzymes are mobile, permitting them to rapidly sample the ER environment. Constant stochastic sampling would allow distinct combinatorial subsets of the machinery to be recruited to different substrates in response to their maturation demands. For this strategy, it would be critical for the ER environment to be maintained in a state where substrate and chaperones have rapid and complete access to each other. Such a highly dynamic state should be robust and remain largely unchanged in the face of large changes in ER activity.
The degree to which these different strategies are used by ER chaperones remains largely unknown. Previous studies have focused primarily on analyzing individual substrate–chaperone interactions. Although this approach has resulted in numerous insights into the substrate specificities, affinities, and functional cycles of individual chaperones, it has not clarified their organization and dynamics in vivo. The organizational state of any ER chaperone in its native environment under either quiescent or substrate-engaged conditions continues to be a matter for speculation (13). In this study, we have analyzed the dynamics of a functional ER chaperone in live cells by using fluorescent fusion proteins combined with photobleaching methods. By monitoring both a model chaperone, Crt, and the global ER environment under different conditions, we have gained insight into the organizational strategies used by the ER to promote protein maturation. These findings reveal a highly dynamic ER environment that is robust to dramatic changes in substrate flux and can tolerate marked disruptions to its folding capacity.
Results and Discussion
The analysis of protein dynamics in vivo requires fluorescent probes whose biophysical properties can be monitored to infer changes in cellular biochemistry (14, 15). In this study, two quantitatively distinct probes were used. The first is a GFP-tagged ER chaperone whose interactions with other cellular machinery could be tracked by changes in its diffusion. The second is an inert probe that can report on more general parameters of the ER environment, such as its crowdedness and interconnectivity, two variables that directly reflect accessibility of ER components to each other.
The inert probe, ER-localized GFP (hereafter termed ER-GFP), is an average-sized protein, has no known interacting partners, and can rapidly sample the entire ER lumen (16). Furthermore, GFP lacks disulfide bridges and glycosylation sites, folds independently of chaperones, and exhibits fluorescence only when properly folded (17). Thus, the diffusion of ER-GFP fluorescence can be used to monitor changes in the capacity of an ER component to sample its environment (16, 18).
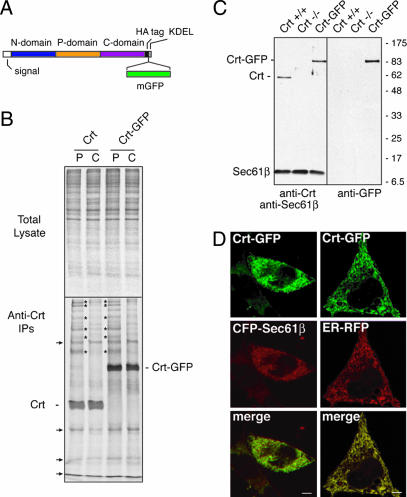
For the chaperone probe, we selected Crt for several reasons: the availability of a knockout cell line (19), chemical inhibitors of its function (20), well characterized biochemical properties (21–26), and structural information (24, 27–29). Based on this information, we generated a GFP-tagged Crt (Crt-GFP) (Fig. 1A) and compared its functional properties relative to untagged Crt upon expression in crt−/− fibroblasts. We found by coimmunoprecipitation analyses of pulse-labeled cells that both Crt and Crt-GFP interacted with an identical subset of newly synthesized polypeptides, most of which were largely released in the following 90-min chase (Fig. 1B). Similar analyses of either untransfected or ER-GFP-transfected crt−/− fibroblasts did not result in immunoprecipitation of these bands (see Fig. 6, which is published as supporting information on the PNAS web site, and data not shown). Importantly, when a conservative point mutation (Y108F) that disrupts lectin activity of Crt (24) was introduced into Crt-GFP [to generate Crt(Y108F)-GFP], it no longer coprecipitated any newly synthesized proteins (Fig. 6). Thus, Crt-GFP displays transient interactions with several newly synthesized proteins, as has been described previously for Crt. Furthermore, when expressed in the identical context of crt−/− fibroblasts, the substrate-specificity of Crt and Crt-GFP are identical. Finally, Crt-GFP interactions with substrates largely, if not entirely, depend on its lectin activity, supporting the argument that most of its substrate associations are direct and glycan-mediated. We conclude that Crt-GFP displays properties comparable with untagged Crt and consistent with its well studied function as a lectin-dependent ER chaperone.
Fig. 1.
Crt-GFP construction and characterization. (A) Illustration of functional domains of Crt and the insertion sites for an hemagglutinin (HA) epitope tag and monomerized GFP. (B) crt−/− cells were transiently transfected with either Crt or Crt-GFP and analyzed 40 h later by pulse–chase labeling and native immunoprecipitation. Pulse labeling (P) with [35S]methionine was for 30 min, followed by chase (C) in unlabeled media for 90 min. Total cell lysates from the samples (Upper) and the anti-Crt immunoprecipitates (Lower) are shown. The positions of Crt and Crt-GFP are indicated. Several coprecipitating proteins that transiently interact with both Crt and Crt-GFP during the pulse and release during the chase period are indicated by asterisks. Background bands that were also seen in control transfected cells (see Fig. 6) are indicated by the arrows. (C) Immunoblots of whole cell lysates from crt+/+, crt−/−, or Crt-GFP cells (crt−/− cells stably expressing Crt-GFP). (Left) Probed with anti-Crt and anti-Sec61β (a loading control). (Right) The same blot stripped and reprobed with anti-GFP. (D) Complete colocalization of Crt-GFP in live cells with cotransfected ER markers: mCFP-Sec61β (Left) or ER-RFP (Right). (Scale bars, 5 μm.)
We therefore stably expressed Crt-GFP in crt−/− fibroblasts at a level corresponding to endogenous Crt levels of matched crt+/+ cells (Fig. 1C). Crt-GFP in this stable line was localized to the ER (Fig. 1D) and transiently interacted with several newly synthesized proteins in a pulse–chase experiment similar to that shown in Fig. 1B (data not shown). Thus, crt−/− cells have been reconstituted with functional Crt-GFP, whose fluorescence can now be used to track its behavior in vivo. In parallel, ER-GFP expressed in crt+/+ cells can be used to monitor general ER properties under comparable experimental conditions.
With these tools, we investigate the dynamics of chaperone–substrate interactions in three stages. First, we establish the properties of ER-GFP and Crt-GFP under conditions where no substrates are actively entering the ER. These experiments provide basic information on chaperone-complex dynamics in the quiescent ER and distinguishes among competing models of ER organization. Second, we ask how these parameters change upon introduction of substrate under normal metabolic conditions, allowing us to assess the impact of chaperone activity on both the chaperone and the ER environment. Third, we investigate acute inhibition of the lectin chaperone system to gain insight into the homeostatic mechanisms that maintain the ER environment.
To deplete the ER of substrate (Fig. 2A), the initiation of new protein synthesis was inhibited with pactamycin (Pact), which also permitted preexisting nascent polypeptides to complete synthesis (≈10–15 min) (30) and subsequent maturation. Based on the results shown in Fig. 1B and previously established rates of maturation and exit of model proteins (23), we reasoned that 1 hour after addition of Pact was sufficient to create a “quiescent” ER (Fig. 2A). To analyze the organizational state of Crt-GFP under quiescent conditions, we performed fluorescence loss in photobleaching (FLIP). For FLIP (Fig. 3A), a small region of the cell Crt-GFP fluorescence was repeatedly photobleached. Over time, Crt-GFP fluorescence in the entire cell was steadily and uniformly lost, whereas fluorescence of adjacent cells was unaffected, as expected for a freely diffusing protein distributed throughout a continuous ER (14, 16, 18).
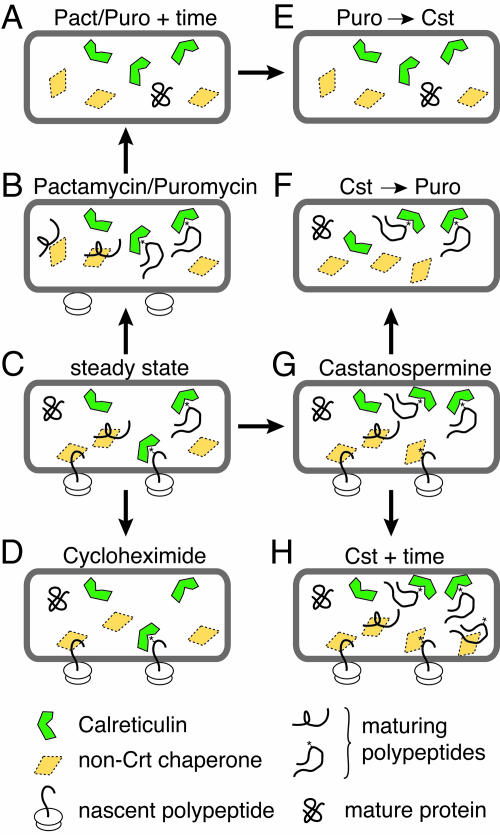
Fig. 2.
Schematic diagram of the experimental conditions analyzed in this study. At steady state (C), the ER contains a mixture of chaperone-bound nascent polypeptides, chaperone-bound maturing polypeptides, free chaperones, and mature proteins. The manners in which these populations are expected to change upon the indicated treatments are illustrated. For example, treatment with Puro or Pact inhibits new protein synthesis, resulting initially in chaperone-bound maturing polypeptides (B) followed by a quiescent ER (A) in which substrate polypeptides have matured and the chaperones are inactive. Treatment with cycloheximide (D) traps nascent proteins in the translocon. Treatment with Cst (G and H) creates pools of glycoproteins that remain associated with Crt-GFP or never associate with Crt-GFP and then associate with alternative chaperones (see Fig. 5). Progressive treatments with Cst and Puro deplete the pool of nascent glycoproteins that do not associate with Crt-GFP (E and F). However, the order of addition affects the pool of nascent proteins bound to Crt-GFP (E and F). Details of each condition are given in Results and Discussion.
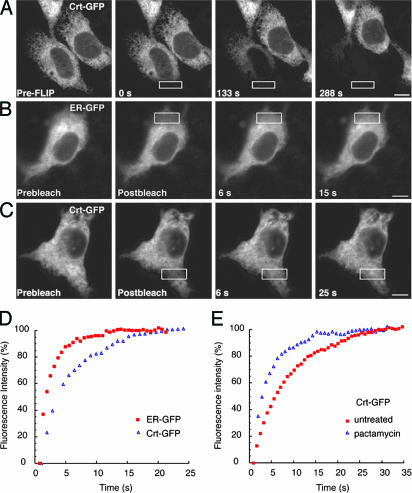
Fig. 3.
Dynamics of Crt-GFP. (A) Images of a cell expressing Crt-GFP before (Left) and at various times during repeated photobleaching in the region outlined by the white box. Fluorescence in the bleached cell was depleted uniformly over time. (B and C) FRAP analysis of quiescent cells (treated with Pact for 1 h; Fig. 2A) expressing ER-GFP or Crt-GFP. Images were captured immediately before (Prebleach), immediately after (Postbleach), and at times after photobleaching in the area outlined by the white box. Both ER-GFP and Crt-GFP are highly mobile, because unbleached fluorescent proteins rapidly diffuse into the bleached regions. [Scale bars, 5 μm (A–C).] (D) Plots of recovery rates reveal that ER-GFP (red squares) diffuses more rapidly than Crt-GFP (blue triangles). (E) FRAP analysis of Crt-GFP in cells actively translating new proteins (untreated, red squares) or under quiescent conditions (Pact-treated, blue triangles) demonstrates that Crt-GFP mobility decreases in the presence of substrates.
These results indicate that Crt-GFP throughout the ER has access to the region of photobleaching, without any significant immobilized population. Therefore, Crt-GFP is unlikely to be part of a stable matrix of chaperones in the ER lumen, as had been suggested on the basis of crosslinking studies (10). This is not to say that Crt-GFP does not interact with other chaperones under these conditions but simply that such interactions would have to be highly dynamic and transient or represent a small fraction of the total population of Crt to be compatible with the diffusional analysis. Furthermore, our data support an argument against confined subpopulations of Crt-GFP within discrete regions of the ER (i.e., a putative “quality control compartment”). Hence, the FLIP result constrains the number of compatible models for Crt organization to those in which it is highly dynamic in the quiescent ER environment.
To reconcile the conclusions from crosslinking studies, we note that crosslinkers irreversibly trap transient interactions (both specific and nonspecific). This trapping can “daisy-chain” proteins that contacted each other only transiently without ever residing in a large complex. Indeed, our observation that Crt-GFP can sample the entire ER within ≈5–7 min (Fig. 3A) suggests that such an effect could easily account for the matrix-like effect after 30 min of crosslinking with 2 mM crosslinker (10).
Next, we quantitated the mobility of ER-GFP and Crt-GFP. The primary mobility parameter is the effective diffusion coefficient, Deff, which can be derived from the rate of fluorescence recovery after photobleaching (FRAP) of a small region of the cell (31). A doubling of the Deff indicates that the molecule can now stochastically sample twice the area in the same period. Parameters that directly influence Deff include viscosity (or crowdedness of the environment), size of the protein complex, protein–protein interactions, or combinations of these variables (14). For example, an increase in protein-complex size by a factor of two would lead to a 2-fold decrease in Deff, all else being equal (see Stokes–Einstein equation in Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Thus, even modest changes in Deff can be due to biologically significant changes. Another commonly measured parameter is the mobile fraction, an operationally defined value indicating the percent of total fluorescent molecules that contribute to the recovery during the time frame of the experiment. We found this parameter to be largely invariant in the subsequent experiments, so it was not analyzed extensively (see Fig. 7, which is published as supporting information on the PNAS web site, and Supporting Materials and Methods for a detailed discussion).
FRAP analysis of ER-GFP in a quiescent ER yielded Deff values between 8.7 and 10 μm2/s (Fig. 3B and Table 1). Essentially identical results were obtained by using other translational inhibitors (Table 1). These values were comparable with the values of 5–10 μm2/s in translating cells, as reported in ref. 16. Based on the Deff for ER-GFP, one can estimate the Deff expected for monomeric and freely diffusing Crt-GFP (14). Deviations from the predicted value could be used to infer interactions between Crt-GFP and other proteins. Based on the Deff for ER-GFP and the Stokes radii for GFP (2.3 nm) (32) and Crt (4.5 nm) (27), we estimated that Crt-GFP should exhibit a Deff of between 3.0 and 3.4 μm2/s in the quiescent ER (see Supporting Materials and Methods). The experimental value proved to be surprisingly close: either 2.4 μm2/s or 3.3 μm2/s, depending on the choice of translational inhibitor [either puromycin (Puro) or Pact] (Table 1, Fig. 3 C and D). This finding is consistent with the majority of Crt-GFP existing as either a monomeric protein or in small and dynamic complexes. Together, these findings demonstrate that, in the quiescent state, both ER-GFP and Crt-GFP can rapidly and efficiently sample the entire ER, as expected for an environment designed for efficient multistep maturation events. Thus, nascent substrates that enter the ER lumen immediately encounter an environment in which at least one maturation enzyme is highly dynamic.
Table 1.
Deff values from FRAP analyses of Crt-GFP and ER-GFP
| Construct | Condition | No. | Deff, μm2/s ± SD |
|---|---|---|---|
| Crt-GFP | (C) Steady state | 28 | 1.3 ± 0.3 |
| (A) Pact | 14 | 2.4 ± 0.7* | |
| (D) Chx | 15 | 1.9 ± 0.6* | |
| (A) Puro | 11 | 3.3 ± 1.4* | |
| ER-GFP | (C) Steady state | 20 | 8.7 ± 2.5 |
| (A) Pact | 14 | 8.7 ± 2.3 | |
| (D) Chx | 17 | 8.2 ± 2.4 | |
| (A) Puro | 11 | 10.0 ± 2.2 |
The panel of Fig. 2 that illustrates each condition is indicated in parentheses to the left. Statistical comparisons of Deff values were carried out by using a two-tailed Student t test.
*Statistically significant change (P < 0.005) relative to the steady-state condition.
Next, we investigated the consequences of substrate engagement for both Crt dynamics and the folding environment of the ER. The substrates for Crt are predominantly newly synthesized secretory and membrane glycoproteins that include two populations: “nascent” and “maturing” polypeptides. Nascent substrates are ribosome-associated polypeptides in the process of being synthesized and translocated through the ER translocon. Maturing substrates are those that have completed synthesis but are in the process of posttranslational maturation events (such as folding and disulfide bond formation). We sought to separately evaluate Crt interactions with each pool. The strategy was to trap nascent polypeptides by treatment with a translational elongation inhibitor cycloheximide (Chx) (Fig. 2D). This situation, in which only nascent substrates were present, could then be compared to a situation where all substrates are present (Fig. 2C) to determine their respective contributions to Crt dynamics.
We observed a slower Deff for Crt-GFP when only nascent substrates were present (i.e., Chx treatment; Table 1). Under these conditions, the potential substrates for Crt-GFP are in the effectively immobile translocon (0.04 μm2/s) (8). Given the low mobility of the substrate, the slower Deff for Crt-GFP together with an unchanged and highly mobile fraction can be interpreted as transient binding and releasing from nascent chains. Hence, transient periods of Crt-GFP immobility interspersed with an otherwise rapid diffusion lead to seemingly slowed diffusion. We can exclude the alternate possibility that a fraction of Crt-GFP is stably bound to these nascent chains. For this situation, we would have expected a lower mobile fraction but an unchanged Deff for the population that remains mobile. We can further rule out a generalized crowding of the ER under these conditions because the diffusion of our inert probe (ER-GFP) was unchanged (Table 1). Thus, Crt-GFP interactions with nascent substrates are highly dynamic in vivo.
In actively metabolizing cells, where both nascent and maturing substrates populate the ER, Crt-GFP displayed an even lower Deff than with nascent substrates alone (Table 1; Fig. 3E). Yet, the mobile fraction for Crt-GFP and both parameters for the inert probe were unchanged relative to a quiescent ER. Thus, despite a markedly changed situation from a largely quiescent ER lumen to one containing numerous substrate–chaperone complexes, no change in ER-GFP diffusion was detectable, indicating that the efficiency of an ER component in sampling the entire lumenal environment is almost completely impervious to large changes in substrate load.
Furthermore, under no condition was there significant evidence for either a matrix of chaperones or for extremely large folding assemblies containing Crt (10, 13, 33). Rather, the data support a more dynamic situation in which folding complexes remain relatively small and/or assemble and disassemble (34) over short time frames. Indeed, this mode of action for chaperones may be the reason for the robustness of the ER environment to changes in substrate flux. If chaperone complexes were to interact stably with either each other or multiple substrates simultaneously, the resulting obstructive barriers to diffusion could impede access of new substrates to the folding machinery.
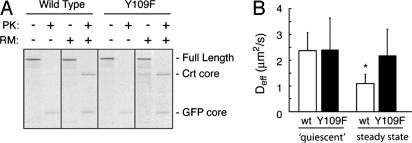
From these data, the precise reason for the changes in Crt-GFP diffusion in a substrate-specific manner could not be conclusively deduced. These changes could either be due to a direct interaction between Crt-GFP and substrates (potentially in combination with other chaperones, such as ERp57), or indirectly through other substrate-bound chaperones that interact with Crt-GFP. To distinguish between these possibilities, we exploited the fact that most (if not all) substrate interactions with Crt are through its lectin activity (23, 33, 35). Crt(Y108F)-GFP is significantly reduced in its interactions with substrates (Fig. 6) but not in its overall folding (24) (Fig. 4A). Under quiescent ER conditions, the Deff of Crt(Y108F)-GFP was identical to Crt-GFP (Fig. 4B). However, in the presence of substrate, Crt(Y108F)-GFP did not show the significant decrease in Deff observed for Crt-GFP (Fig. 4B). Thus, essentially all substrate-specific changes in Crt-GFP dynamics are due to lectin-mediated interactions with maturing polypeptides. Furthermore, the lack of change in Deff for Crt(Y108F)-GFP in the presence or absence of substrate reinforces our conclusion (based on the ER-GFP probe) that the general diffusional environment of the ER is largely impervious to substrate load.
Fig. 4.
Changes in Crt-GFP dynamics depend on its lectin activity. (A) Crt-GFP or Crt(Y109F)-GFP were translated in vitro in the absence or presence of rough microsomal membranes (RM). The samples were adjusted to 0.5% Triton X-100 in physiological salt buffer and digested with proteinase K (PK) as indicated. Correctly folded Crt and GFP contain core domains that resist digestion by PK under these conditions. This “Crt core” band is observed for the wild type and Y109F mutant only when synthesized in the presence of RM. The GFP domain is observed in all PK-digested lanes to the same extent and serves as a control. (B) Deff values of Crt-GFP (n = 14) and Crt(Y109F)-GFP (n = 26) under quiescent conditions (Pact-treated for 1 h) showed no difference. In actively metabolizing cells, Crt-GFP (n = 24) Deff significantly decreases (P < 0.005) (asterisk) from the quiescent state, whereas Crt(Y109F)-GFP (n = 14) is unchanged.
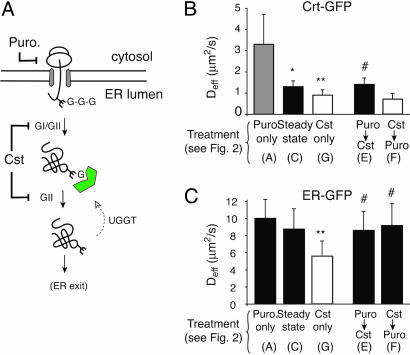
The remarkable ability of the ER to maintain an essentially unchanged environment (as judged by ER-GFP diffusion), despite marked changes to substrate flux, indicates a highly robust system. To investigate the limits of the chaperone systems in maintaining homeostasis of the ER environment, we acutely perturbed the lectin chaperones (Crt and its membrane-bound homologue calnexin by disrupting their well characterized substrate-binding cycles. When substrates are first N-glycosylated, their core glycan contains three terminal glucose residues, which are successively trimmed by glucosidases I and II, leaving a single glucose (36). The monoglucosylated glycan is the substrate for Crt and interacts with Crt until the terminal glucose is further trimmed by glucosidase II, and the substrate is no longer capable of binding Crt. Thus, inhibiting glucosidases with castanospermine (Cst) will prevent new glycoproteins from interacting with lectin chaperones (20) and maintain the association between the lectin chaperones and already monoglucosylated substrates (37) (Fig. 5A).
Fig. 5.
Consequences of acute inhibition of the lectin chaperones. (A) Diagram illustrating the effect of Cst on Crt–substrate interactions. Cst inhibits both glucosidase I and II (GI/GII), preventing glucose trimming of glycosylated substrates. Newly synthesized glycoproteins (with three glucoses) cannot bind to Crt (indicated in green), whereas preexisting monoglucosylated glycoproteins remain substrates for Crt binding. The site of Puro action is also indicated. (B and C) FRAP analyses of Crt-GFP and ER-GFP were used to determine the Deff under the conditions indicated below the graph. The panels of Fig. 2 that correspond to each treatment condition are indicated in parentheses below the graph. Symbols above the bars indicate statistical significance (P < 0.005) between the following comparisons: ∗, Puro only versus Steady state; ∗∗, Cst only versus Steady state; #, Cst only versus dual treatments.
Upon treatment of cells for between 1–2 h with Cst, we observed a significant ≈30% decrease in Deff (without any change in mobile fraction) for Crt-GFP relative to untreated metabolically active cells (Fig. 5B, Cst only compared with Steady state; see Table 2). This decrease could be anticipated, because Crt disengagement from substrate is reduced in the presence of Cst. Consistent with this interpretation, Cst did not cause a decrease in Crt diffusion if protein synthesis was inhibited with Puro before (and during) Cst treatment (Fig. 5B, Puro → Cst and Table 2). Remarkably, however, the diffusion of ER-GFP was noticeably reduced upon acute treatment with Cst (Fig. 5C, Cst only compared with Steady state; see Table 2). Because the mobile fraction of ER-GFP remained unchanged (see Supporting Materials and Methods), its decreased mobility was uniform and reflects a generalized change in the diffusional properties of the ER consistent with increased crowdedness. This change in ER environment depended absolutely on substrate synthesis, because no effect was observed if protein synthesis was blocked before and during Cst treatment (Fig. 5C, Puro → Cst and Table 2). Thus, acute inhibition of the lectin chaperone system leads to a marked change in the diffusional properties of the ER environment that far surpasses any changes that occur during simple changes in substrate flux.
Table 2.
Deff values of Crt-GFP and ER-GFP after treatment with Cst and Puro
| Construct | Condition | No. | Deff, μm2/s ± SD |
|---|---|---|---|
| Crt-GFP | (C) Steady state | 28 | 1.3 ± 0.3 |
| (H) Cst | 25 | 0.9 ± 0.2* | |
| (E) Puro → Cst | 24 | 1.4 ± 0.4† | |
| (F) Cst → Puro | 10 | 0.7 ± 0.2* | |
| ER-GFP | (C) Steady state | 20 | 8.7 ± 2.5 |
| (H) Cst | 20 | 5.6 ± 1.7* | |
| (E) Puro → Cst | 8 | 8.5 ± 2.3† | |
| (F) Cst → Puro | 11 | 9.1 ± 2.6† |
The panel of Fig. 2 that schematically depicts each condition is indicated in parentheses to the left. Statistical comparisons of Deff values were performed by using two-tailed Student t tests.
*Statistically significant change (P < 0.005) relative to the steady-state condition.
†Statistical significance (P < 0.005) relative to the Cst treatment.
The change in the ER environment could be due to a combination of two consequences of Cst treatment. One possibility is that Crt–substrate complexes that become trapped in the presence of Cst could be large and occlusive. Alternatively, newly synthesized substrates that are now prevented from binding Crt could form large and occlusive complexes with alternative chaperones. To distinguish between these possibilities, we inhibited new protein and Table 2 synthesis after treatment of cells with Cst for 1 h. Here, Crt–substrate complexes should still be formed and trapped, but the complexes with other chaperones should be minimized (see Fig. 2F). Remarkably, the ER environment returned to normal levels as judged by the Deff of ER-GFP (Fig. 5C, Cst → Puro and Table 2). That Crt–substrate complexes were still formed and maintained under these conditions was confirmed by the low Deff for Crt-GFP (Fig. 5B, Cst → Puro and Table 2). Thus, the ER environment is relatively unaffected by Crt–substrate complexes but becomes occlusive to ER-GFP only upon continued substrate generation under conditions where the substrates cannot engage the lectin chaperone system. Furthermore, the reversal of the effects of Cst on ER-GFP simply by inhibiting protein synthesis for 1 h suggests that the nonlectin chaperone systems are not irreversibly bound in occlusive complexes. Rather, they are readily resolved if provided an opportunity to do so under conditions of reduced load of additional substrate.
Conclusions
The results represent the direct measurements of the dynamics of a functional ER chaperone in vivo. These experiments lead to a view of Crt as a highly dynamic chaperone and the ER as a highly robust folding environment. Despite high concentrations of numerous maturation enzymes (9, 38), Crt remained highly mobile. Even when a population of nascent substrate is effectively immobilized (with Chx; see Fig. 2D), Crt-GFP shows an unchanged mobile fraction but a reduced Deff. Thus, interactions between Crt and its substrates are dynamic, consistent with the weak affinity of Crt (Kd = 2 μM) (25). Similar dynamic interactions are likely to occur with maturing polypeptides and Crt-GFP, because the introduction of this population further reduces Deff. The mobility of Crt and the transient nature of its interactions is consistent with the finding that chaperone-associated misfolded proteins are also highly mobile (18). In addition, ternary interactions among substrate, Crt, and other chaperones (39) with which it cooperates during protein maturation are also likely to be highly dynamic. Were these components to form stable, large, multiprotein complexes, their diffusion would be markedly slowed or immobilized (8, 18, 40). Thus, it is unlikely that substrate proteins encounter a matrix of chaperone complexes on which they fold, nor does such a matrix form around nascent substrates. Instead, Crt continues to constantly sample the entire ER lumen.
The observation that changes in substrate flux through the ER does not significantly alter the diffusional environment revealed a surprisingly robust maturation system. This suggests that newly synthesized polypeptides have essentially equal capacity for diffusive encounters with the maturation machinery. Thus, saturation of the maturation capacity simply by changes in expression levels over a wide range is unlikely to occur. Furthermore, even when ER homeostasis was perturbed during acute lectin chaperone inhibition, the remaining machinery could quickly reverse the effects, provided that its substrate burden is minimized, illustrating that the nonlectin machinery can handle, to a reasonable extent, glycoproteins that were destined for the Crt/calnexin (Cnx) system. Given that Crt/Cnx interactions are through substrate glycans (e.g., Figs. 4B and 6), Crt/Cnx may be unable to display the same compensatory behavior in the absence of other chaperones. This inability may explain why a decrease in BiP availability leads to rapid induction of the unfolded protein response (including translational attenuation) (41), whereas a prevention of access to the lectin chaperones does not induce translational inhibition or cell death associated with an unfolded protein response (42).
The view of a highly dynamic and robust folding environment for one chaperone of the ER provides an initial framework to evaluate the organizational properties of both other ER components and changes that occur under various conditions. The finding that some maturation enzymes (such as the OST) are relatively immobile (8), whereas others (such as Crt) are highly dynamic, indicates that different components are organized differently. The spatial organization and dynamic properties of any given maturation factor presumably reflect when and how it is used during protein biogenesis. This information on dynamics has significant implications for the strategy used by the ER to both maximize maturation efficiency and minimize the capacity for misfolding. It will be important to analyze, by fluorescence-based methods in live cells, other classes of maturation factors. Being able to detect and analyze subtle changes to ER homeostasis in live cells should help elucidate mechanisms that contribute to the development or exacerbation of the growing list of protein-folding diseases of the ER.
Materials and Methods
Crt-GFP (43) and mCFP-Sec61β (44) have been described. ER-GFP and ER-RFP contain fusions of the bovine prolactin signal sequence, mRFP or mGFP, and a KDEL ER retention sequence in the pCDNA3.1 vector. The Crt(Y108F)-GFP was created by site-directed mutagenesis by using Quickchange (Stratagene). Antibodies used were rabbit anti-Crt (Affinity BioReagents), monoclonal anti-GFP (Clontech), and horseradish peroxidase-conjugated anti rabbit and mouse IgG antibodies (Amersham Pharmacia Biosciences). Pact was a generous gift from E. Steinbrecher (Amersham Pharmacia, Peapack, NJ) and was used at 0.2 μM for at least 30 min. Cells were treated for 30 min with Puro (1 mM; Sigma) or Chx (0.5 mM; Calbiochem) and 1 h with 1 mM Cst (Sigma).
Wild-type Crt-expressing (K41) cells and Crt knockout cells (K42) (19, 45) were generously provided by Marek Michalak (University of Alberta, Edmonton, Canada). Culturing and creation of stable clones of crt−/− cells expressing Crt-GFP were described in ref. 43. Transient transfections, immunoblotting, pulse–chase analyses, the in vitro limited protease digestions, and imaging were performed as described in ref. 43. Image analysis was performed by using nih image 1.63 and lsm image examiner software. Composite figures were prepared by using photoshop 7.0 and illustrator cs software (both from Adobe). Fluorescence recovery curves were plotted by using kaleidagraph 3.5 (Synergy Software). We used a two-tailed Student t test (excel, Microsoft) to compare the different conditions. Additional details of the methodology are in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the R.S.H. and J.L.-S. laboratories for support and helpful discussions. This work was supported by the National Institutes of Health, National Institute of Child Health and Human Development Intramural Research Program. E.L.S. is an Ellison Medical Foundation New Scholar in Aging.
Abbreviations
- Chx
cycloheximide
- Crt
calreticulin
- Cst
castanospermine
- Deff
effective diffusion coefficient
- ER
endoplasmic reticulum
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- OST
oligosaccharyl transferase
- Pact
pactamycin
- Puro
puromycin.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hellman R., Vanhove M., Lejeune A., Stevens F. J., Hendershot L. M. J. Cell Biol. 1999;144:21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helenius A., Marquardt T., Braakman I. Trends Cell Biol. 1992;2:227–231. doi: 10.1016/0962-8924(92)90309-b. [DOI] [PubMed] [Google Scholar]
- 3.Osborne A. R., Rapoport T. A., van den Berg B. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 4.Schnell D. J., Hebert D. N. Cell. 2003;112:491–505. doi: 10.1016/s0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 5.Anken E., Braakman I., Craig E. Crit. Rev. Biochem. Mol. Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y., Hendershot L. M. J. Chem. Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Chavan M., Yan A., Lennarz W. J. J. Biol. Chem. 2005;280:22917–22924. doi: 10.1074/jbc.M502858200. [DOI] [PubMed] [Google Scholar]
- 8.Nikonov A. V., Snapp E., Lippincott-Schwartz J., Kreibich G. J. Cell Biol. 2002;158:497–506. doi: 10.1083/jcb.200201116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guth S., Volzing C., Muller A., Jung M., Zimmermann R. Eur. J. Biochem. 2004;271:3200–3207. doi: 10.1111/j.1432-1033.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- 10.Tatu U., Helenius A. J. Cell Biol. 1997;136:555–565. doi: 10.1083/jcb.136.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevet E., Jakob C. A., Thomas D. Y., Bergeron J. J. Semin. Cell Dev. Biol. 1999;10:473–480. doi: 10.1006/scdb.1999.0316. [DOI] [PubMed] [Google Scholar]
- 12.Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. Mol. Biol. Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleizen B., Braakman I. Curr. Opin. Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Lippincott-Schwartz J., Snapp E., Kenworthy A. Nat. Rev. Mol. Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 15.Wouters F. S., Verveer P. J., Bastiaens P. I. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 16.Dayel M. J., Hom E. F. Y., Verkman A. S. Biophys. J. 1999;76:2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ormo M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 18.Nehls S., Snapp E. L., Cole N. B., Zaal K. J., Kenworthy A. K., Roberts T. H., Ellenberg J., Presley J. F., Siggia E., Lippincott-Schwartz J. Nat. Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- 19.Mesaeli N., Nakamura K., Zvaritch E., Dickie P., Dziak E., Krause K. H., Opas M., MacLennan D. H., Michalak M. J. Cell Biol. 1999;144:857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond C., Braakman I., Helenius A. Proc. Natl. Acad. Sci. USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J. X., Braakman I., Matlack K. E., Helenius A. Mol. Biol. Cell. 1997;8:1943–1954. doi: 10.1091/mbc.8.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson J. R., Helenius A. J. Cell Sci. 1999;112:2775–2784. doi: 10.1242/jcs.112.16.2775. [DOI] [PubMed] [Google Scholar]
- 23.Peterson J. R., Ora A., Van P. N., Helenius A. Mol. Biol. Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor M., Ellgaard L., Gopalakrishnapai J., Schirra C., Gemma E., Oscarson S., Helenius A., Surolia A. Biochemistry. 2004;43:97–106. doi: 10.1021/bi0355286. [DOI] [PubMed] [Google Scholar]
- 25.Kapoor M., Srinivas H., Kandiah E., Gemma E., Ellgaard L., Oscarson S., Helenius A., Surolia A. J. Biol. Chem. 2003;278:6194–6200. doi: 10.1074/jbc.M209132200. [DOI] [PubMed] [Google Scholar]
- 26.Ellgaard L., Helenius A. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 27.Bouvier M., Stafford W. F. Biochemistry. 2000;39:14950–14959. doi: 10.1021/bi0019545. [DOI] [PubMed] [Google Scholar]
- 28.Ellgaard L., Riek R., Herrmann T., Guntert P., Braun D., Helenius A., Wuthrich K. Proc. Natl. Acad. Sci. USA. 2001;98:3133–3138. doi: 10.1073/pnas.051630098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. Mol. Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 30.Snapp E. L., Reinhart G. A., Bogert B. A., Lippincott-Schwartz J., Hegde R. S. J. Cell Biol. 2004;164:997–1007. doi: 10.1083/jcb.200312079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snapp E., Altan-Bonnet N., Lippincott-Schwartz J. In: Current Protocols in Cell Biology. Bonafacino J. S., Dasso M., Harford J., Lippincott-Schwartz J., Yamada K., editors. New York: Wiley; 2003. Unit 21.1. [Google Scholar]
- 32.Hink M. A., Griep R. A., Borst J. W., van Hoek A., Eppink M. H., Schots A., Visser A. J. J. Biol. Chem. 2000;275:17556–17560. doi: 10.1074/jbc.M001348200. [DOI] [PubMed] [Google Scholar]
- 33.Rizvi S. M., Mancino L., Thammavongsa V., Cantley R. L., Raghavan M. Mol. Cell. 2004;15:913–923. doi: 10.1016/j.molcel.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Kuznetsov G., Chen L. B., Nigam S. K. J. Biol. Chem. 1997;272:3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- 35.Danilczyk U. G., Williams D. B. J. Biol. Chem. 2001;276:25532–25540. doi: 10.1074/jbc.M100270200. [DOI] [PubMed] [Google Scholar]
- 36.Helenius A., Aebi M. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., White A. L. Biochemistry. 2000;39:8993–9000. doi: 10.1021/bi000027v. [DOI] [PubMed] [Google Scholar]
- 38.Gerner C., Vejda S., Gelbmann D., Bayer E., Gotzmann J., Schulte-Hermann R., Mikulits W. Mol. Cell. Proteomics. 2002;1:528–537. doi: 10.1074/mcp.m200026-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Zapun A., Darby N. J., Tessier D. C., Michalak M., Bergeron J. J., Thomas D. Y. J. Biol. Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- 40.Marguet D., Spiliotis E. T., Pentcheva T., Lebowitz M., Schneck J., Edidin M. Immunology. 1999;11:231–240. doi: 10.1016/s1074-7613(00)80098-9. [DOI] [PubMed] [Google Scholar]
- 41.Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehrman M. A. J. Biol. Chem. 2001;276:8623–8626. doi: 10.1074/jbc.R100002200. [DOI] [PubMed] [Google Scholar]
- 43.Shaffer K. L., Sharma A., Snapp E. L., Hegde R. S. Dev. Cell. 2005;9:545–554. doi: 10.1016/j.devcel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura K., Bossy-Wetzel E., Burns K., Fadel M. P., Lozyk M., Goping I. S., Opas M., Bleackley R. C., Green D. R., Michalak M. J. Cell Biol. 2000;150:731–740. doi: 10.1083/jcb.150.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.