Abstract
Sonic hedgehog (Shh) produced in the zone of polarizing activity is the major determinant of anteroposterior development of the amniote limb. The mature and active Shh protein is cholesterol-modified at its C terminus, and the hydrophobic nature of the modification requires the function of Dispatched (mDispA), a seven-pass transmembrane protein, for Shh release from its source. The current model suggests that the cholesterol moiety promotes the spread of Shh gradient in the limb bud. However, this model is inconsistent with findings in Drosophila and not in line with current thoughts on the role of the cholesterol moiety in Shh multimerization. Therefore, it remains unclear how the cholesterol moiety affects the postrelease extracellular behavior of Shh that relates to the shape of its activity gradient in responsive tissues. Here, we report functional analyses in mice showing that Shh lacking cholesterol modification (ShhN) has an increased propensity to spread long-distance, eliciting ectopic Shh pathway activation consistent with target gene expressions and modulating the level of Gli3 processing in the anterior limb mesoderm. These molecular alterations are reflected in the mispatterning of digits in ShhN mutants. Additionally, we provide direct evidence for the long-distance movement of ShhN across the anteroposterior axis of the limb bud. Our findings suggest that the cholesterol moiety regulates the range and shape of the Shh morphogen gradient by restricting rather than promoting the postrelease spread of Shh across the limb bud during early development.
Keywords: cholesterol moiety, limb patterning, lipid modification, mouse genetics
Secreted molecules encoded by the Hedgehog (Hh) gene family are key signals in regulating the growth and patterning of invertebrate and vertebrate embryos (1–3). One of the prominent features of Hedgehog ligands is the ability to act as a classical morphogen in developing tissues. In vertebrates, this feature is highlighted by the apparent requirement of Shh signaling in the specification of positional information along the anteroposterior axis of the limb bud (4, 5). The active Shh protein ShhNp (p stands for “processed”) is doubly lipid-modified, with a cholesterol moiety at its C terminus and a palmitate at its N terminus (6, 7). The cholesterol moiety of ShhNp is acquired as the Shh precursor undergoes intramolecular cleavage reaction (8), and palmitate addition is catalyzed by putative acyltransferase encoded by the Skinny hedgehog (Ski) gene (9, 10). The palmitoylation of Shh appears to be important to augment Shh activity, because mouse embryos exclusively expressing Shh lacking palmitoylation (ShhC25S) have significantly reduced patterning activity in the neural tube and the limb (11). Similarly, embryos lacking Ski function also show phenotypes resembling those of Hh loss-of-function in both Drosophila and mouse (9–11).
ShhNp is tightly associated with cell membrane in tissue culture cells (8, 12), and the hydrophobic nature of the cholesterol modification requires the function of mouse Dispatched (mDisp), a 12-pass transmembrane protein with a sterol-sensing domain, in order for Shh to be released from its site of synthesis (13–15). Work by others has indicated that Shh lacking cholesterol moiety (ShhN) displays surprisingly restricted distribution and activity in the limb bud and concluded that the cholesterol moiety is required for Shh to move away from cells close to the zone of polarizing activity (ZPA) for long-distance signaling (16, 17). However, these findings are at odds with studies performed in Drosophila imaginal discs (6, 18), where Hh lacking cholesterol has extended range of movement and signaling. Studies by others may have been hampered by the significantly lower Shh transcript level in the ZPA of ShhN mutants, compared with wild type, which is apparent even at an early limb bud stage (16). Therefore, it remains unclear how the cholesterol moiety affects the postrelease extracellular behavior of Shh and how this builds the range and shape of the Shh activity gradient during anteroposterior patterning of the limb. Here, we show that cholesterol modification of Shh is essential to the spatial regulation of Shh signaling gradient during limb development. In contrast to previous reports, we provide evidence that, in the absence of cholesterol moiety, Shh spreads far from its site of synthesis, eliciting ectopic pathway activation in the anterior limb margin. The long-range effects of ShhN are completely refractory to the absence of mDispA function. Our data also indicate that the long-range spreading of ShhN reduces local concentration that significantly affects Shh pathway activity in the posterior limb mesoderm. Our study reveals an essential role for cholesterol moiety in restricting rather than promoting the Shh activity gradient during limb development.
Results
Shh Lacking Cholesterol Moiety Has Extended Range of Signaling.
In an effort to address the function of the Shh cholesterol moiety in regulating Shh gradient, we generated a line of conditional mice expressing ShhN in a Cre recombinase-dependent manner. The design of these mice is aimed at circumventing embryonic lethality and RNA instability potentially associated with nonsense-mediated RNA decay caused by premature translational termination (19). The modified Shh locus (referred to as Shhfloxc) contains an upstream loxp sequence in-frame with the Shh processing domain, followed by a downstream loxp sequence soon after the termination codon (see Fig. 7, which is published as supporting information on the PNAS web site). Therefore, in the absence of Cre recombinase, the cholesterol-modified ShhN will contain additional loxp-derived residues at the C terminus. Because Shhfloxc/+ is fully viable with no discernable phenotypes, it is unlikely that the presence of these additional loxp-derived residues will alter ShhN behavior or exert dominant effect on wild-type Shh protein. Moreover, limb skeletal patterns of homozygous Shhfloxc embryos remain relatively unaffected (see Fig. 8, which is published as supporting information on the PNAS web site), although they die shortly after birth. The presence of an upstream loxp sequence between the signaling and processing domains appears to reduce the efficiency of Shh precursor processing, as noted, with an insertion of a gfp sequence at a similar location in Drosophila Hh (20).
To generate embryos expressing ShhN, we intercrossed Shhfloxc and Sox2-Cre mice that express Cre uniformly in the epiblast and all embryonic derivatives under the control of Sox2 enhancer (21). The Sox2-Cre line has been successfully used to globally delete gene functions, including Shh (21, 22). Complete excision of ShhC in early limb buds was confirmed by crossing to ZEG reporter mice as well as by PCR analysis (Fig. 7).
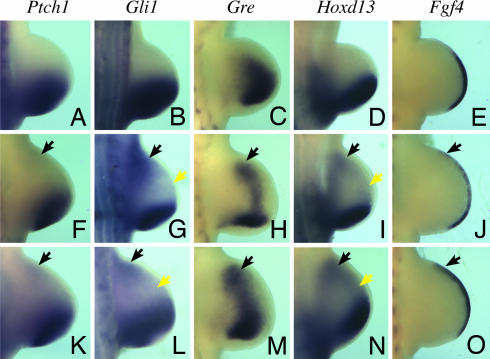
To determine whether ShhN is capable of long-range signaling, we examined patterns of Shh target gene expressions in ShhN limbs. The expressions of Ptch1 and Gli1 provide direct and sensitive readouts of Shh pathway activation (23, 24). In contrast to wild-type limbs, we observed ectopic expressions of Gli1 and weak but detectable Ptch1 in the anterior limb margin of both fore- and hindlimb buds expressing ShhN (Fig. 1, black arrows and Fig. 9, which is published as supporting information on the PNAS web site), in either ShhN−or ShhN+ background, regardless of the absence or presence of ShhNp. Furthermore, noticeable Gli1 expression is also activated in a diffuse pattern across the ShhN mutant limb mesoderm (Fig. 1G, yellow arrow). As an additional readout of Shh signaling, we examined patterns of other Shh-regulated genes that are normally expressed in posterior limb margins. Consistent with ectopic Shh pathway activation, the expressions of Gremlin (Gre), Hoxd13, and Fgf4 are also activated in anterior limb mesoderm and the apical ectodermal ridge (AER) at the distal tip of the limb bud (Fig. 1). Thus, these findings suggest that ShhN is capable of long-range signaling.
Fig. 1.
ShhN elicits ectopic Shh pathway activation and target gene expressions in the hindlimb bud. (A–O) Whole-mount in situ hybridization of E10.5 wild-type (A–E), ShhN+ (K–O), and ShhN− (F–J) hindlimb buds, examining the expression of Shh and its target genes Ptch1, Gli1, Gre, Hoxd13, and Fgf4, as indicated. Black arrows denote ectopic activation of Shh target genes in the anterior limb margins. Also note that the expressions of Gli1 and Hoxd13 (G and I) can be detected in a diffused pattern in the midregion of ShhN+ and ShhN− limb-bud mesoderm (yellow arrows).
The Establishment of Ectopic Shh-Expressing Cells in the Anterior ShhN Limb Margin.
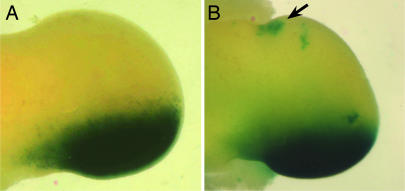
The relatively enhanced Shh pathway activation in the anteriormost margin (Fig. 1, black arrows) is likely attributed to subsequent maintenance of long-range ShhN signal through establishment of an ectopic Shh-expressing domain via an Shh-Fgfs positive-feedback loop (25, 26). Because RNA in situ hybridization is not sensitive enough to detect a small population of Shh-expressing cells, we used a Shh-Cre line that we had generated (Y. Litingtung and C.C., unpublished work), which expresses Cre under the control of endogenous Shh promoter and also abolishes Shh function. Accordingly, the phenotype of the homozygous Shh-Cre line is indistinguishable from Shh−/− embryos. A similar Shh-Cre line was used to follow patterns of Shh-expressing cells and their descendents in the limb by lacZ reporter expression by using R26R mice (27). Consistent with higher Shh pathway activity, we find that Shh expression has, indeed, been activated in the anterior margin of Shhfloxc/ShhCre limbs at embryonic day (E)11.5 (Fig. 2, arrow).
Fig. 2.
Ectopic Shh expression in the anterior margin of the limb bud at E11.5. The Shh-expressing cells and their descendants are marked by lacZ reporter gene in ShhCre;Rosa26R (A) and ShhCre/Shhfloxc;Rosa26R (B) embryos. Note the presence of ectopic Shh-expressing cells only in the anterior margins of ShhN mutant limb buds (arrow).
ShhN Spread Far from the ZPA and Affects Shh Pathway Activity in the Limb.
In addition to ectopic Shh pathway activation, we observe a clear reduction in the normal levels of Shh target gene expressions in the posterior limb mesoderm, closer to the site of ShhN synthesis (Fig. 1). The AER of ShhN− limbs also shows reduced and discontinuous Fgf4 expression (Fig. 1J), consistent with reduced Gre expression, which is required to stabilize the expression of Fgf4 by antagonizing Bmp signaling (28). In forelimb development, which precedes that of the himblimb, reduced expressions of Shh target genes in the posterior limb buds are more pronounced (Fig. 9), as expected from prolonged disruption of the Shh-Fgfs positive-feedback loop (25, 26). The reduction in target gene expressions is not due to low Shh transcript level, because significant Shh expression is detected in the ZPA (see Fig. 3). Thus, these reductions are likely attributed to lower Shh protein concentration in the ZPA as a result of long-range spreading of Shh.
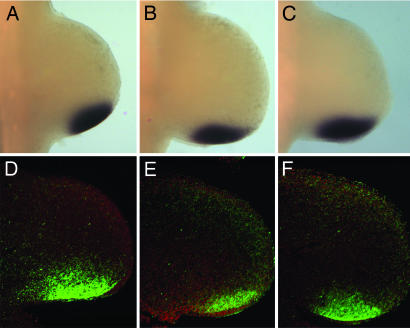
Fig. 3.
ShhN has increased propensity to spread across the anterior/posterior axis of the limb bud. Shh RNA (A–C) and protein (D–F, green) distributions in wild type (A and D), ShhN− (B and E), and ShhN+ (C and F) limb buds at E10.5. The limb buds in D–F were counterstained with Hoescht's dye (red).
To provide direct evidence for long-range ShhN movement, we examined Shh protein distribution in the limb bud. As reported, ShhNp protein is distributed in a graded fashion with highest level in the ZPA, followed by a gradual decline as the protein spreads anteriorly up to one-third of the limb bud (29) (Fig. 3D). By contrast, we find that in ShhN limbs, ShhN protein is detected across the anterior/posterior axis up to the anterior limb margin (Fig. 3 E and F), suggesting that ShhN has increased propensity to spread far from its site of synthesis. Indeed, we observe reduction of ShhN protein in the ZPA of ShhN− limbs, consistent with reduced Shh pathway activation in the posterior limb mesoderm. Interestingly, long-range ShhN protein is preferentially distributed in the distal limb mesoderm close to the ridge, suggesting that this region may be more permissive to ShhN spreading or that ShhN is rapidly cleared from the proximal mesoderm. Our findings provide strong evidence that ShhN has extended range of movement across the limb bud.
Reduction of Gli3R Formation in ShhN-Expressing Limb Buds.
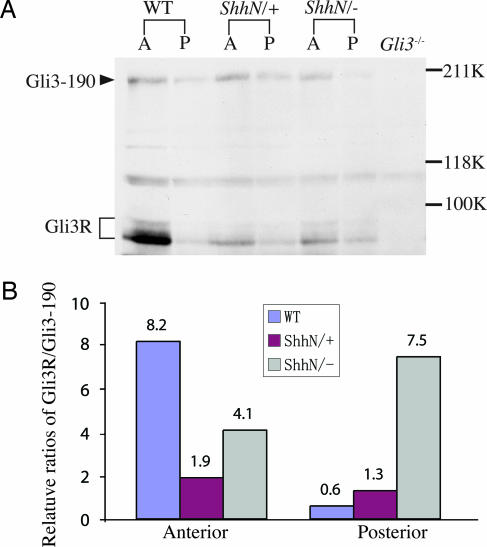
Control of Gli3 proteolytic processing by Shh signal is a critical determinant of anterior/posterior limb polarity (30–32). In early limb buds, cells away from the source of Shh signal respond by generating Gli3 truncated forms (Gli3R), which are known to repress transcription; in cells close to the source, generation of Gli3R is actively inhibited (33). To determine the effects of ShhN on Gli3 processing, we examined the relative distribution of Gli3 full-length (Gli3–190) and Gli3R in anterior and posterior halves of ShhN limbs by Western blot analysis using a Gli3-specific antibody (30). Consistent with long-range movement of ShhN, a high Gli3R/Gli3190 level, normally observed in anterior halves, is reduced by as much as 50% in ShhN− and 75% in ShhN+ mutants (Fig. 4). By contrast, posterior halves of ShhN− limbs show a clear reduction of Gli3190 concomitant with a moderate increase in Gli3R level, in accord with the reduced ShhN level and target gene expressions near the ZPA.
Fig. 4.
Long-range ShhN signal regulates Gli3 processing in the limb. (A) Protein extracts from E10.5 wild-type, ShhN+, and ShhN− limb buds of anterior (A) and posterior (P) halves were immunoblotted and incubated with a Gli3-specific antibody recognizing full-length (Gli3–190) and repressor forms (Gli3R) of Gli3. (B) Histograms showing relative Gli3R/Gli3–190 ratio in anterior and posterior halves of wild-type and ShhN limb buds. Levels of Gli3–190 and Gli3R were normalized to an internal nonspecific control band (asterisk). Note significant reduction of Gli3R in the anterior halves of ShhN limbs compared with wild type.
Mispatterning of Digits in ShhN Embryos.
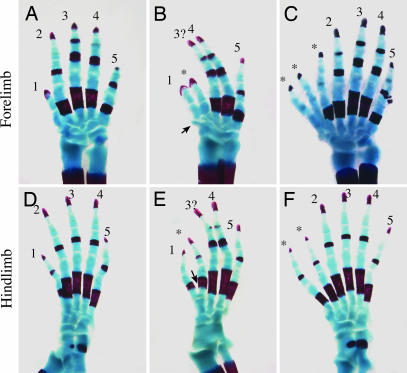
We next examined the effects of ShhN on the specification of anteroposterior digit identities. Despite significant reduction of Gli3R/Gli3190 levels in anterior halves of the ShhN− limb mesoderm, the limbs develop only five digits with clearly identifiable digits 4 and 5 (Fig. 5B and E). The identity of the central digit cannot be unambiguously assigned because of defective development of carpal and tarsal bones that are normally associated with the central digit; however, digit length and condensation pattern are suggestive of digit 3 (see Figure 10, which is published as supporting information on the PNAS web site). In the forelimb, digit 2 is replaced with a biphalangeal digit characteristic of digit 1. In the hindlimb, the identity of the digit immediately anterior to the central digit is suggestive of digit 2, based on the ossification pattern of its tarsal bone (Fig. 5E, arrowhead), although it lacks the characteristic triphalanges. The lack of preaxial polydactyly and defective digit 2 in ShhN− limbs are likely attributed to the reduction of Shh protein near the ZPA, which, in turn, destabilizes the posterior AER. Because palmitoylation is required for Shh activity, we cannot exclude the possibility that ShhN is less efficiently palmitoylated and, thus, also contributes to the defective AER or that additional C-terminal residues could render ShhN less potent. However, it should be noted that fusion of a foreign protein to the C terminus of ShhN did not appear to diminish its biological activities (34, 35).
Fig. 5.
The expression of ShhN causes abnormal development of digits. Skeletal analysis of E18.5 wild-type (A and D), ShhN− (B and E), and ShhN+ (C and F) digits. (B and E) ShhN− limbs develop five digits with defective digit 2. A small cartilaginous outgrowth adjacent to the presumptive forelimb digit 1 is evident (B, arrow). (C and F) ShhN+ limbs develop six to seven digits with complete formation of digits 2–5. Note that digit 1 in the hindlimb is replaced by digits with more posterior characteristics (F, asterisks).
The long-range effect of ShhN on limb patterning becomes more evident when the AER is stabilized in ShhN+ limbs, which develop six to seven digits per limb with complete formation of digits 2–5 (Fig. 5 C and F). The identity of ectopic digits can be difficult to assign; however, the presence of triphalanges and associated metatarsal elements in the anterior hindlimb digits indicates that the ectopic digits possess identities that are more posterior than digit 1, the anteriormost digit.
The Long-Distance Effect of ShhN Is Independent of ShhNp Activity and mDispA Function.
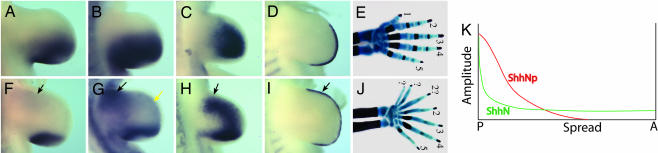
Although preaxial polydactyly observed in ShhN+ limbs is consistent with an extended range of ShhN signal, we cannot rule out the possibility that the formation of ectopic digits is due to long-range ShhNp activity. For example, ShhNp may compete with ShhN for long-range movement, as previously proposed to explain the dominant phenotype of ShhN+ limbs (16). To resolve this issue, we took advantage of mDispA−/− mutants that are deficient in releasing ShhNp to target fields. Therefore, in ShhN+;mDispA−/− mutant limbs, long-range patterning effects of Shh will be primarily attributed to ShhN activity. Although previous studies showed that mDispA is dispensable for ShhN activity in the spinal cord, addressing its role in the release of ShhN in the limb has been hindered by the inability of ShhN to signal long-range from the ZPA in the presence or absence of mDispA (17). Therefore, to shed light on this intriguing observation, we first examined the effects of ShhN on patterns of Shh target gene expressions in mDispA−/− mutant limbs. As shown in Fig. 6, the expression patterns of Shh target genes in ShhN+;mDispA−/− limbs resemble those of ShhN−. Ptch1, Gli1, and Gre all show ectopic expressions in the anterior limb mesoderm, whereas their posterior expression domains are reduced (Fig. 6; and see Fig. 11, which is published as supporting information on the PNAS web site). Unlike in ShhN− limbs, Fgf4 shows sustained expression in the AER (Fig. 6I), even though ShhNp is restricted to the ZPA. Thus, the previously reported dominant effect of ShhN+ is likely attributed to the long-range signaling effect of ShhN. In sum, these results demonstrate that long-range signaling by ShhN can occur without mDispA function and further establish that mDispA is dedicated to the regulated release of cholesterol-modified Shh during normal limb-bud development.
Fig. 6.
Long-range signaling capacity of ShhN is independent of mDispA function. Whole-mount in situ hybridization of E10.5 wild-type (A–D) and ShhN+;mDispA−/− (F–J) forelimb buds, showing the expression of Ptch1 (A and F), Gli1 (B and G), Gre (C and H), and Fgf4 (D and I). Note ectopic expressions of Shh target genes in the anterior margins of ShhN+;mDispA−/− limb buds (arrows). (E and J) Skeletal analysis of E18.5 wild-type (E) and ShhN+;mDispA−/− (J) forelimb digits. Note that digit numbers and patterns resemble those of ShhN+. (K) A model depicting ShhNp and ShhN gradients in early limb buds.
To determine whether ShhN is sufficient to elicit ectopic digits in the absence of long-range ShhNp activity, we examined skeletal patterns of ShhN+;mDispA−/− embryos. mDispA−/− single mutants normally die at or before E9.5, with phenotypes resembling Smo−/− embryos (13–15). In contrast ShhN+;mDispA−/− embryos survive to at least E18.5 and develop six to seven digits per limb (Fig. 6 E and J), similar to ShhN+ limbs. All ShhN+;mDispA−/− digits show segmentation and calcification defects characteristics of Ihh mutants (36), implicating long-range function of Ihh in skeletal development. Collectively, our results indicate that ectopic digits in ShhN+ limbs can be attributed primarily to long-range signaling activity of ShhN.
Discussion
We have shown that cholesterol modification of Shh is essential to the spatial regulation of Shh gradient and signaling in the limb bud. The cholesterol moiety is thought to be required to promote the spread of Shh in the limb bud because of the inability of ShhN to signal beyond a few cells away from the ZPA (16, 17). In contrast, our results clearly indicate that the cholesterol moiety is necessary to limit rather than promote the spread of Shh across the anteroposterior axis of the limb bud. This conclusion is based on several observations in ShhN-expressing limbs: (i) Shh target genes are ectopically expressed in the anterior margin of the limb bud; (ii) Gli3 repressor formation is significantly attenuated in anterior halves of limb buds; (iii) Shh expression is ectopically activated in the anterior limb margin via the Fgf-Shh positive-feedback loop; (iv) ShhN+ limbs display profound preaxial polydactyly; and (v) ShhN protein is detected across the anterior/posterior axis up to the anterior limb margin. Therefore, the cholesterol moiety is critical in generating the defined Shh spatial gradient, target gene expression domains, and Gli3R levels for the correct specification of digit numbers and identities. The observation that ShhN protein does not accumulate at high level near the ZPA suggests that extended anterior spreading of ShhN also significantly alters its local concentration and, thus, the shape of the gradient (Fig. 6K).
The major difference between our findings and those reported by Lewis et al. (16) is likely attributed to the level of ShhN expression in the ZPA. Previously, the effect on limb patterning was studied in embryos expressing ShhN from a gene-targeted allele that harbored a stop codon at the cleavage site; heterozygous ShhN embryos died soon after birth, and no stable line could be generated (16). Eukaryotes ranging from yeast to human use a surveillance mechanism, known as nonsense-mediated mRNA decay, to detect and degrade mRNAs with premature translation termination codons (19). Hence, previous studies may have been hampered by the significantly lower Shh transcript level in the ZPA of ShhN− mutants compared with wild type, that is apparent even in early limb buds (16). In contrast, we generated the ShhN allele by conditionally deleting the C-terminal processing domain of Shh protein that is necessary for cholesterol modification of ShhN. This strategy not only permits us to generate a stable ShhN conditional allele but also circumvents potential RNA instability associated with nonsense-mediated RNA decay. Indeed, we observed more robust ShhN transcript levels in the ZPA of early ShhN− mutant limbs (Fig. 3). The difference in the level of ShhN may also explain the observation that limb phenotypes of our ShhN− and ShhN+ mutants are nearly identical to those reported by Lewis et al. when ShhN alleles are expressed in a reduced Ptch1 background.
How does the cholesterol moiety restrict Shh spreading upon its release by mDispA? Previous tissue culture studies identified an active soluble mutimeric form of ShhNp, which was proposed to mediate intercellular movement of Shh in embryos (37). The structure of ShhNp multimer was thought to be analogous to micelles, with hydrophobic lipid moieties buried internally. Accordingly, assembly of the Shh multimer depends critically on the cholesterol moiety (11, 37). Although the biological relevance of the multimer in vivo remains to be determined, we speculate that multimerization of ShhNp not only renders ShhNp soluble but also functions to increase local Shh concentration and to limit ligand availability for long-range movement. This notion is consistent with our observation that ShhN displays increased capacity to spread across the anteroposterior axis of the limb bud at the expense of reduced local concentration, because the assembly of the Shh mutimeric complex would be inhibited in the absence of the cholesterol modification.
Other means by which the cholesterol moiety could restrict Shh spreading are through its interaction with receptors and components of extracellular matrix. It was first demonstrated in Drosophila that Ptc receptor not only functions to inhibit Hh pathway activation but also to sequester Hh movement (38). The latter property of Ptc appears to be associated with internalization and degradation of Hh upon its binding to Ptc (20). The observations that ShhN and ShhNp have comparable binding affinities to Ptch (7) and that ShhN is readily endocytosed upon Ptch binding (39) in vitro suggest that less efficient internalization by responsive cells is unlikely to be the primary mechanism for the long-range spreading of ShhN. In support of this argument, we observed enhanced ShhN signaling capacity by reducing the dosage of Ptch alleles as in ShhN−;Ptch+/− limb buds (data not shown), presumably because of attenuation of Ptch-mediated sequestration.
Recent findings in Drosophila have implicated the involvement of large macromolecular complexes known as lipoproteins in the intercellular transport of Hh (40). The central core of the lipoprotein consists of cholesterol and apoprotein molecules encased by an outer shell of phospholipids. Although the involvement of similar lipoproteins in transporting Shh signaling remains to be investigated, the low-density lipoprotein receptor 2 (Lrp2, megalin) has been shown to function as an endocytic receptor for Shh in tissue culture cells (41). However, Lrp2 has also been shown to bind other molecules including Bmp; thus, its function is not limited to the Shh signaling pathway (42). Intriguingly, the Lrp2 loss-of-function mutant does not display a global alteration of Shh signaling but shows defects only in ventral forebrain development (42). This finding suggests that either lipoproteins have limited function in Shh signaling, or other related receptors may compensate for Lrp2 function analogous to Lrp5 and Lrp6 in the Wnt signaling pathway (43). In any case, our ShhN mice will serve as an important reagent for determining the role of cholesterol moiety in lipoprotein-mediated Shh transport and endocytosis during embryonic development.
Materials and Methods
Generation of Shhflox Mutant Mice.
To generate conditional Shhflox mice, loxp sites were inserted flanking the Shh processing domain, which is situated in exon 3. To facilitate the insertion of the 5′ loxp sequence, we made a C750A nucleotide change at a location two amino acids before the intramolecular cleavage site of the Shh precursor by site-directed mutagenesis. This change resulted in a silent amino acid substitution and the generation of a BspE1 site. The BspEI/BamHI fragment of exon 3 was subsequently replaced in-frame with a synthetic BspEI/BamHI fragment containing a loxp sequence in addition to all of the original sequences. This replacement generated 13 additional amino acids between the N-terminal Shh signaling domain and the Shh processing domain. A PacI fragment, containing the second loxp site, immediately followed by a Myc epitope tag with stop codon and a frt-PGKneo-frt cassette, were then inserted 10 nucleotides downstream of the Shh stop codon in exon 3. A Myc epitope tag was added in-frame with the second loxp sequence to facilitate detection of ShhN after Cre-mediated recombination. However, we failed to detect Myc expression with a single Myc epitope tag, even though ShhN is expressed (see Fig. 3). To facilitate homologous recombination in ES cells, a diphtheria toxin gene under the control of PGK promoter (PGK-dt) was inserted upstream of the 4.7-kb 5′ homologous arm. The 3′ homologous arm is 4.5 kb in length. The resulting targeting construct was electroporated into RI ES cells, and 400 neo-resistant clones were selected. Southern blot analysis using a 1-kb XbaI/XhoI probe situated outside of the targeting vector indicated that ≈5% of the clones underwent homologous recombination events with correct gene targeting (Fig. 7). Two of these clones were microinjected into blastocysts to generate chimeras. Two independent Shhfloxneo lines were subsequently established from chimeras.
To remove the neo cassette, Shhfloxneo mice were bred to a flpe mouse (44), which carries an enhanced flp recombinase under the control of an actin promoter. The absence of neo cassette was confirmed by Southern blotting and PCR (data not shown). The resulting mice, referred to as Shhflox, were maintained as heterozygotes, because homozygotes die soon after birth. To generate embryos exclusively expressing ShhN, Shhfloxc/+ mice were bred to Shh+/−;Sox2Cre mice (21, 45). Complete excision of the Shh processing domain was confirmed by three primers (P1–P3) located within or outside of the processing domain (Fig. 7). The sequence of these primers is: P1, 5′-GTGTACCTGTCTCCTTTGGCACTC-3′; P2, 5′-TCTAGAGCGGCCATTCTCACTATT; P3, TACCCGCTTCCATTGCTCAG-3′.
Staining and RNA in Situ Hybridization.
Cartilage and bones were stained with Alcian blue and Alizarin red (46). Whole-mount RNA in situ hybridizations were performed as described in ref. 30. For Shh in situ hybridization, a probe that encompasses exons 1 and 2 of Shh cDNA was used; thus, we cannot rule out the possibility that null allele RNA may contribute to the in situ signal in ShhN− limb buds.
Immunohistochemistry and Western Analysis.
Immunohistochemistry on limb sections was performed as described in ref. 29, with the following modifications: dissected E10.5 embryos were fixed at 4°C for 8 h in EFA solution, which consists of 6 vol of 100% ethanol, 3 vol of 37% formaldehyde and 1 vol of 100% acetic acid. A series of 8-μm paraffin sections from limbs of different genotypes (n = 3 for each genotype) were collected onto slides, which were then incubated in PBS containing 10% goat serum, 0.2% BSA, and 0.1% Triton X-100 for 1 h at room temperature before an overnight incubation with anti-Shh antibody at 1:1,500 dilution (H-160; Santa Cruz Biotechnology). Detection was performed by using biotinylated goat anti-rabbit antibody (Vector Laboratories) and a Tyramide Signal Amplification kit (PerkinElmer) according to manufacturers protocols.
Protein lysate samples, 150 μg each, collected from anterior and posterior limb-bud halves, were resolved on 7.5% SDS-polyacrylamide gels. Gli3190 and Gli3R species were detected by using a Gli3 N-terminal antibody as described in ref. 30.
Supplementary Material
Acknowledgments
We thank P. A. Beachy (The Johns Hopkins University School of Medicine, Baltimore) for providing the mDispA mice and D. Threadgill (University of North Carolina, Chapel Hill, NC) for the frt-neo-frt cassette; J. F. Fallon and G. Gu for critical reading of the manuscript; and P. A. Beachy and C. V. E. Wright for helpful discussion. Support from the Transgenic Mouse/ES Cell Shared Resource, Cell Imaging Core and Sequencing Core at Vanderbilt University is acknowledged. This work was funded by grants from the National Institutes of Health and the March of Dimes Foundation.
Abbreviations
- AER
apical ectodermal ridge
- E(n)
embryonic day n
- ZPA
zone of polarizing activity.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ingham P. W., McMahon A. P. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Lum L., Beachy P. A. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 3.Hooper J. E., Scott M. P. Nat. Rev. Mol. Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 4.Johnson R. L., Tabin C. J. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- 5.Tickle C. Dev. Cell. 2003;4:449–458. doi: 10.1016/s1534-5807(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 6.Porter J. A., Ekker S. C., Park W. J., von Kessler D. P., Young K. E., Chen C. H., Ma Y., Woods A. S., Cotter R. J., Koonin E. V., Beachy P. A. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 7.Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K., et al. J. Biol. Chem. 1998;273:14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- 8.Porter J. A., Young K. E., Beachy P. A. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 9.Chamoun Z., Mann R. K., Nellen D., von Kessler D. P., Bellotto M., Beachy P. A., Basler K. Science. 2001;293:2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 10.Lee J. D., Treisman J. E. Curr. Biol. 2001;11:1147–1152. doi: 10.1016/s0960-9822(01)00323-2. [DOI] [PubMed] [Google Scholar]
- 11.Chen M. H., Li Y. J., Kawakami T., Xu S. M., Chuang P. T. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bumcrot D. A., Takada R., McMahon A. P. Mol. Cell. Biol. 1995;15:2294–2303. doi: 10.1128/mcb.15.4.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami T., Kawcak T., Li Y. J., Zhang W., Hu Y., Chuang P. T. Development (Cambridge, U.K.) 2002;129:5753–5765. doi: 10.1242/dev.00178. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y., Erkner A., Gong R., Yao S., Taipale J., Basler K., Beachy P. A. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 15.Caspary T., Garcia-Garcia M. J., Huangfu D., Eggenschwiler J. T., Wyler M. R., Rakeman A. S., Alcorn H. L., Anderson K. V. Curr. Biol. 2002;12:1628–1632. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- 16.Lewis P. M., Dunn M. P., McMahon J. A., Logan M., Martin J. F., St. Jacques B., McMahon A. P. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- 17.Tian H., Jeong J., Harfe B. D., Tabin C. J., McMahon A. P. Development (Cambridge, U.K.) 2005;132:133–142. doi: 10.1242/dev.01563. [DOI] [PubMed] [Google Scholar]
- 18.Burke R., Nellen D., Bellotto M., Hafen E., Senti K. A., Dickson B. J., Basler K. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfuss G., Kim V. N., Kataoka N. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 20.Torroja C., Gorfinkiel N., Guerrero I. Development (Cambridge, U.K.) 2004;131:2395–2408. doi: 10.1242/dev.01102. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi S., Lewis P., Pevny L., McMahon A. P. Mech. Dev. 2002;119:S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 22.Lu C. C., Robertson E. J. Dev. Biol. 2004;273:149–159. doi: 10.1016/j.ydbio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich L. V., Johnson R. L., Milenkovic L., McMahon J. A., Scott M. P. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 24.Ahn S., Joyner A. L. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Niswander L., Jeffrey S., Martin G. R., Tickle C. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- 26.Martin G. R. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 27.Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P., Tabin C. J. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Zuniga A., Haramis A. P., McMahon A. P., Zeller R. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 29.Gritli-Linde A., Lewis P., McMahon A. P., Linde A. Dev. Biol. 2001;236:364–386. doi: 10.1006/dbio.2001.0336. [DOI] [PubMed] [Google Scholar]
- 30.Litingtung Y., Dahn R. D., Li Y., Fallon J. F., Chiang C. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 31.Huangfu D., Anderson K. V. Proc. Natl. Acad. Sci. USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu A., Wang B., Niswander L. A. Development (Cambridge, U.K.) 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 33.Wang B., Fallon J. F., Beachy P. A. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y., Drossopoulou G., Chuang P. T., Duprez D., Marti E., Bumcrot D., Vargesson N., Clarke J., Niswander L., McMahon A., Tickle C. Development (Cambridge, U.K.) 1997;124:4393–4404. doi: 10.1242/dev.124.21.4393. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro R. I., Wen D., Levesque M., Hronowski X., Gill A., Garber E. A., Galdes A., Strauch K. L., Taylor F. R. Protein Expr. Purif. 2003;29:272–283. doi: 10.1016/s1046-5928(03)00062-7. [DOI] [PubMed] [Google Scholar]
- 36.St. Jacques B., Hammerschmidt M., McMahon A. P. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng X., Goetz J. A., Suber L. M., Scott W. J., Jr., Schreiner C. M., Robbins D. J. Nature. 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Struhl G. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 39.Incardona J. P., Lee J. H., Robertson C. P., Enga K., Kapur R. P., Roelink H. Proc. Natl. Acad. Sci. USA. 2000;97:12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panakova D., Sprong H., Marois E., Thiele C., Eaton S. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy R. A., Barth J. L., Chintalapudi M. R., Knaak C., Argraves W. S. J. Biol. Chem. 2002;277:25660–25667. doi: 10.1074/jbc.M201933200. [DOI] [PubMed] [Google Scholar]
- 42.Spoelgen R., Hammes A., Anzenberger U., Zechner D., Andersen O. M., Jerchow B., Willnow T. E. Development (Cambridge, U.K.) 2005;132:405–414. doi: 10.1242/dev.01580. [DOI] [PubMed] [Google Scholar]
- 43.Holmen S. L., Giambernardi T. A., Zylstra C. R., Buckner-Berghuis B. D., Resau J. H., Hess J. F., Glatt V., Bouxsein M. L., Ai M., Warman M. L., Williams B. O. J. Bone Miner. Res. 2004;19:2033–2040. doi: 10.1359/JBMR.040907. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 45.Chiang C., Litingtung Y., Lee E., Young K. E., Corden J. L., Westphal H., Beachy P. A. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 46.Chiang C., Litingtung Y., Harris M. P., Simandl B. K., Li Y., Beachy P. A., Fallon J. F. Dev. Biol. 2001;236:421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.