Abstract
The Cuatro Cienegas basin in the Chihuahuan desert is a system of springs, streams, and pools. These ecosystems support >70 endemic species and abundant living stromatolites and other microbial communities, representing a desert oasis of high biodiversity. Here, we combine data from molecular microbiology and geology to document the microbial biodiversity of this unique environment. Ten water samples from locations within the Cuatro Cienegas basin and two neighboring valleys as well as three samples of wet sediments were analyzed. The phylogeny of prokaryotic populations in the samples was determined by characterizing cultured organisms and by PCR amplification and sequencing of 16S rRNA genes from total community DNA. The composition of microbial communities was also assessed by determining profiles of terminal restriction site polymorphisms of 16S rRNA genes in total community DNA. There were 250 different phylotypes among the 350 cultivated strains. Ninety-eight partial 16S rRNA gene sequences were obtained and classified. The clones represented 38 unique phylotypes from ten major lineages of Bacteria and one of Archaea. Unexpectedly, 50% of the phylotypes were most closely related to marine taxa, even though these environments have not been in contact with the ocean for tens of millions of years. Furthermore, terminal restriction site polymorphism profiles and geological data suggest that the aquatic ecosystems of Cuatro Cienegas are hydrologically interconnected with adjacent valleys recently targeted for agricultural intensification. The findings underscore the conservation value of desert aquatic ecosystems and the urgent need for study and preservation of freshwater microbial communities.
Keywords: Cuatro Cienegas, terminal restriction site polymorphism, 16S clone library, water conservation, microbial ecology
Conservation efforts often focus on landscapes of scenic value or habitats with endangered or charismatic animals and plants. However, ecosystems also harbor a myriad of microorganisms that not only play a critical role in ecosystem functioning but also contain a remarkable record of their evolutionary history within their genomes. Freshwater aquatic ecosystems face increasing anthropogenic pressures worldwide (1–4), especially in arid regions (5–7), where we risk losing unique aquatic habitats without even knowing the nature and extent of their biodiversity. Here we report findings regarding microbial biodiversity in an endangered desert aquatic ecosystem in Mexico [the Cuatro Cienegas basin (CCB)] indicating that the composition of modern microbial assemblages may reflect their distant geological past and that there is significant subsurface interconnection of the ecosystems in adjacent sedimentary basins. Understanding the spatial and evolutionary relationships of the microbiota of the CCB is regarded as a critical step toward implementing an effective conservation strategy to protect the ecosystems found there.
The CCB is in central Mexico, in the state of Coahuila and is a valley measuring ≈30 km by 40 km located at ≈740 m above sea level and surrounded by high mountains (>3,000 m). The CCB is an enclosed evaporitic basin that receives ≈150 mm of annual precipitation. Despite the arid climate, the CCB harbors an extensive system of springs, streams, and pools of significant scientific interest (8). Documented biodiversity includes >70 endemic species of aquatic vertebrates, distributed among a wide variety of aquatic and terrestrial ecosystems. Other remarkable features of the aquatic ecosystems include the living stromatolites and other microbial communities that form the basis of complex food webs (8–11). From this perspective, CCB is widely regarded as a biodiversity oasis within the Chihuahuan desert. Although there is ample evidence that prokaryotes form the basis of food webs in this unique setting, we still know very little about the microbial diversity of these ecosystems. Given the high levels of endemism and biodiversity of higher organisms at the site, an 85,000-ha area is currently designated as a federal “Area for the Protection of Flora and Fauna” (12). Such a designation conceptually accommodates conservation of natural systems alongside sustainable development activities. The World Wildlife Federation, Mexico’s National Commission on Biodiversity (CONABIO), and nongovernment organizations, such as PRONATURA and the Nature Conservancy, have all classified the Cuatro Cienegas valley as globally outstanding because of its high species endemism and recent history of evolutionary radiations.
The high mountains surrounding the CCB expose upper Jurassic to lower Cretaceous limestones, sulfate-rich evaporites, sandstones, and conglomerates of the San Marcos and Cupido formations (8, 13) (Fig. 3, which is published as supporting information on the PNAS web site). Diverse surface habitats, including marshes, ponds, springheads, spring-fed streams, and playa lakes are interconnected with subsurface caverns, sinkholes, and other limestone and evaporite karst features (10). Between locations, environmental conditions can differ dramatically in water chemistry, flow rate, and size/volume of spring discharge (10, 14). Radiocarbon dating of sediment cores taken from pools indicates that some have existed for thousands of years, perhaps as long as 31,000 (15). Older travertine hot-spring deposits and lower-temperature tufa mounds are found in association with some active springs but also occur in the older, dry portions of the basin floor, suggesting the long-term persistence of aquatic habitats in the basin (10). Although preliminary models have been proposed (16), the hydrology of the region is still poorly understood, and, before now, its subsurface microbiota has not been characterized, despite the fact that current evidence argues for a unique evolutionary history.
Results
From the clone library, we obtained a total of 98 16S rRNA partial gene sequences (see Table 1, which is published as supporting information on the PNAS web site, for GenBank accession numbers). These sequences represented 38 unique phylotypes from ten major lineages of Bacteria and one of Archaea (Fig. 1). A total of 40% of the clones were γ-Proteobacteria. Among the 350 cultivated strains, there were 250 distinct and diverse phylotypes with the most common ones being related to Bacillus, Pseudomonas, Vibrio, Rhizobiace, Planctomyces, γ-Proteobacteria, and Gram-positive bacteria. The differences between these two results are due to the enormous bias that a limited number of cultivation media have over the microbial diversity of a site.
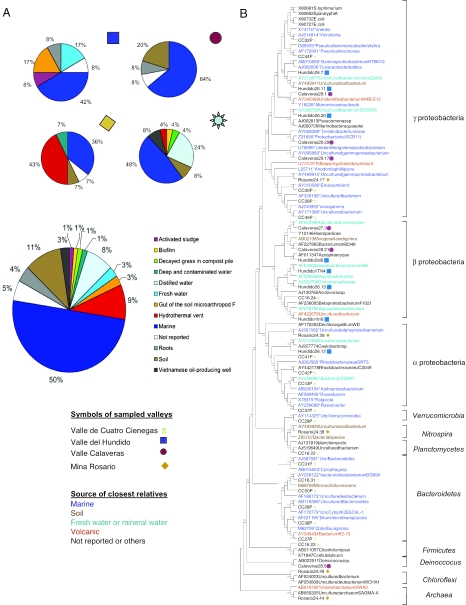
Fig. 1.
Molecular data from rRNA gene sequences in samples from the region of Cuatro Cienegas. (A) Proportion of habitat affiliations of the sequences based on comparison of our 16S rRNA clone library sequences with the Ribosomal Database Project. The large pie diagram illustrates the habitat affiliations for all of the samples, and the smaller diagrams indicate the distributions for each of the sampled valleys. The meaning of each color in the diagrams is shown to the right of the total sample. (B) The phylogenetic tree for microorganisms of the CCB was obtained as a maximum-parsimony tree with 1,000 bootstraps of the 690-bp common fragment of the 16S rRNA gene sequences. The bootstrap value is shown in the branches; none of the bootstrap values is <86%. The symbols show the sampling site for each sequence, and the color of each sequence shows its habitat affiliation (dark blue for marine, light blue for fresh water, brown for soils and sediments, and red for hydrothermal- and magma-associated taxa).
Published data on nine libraries prepared from aquatic or terrestrial environmental samples were used to evaluate whether the known habitats of “nearest neighbors” could be used to predict the habitat of a new phylotype. The habitat affiliations of organisms and clones that were closely related to those from the CCB were determined based on annotated information found in DNA sequence databases (Tables 2 and 3, which are published as supporting information on the PNAS web site). We found there was good accordance between the actual habitats from which the clones were derived and those that were predicted based on the source of the reference sequence (P < 0.01, Table 2). Using this approach, we found that nearly 50% of phylotypes from the CCB were closely related (90–99% sequence similarity) to organisms or cloned 16S rRNA gene sequences from marine environments (Fig. 1 A and B). Soil bacteria constituted the next largest group, followed by thermophilic organisms. At individual sites, these proportions varied. For example, at the Rosario mine, thermophilic bacteria predominated, whereas marine bacteria predominated at the remainder of the sampling locations. This finding was consistent with the identities of cultivated bacteria, which included strains 95–98% similar to marine organisms, such as Bacillus aquamaris, Halomonas elongata, Chromohalobacter canadensis, Exiguobacterium spp., Marinococcus halophilusi, and marine Rhizobiacea. Notably, B. aquamaris was very abundant in habitats at the CCB. Based on these results, we conclude that aquatic ecosystems at the CCB harbor a mixed microbial community, where part of the microbiota consists of organisms that are typically found in soil and freshwater environments but where a surprisingly large number of others are more commonly found in marine environments, such as the cold northern Pacific, the Arctic, Baltic sea, and hydrothermal vents (Table 2).
Cluster analyses of terminal restriction site polymorphism (T-RFLP) data showed there was a high degree of similarity between samples from adjacent valleys (Calaveras and Hundido) and two spring sites in the CCB (Escobedo and Churince), confirming the presence of very closely related lineages of Archaea and Bacteria in the Hundido, Calaveras, and the CCB sites. This high degree of similarity suggests that there is a hydrological connection between the different valleys that maintains a high level of gene flow. T-RFLP data from various sites showed that at least four phylotypes were abundant and common to the CCB, Calaveras, and El Hundido valleys, even if they are >50 kilometers apart in springs of deep wells. Based on existing T-RFLP databases (University of Idaho and Michigan State Microbial Center), one phylotype corresponds to a marine uncultured proteobacterium (EBAC28E03), another corresponds to the aquatic bacterium Planctomyces (which was also abundant in our cultivated strains), a third corresponds to a taxon that has not been previously described and was retrieved in neither the clone libraries nor in the cultivated strains, and a fourth phylotype was related to Lutibacterium anuloederans (a marine α-proteobacterium from the Adriatic Sea) and to Novosphingobium subantarcticum (a taxon found in permafrost sediments).
Discussion
A Microbial Oasis.
Our data show that the CCB is a “desert oasis” of microbial life that includes highly diverse aquatic communities. The high genotypic diversity found at CCB includes 38 unique phylotypes from 10 major prokaryotic lineages. This high diversity is surprising, given that the oligotrophic waters of the CCB represent sometimes extreme aquatic environments with high concentrations of magnesium, calcium, carbonate, and sulfate (14). The bacterial diversity found in our study is comparable with or greater than the species (or phylotype) richness reported for rhizosphere and soil samples (17, 18, 19). This finding is unexpected, because aquatic systems are generally less species-rich than soils and sediments from temperate regions (20), and it has been reported that, in other extreme environments, such as desiccation lagoons and salty marshes, the community diversity can be as low as 7 phylotypes (21). Although the number of phylotypes per local site at CCB was similar to that reported in some studies performed in extreme sediment or aquatic habitats (22, 23), the high heterogeneity among the individual sampling sites makes the overall diversity at the CCB much higher.
Various mechanisms have been proposed for how high diversity can be maintained in a given locality. For example, high local diversity might occur in an environment that has been stable for a sufficient period to allow for niche differentiation (24) or in instances where there is resource heterogeneity, a superabundance of resources, spatial isolation, or nonequilibrium conditions (25). In addition, periodic physical disturbances have been proposed to result in the maintenance of high diversity in many communities, such as rain forests and coral reefs, where there appears to be an association of high diversity with intermediate levels of disturbance (24). Given the characteristics of the CCB ecosystem, the occurrence of nonequilibrium conditions and resource heterogeneity may be important to sustaining high microbial diversity. Examples of this situation, as discussed earlier, are the older travertine hot-spring deposits and lower-temperature tufa mounds that can be found in dry parts of the basin floor but are also associated with currently active springs, suggesting a very dynamic process of opening and drying of the springs (10). This finding highlights the value of CCB as a convenient natural laboratory to study biodiversity drivers and ecosystem dynamics.
Marine Affinity of CCB Microbiota.
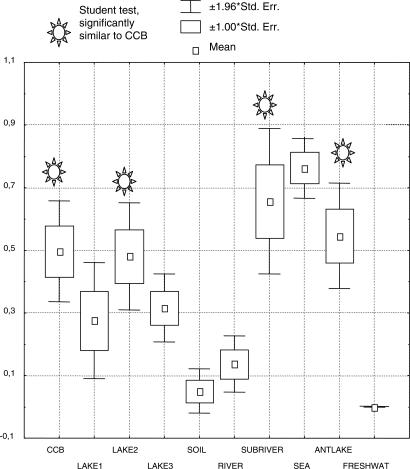
We have found a remarkable, yet unexpected, predominance of taxa with marine microbial affinities in our samples. This finding was surprising, because the chemical composition of the water does not resemble seawater, and the CCB is located 800 km from the Gulf of Mexico. This observation leads us to the hypothesis that some portion of the biota and water of CCB may be derived from microbes and water entrapped when the Mesozoic strata that underlie the CCB were formed and which have been released more recently during active and ongoing subsurface karstification of limestones and evaporites. It is important to note that we have successfully cultured several of the taxa having marine affinities, indicating that our methods have detected live bacteria and not just fossilized DNA sequences. A precedent for this “marine isolation hypothesis” is the observation of long-term survival of bacteria entrapped in Permian-aged evaporites (26, 27) as well as the report of Inagaki et al. (28) on the “Paleome” of DNA from anoxic black shale marine sediments formed 100 million years ago. Supporting the inference of marine affiliation of the microbes of CCB, the results from nearest-neighbor analysis of data from published studies of microbial diversity in diverse environments showed that clones obtained from seawater samples were most closely related to known marine bacteria, whereas samples from continental freshwater had nonmarine prokaryotes, independent of their salinity (see Table 4, which is published as supporting information on the PNAS web site). The analysis of clones from the CCB using the same procedure indicated that a large number of the bacteria from habitats in the CCB were most closely related to marine bacteria. Interestingly, joining CCB as an exception to the general pattern of continental waters being dominated by microbial taxa with freshwater affiliation (Fig. 2) were clones from a saline lake in the Atacama desert of Chile (29). This finding is notable because stratigraphic analysis of the back-arc basin of the Atacama region shows that, as at CCB, there were numerous marine transgressions that isolated the magmatic arc from the mainland of South America during the Jurassic period. At this Chilean site, successive marine regressions were marked by the deposition of widespread evaporites, very similar to the situation in the CCB.
Fig. 2.
Habitat affiliation was tested with a nearest-neighbor analysis, where each cloned sequence was compared with those reported for the most closely related reference sequences in gene sequence databases. The habitat of origin of the reference sequence was assigned to the undescribed population from eight studies from the literature (17, 29, 35, 40–44) as well as our study. Microbial samples were from the following habitats: CCB (springs and deep water from Cuatro Cienegas Coahuila); sulfurous lakes (Cisó and Valar lakes, Spain); saline Atacama lake (a lake in Atacama desert, Northern Chile); saline Mono Lake (California); soil (soils of the arid southwestern United States); Changjiang River (China); a subterranean river (Sulfur River, Parker Cave, Kentucky); sea (marine waters of the Cariaco Basin, Pacific Ocean); hypersaline lakes (Antarctica); and freshwater (water in a drinking water distribution system) (More data available in Table 4).
At the end of the Paleozoic, the supercontinent, Pangea, fragmented to form two great landmasses: Laurasia (north) and Gondwanaland (south). With the separation of Laurentia (North America) from Eurasia during the Jurassic, the North Atlantic and Gulf of Mexico began to open, eventually connecting to the ancient Tethys sea, through the Mediterranean to the Pacific. We believe that most of the marine microbiota of CCB pozas are likely to be relicts from those times, including the microbial mats and the very diverse living estromatolites. In the CCB region, a regional uplift called the Coahuila Island was present throughout the late Jurassic (Tithonan) to early Cretaceous (Neocomian) periods. The sedimentary sequences exposed in the surrounding mountains represent fluvial and shallow marine sediments that accumulated along the margins of Coahuila Island and were deposited on older (pre-Jurassic period) igneous and metamorphic basement rocks during this time, entrapping interstitial marine waters. This depositional history records pulses of uplift and erosion, followed by brief transgressive intervals that deposited shallow marine limestones and sulfates, which comprise the San Marcos formation. Subsurface dissolution of these formations appear to be the source of high sulfate and bicarbonate concentrations of CCB’s surface waters. We also believe that the ancient water composition changed because of the ionic exchange with the surrounding rocks, forcing the ancient marine microorganisms to adapt to this new environment and diverge from their ancestors.
However, other potential hypotheses for the origins of marine taxa at CCB should be considered. Direct subterranean flow from the Gulf of Mexico can be excluded, because the valley is >700 m above sea level. Alternatively, bacteria could immigrate into the CCB by atmospheric transport (via deposition of water droplets or airborne particulate matter), but the extremely low rainfall in the Chihuahuan desert decreases the probability of this means of dispersal, in actuality. However, it should be noted that all indications suggest that the climate was wetter during the late Pleistocene and Holocene. The low probability of long-distance transport of aquatic organisms is also supported by the low number of taxa typical of freshwater environments that were recovered in our samples (Table 2). Previous studies have shown that microbial strains endemic to different continents and, thus, long separated by continental drift, still have closely related 16S rRNA gene sequences (30). The small differences between the 16S rRNA gene sequences from the CCB and reference sequences found in GenBank may simply reflect the amount of time that the CCB waters have been apart from the sea, allowing the marine and CCB taxa to diverge slightly from their closest relatives. Future studies that include more extensive sampling of freshwater and marine environments in Mexico and the southwest USA, coupled with detailed molecular clock and substitution-rate analyses and with dating of fossil water in the CCB, will shed light on the possibility that, indeed, the modern microbiota at the CCB reflect a biotic imprint of an ancient marine past.
Hydrological Connections Between the CCB and Adjacent Valleys and Geological Data.
Another surprising aspect of our data was the very high degree of similarity of lineages of Archaea and Bacteria from adjacent valleys, as indicated by cluster analyses of T-RFLP data showing that 90% of the numerically abundant phylotypes were common to all sites. These findings suggest that lineages were derived from a common source or that there has been migration among the three valleys that share a deep aquifer. The geologic history of the region supports the notion of a possible hydrological connection between adjacent valleys and the CCB. Recent studies on the geohydrology of the region suggest that, although a considerable fraction of the CCB groundwater may originate from local recharge in mountains surrounding the valley, an additional fraction is derived from interbasin flows of older (fossil) water (K. H. Johannesson, personal communication). Most of the warm pozas in the CCB are distributed around the San Marcos Sierra and have very similar water levels and temperature (31), suggesting that they are fed by a common water source. Differences in water chemistry among them are likely due to differences in the local environment and patchy contact with deep hydrothermal water, an aspect that warrants further research confirmation.
The potential for subsurface hydrologic connections suggested by the molecular data are reasonable in light of the known geology of the CCB region, which is part of the southern Basin and Range province of North America. This is basically an extensional tectonic regime characterized by dominantly northwest-trending mountain ranges and valleys bounded by listric normal faults. In central Mexico, three major fault systems are dominant: the Mojave–Sonora megashear (south), the La Babia (north) and the San Marcos (central). CCB lies between the Mojave and Sonora megashear and the San Marcos fault systems. Thus, CCB has experienced a prolonged tectonic history, with a period of faulting that extends back to the Mesozoic to produce a complex system of fractures (13) that likely provide major flow paths for the regional hydrological system that exists in the Coahuila region today. Indeed, under the recent arid climatic regime, karstification may have been especially effective in expanding these pervasive subsurface fault and fracture systems to produce an open hydrological system that interconnects adjacent basins.
Milk, Conservation, and the Future of the CCB.
The possible hydrological interconnectedness and common fossil water between the CCB and the adjacent valleys may hold special significance for the future of the CCB’s surface biota. Similar to situations occurring with increasing frequency in various arid regions of the world (5, 32, 33), agricultural development and associated water extraction in the region have placed new pressures on the ecological integrity of the unique ecosystems of Cuatro Cienegas. In 2001, ranchers associated with two dairy consortia abandoned operations near Torreón because of shortage of water and the presence of arsenic and heavy metals in the dwindling groundwaters. In 2003, the ranchers began operations in the Valle el Hundido (Fig. 3 B), close to the CCB protected area, proceeding without environmental impact assessments, as required by Mexican law. Ten thousand hectares of alfalfa fields with 106 wells were established, based on a claim that there was no relation between the aquifers of the CCB and El Hundido valleys. Concern about the environmental impacts of this water extraction on the CCB protected area has attracted considerable media attention (press releases and articles in Milenio Torreon 1/28/04, La Palabra 3/12/04, and El Universal 4/3/04 and 4/4/04). Consequently, for the first time in the history of Mexican environmental policy, legal injunctions halting this water use were issued. This decision is now in abeyance because of the proposal of a presidential veto (Diario Oficial de la Federacion 11/3/03) that will permit agricultural interests to extract 20 million cubic meters of water a year. These recent events add to the impact of agricultural development over the last 10 years in the northern valley of Calaveras that appear to have dramatically decreased surface water flows into the Cuatro Cienegas valley. Our microbiological data, along with the low hydrologic recharge of the superficial aquifers and geologic structure of the region indicate that serious concerns are warranted regarding the impacts of regional water extraction on the unique ecosystems in the CCB and nearby valleys. Our results also highlight the need to incorporate a regional perspective in legal designations that seek ecosystem conservation (34), because the critical processes and environmental factors sustaining local habitats and biotas can often be distant and complex, a principle applicable to numerous freshwater systems worldwide.
Methods
In recent times, clone libraries of 16S rDNA have been the gold standard to describe microbial communities in many sites around the planet (17, 29, 35–41), however, other techniques, such as T-RFLP and denaturing gradient gel electrophoresis have been proposed as a fast way to evaluate and compare the diversity of a microbial community (42). Both methods give a fingerprint of the community based on differential migration of the different phylotypes in a gel matrix, however, we consider that T-RFLP overcomes most of the problems concerning this type of method because of its high resolution and replicability (43, 44). In this work, three approaches were followed to characterize the prokaryotic diversity of the CCB: (i) a clone library of 16S rRNA genes was constructed from total community DNA, (ii) strains were cultured from samples and characterized, and (iii) T-RFLP profiles of microbial communities were determined.
Sampling.
During 2002, 10 2-liter water samples from eight locations within the three neighboring valleys (two in the CCB and the Rosario mine, three in El Hundido and three in Calaveras; Fig. 3) were filtered by using a standard 0.45-μm filter that will trap most known Eubacteria and Archaea and processed for DNA extraction and analysis (see below). These samples were used to test the hypothesis of hydraulic interconnection among these valleys (by comparing community composition) and to characterize regional genetic diversity. Unfiltered samples were used in the isolation and cultivation of particular organisms from the samples (see below). To assess the microbial diversity in the sediments as well as their origin, 10 g of wet sediments from three Cuatro Cienegas spring pools (pozas) were obtained by sampling 10 cm beneath the sediment–water interface. The global positioning system coordinates of the sampling locations are given in Table 1. The CCB sites are 10 km from each other, whereas Calaveras sites are <10 km from each other and 25 km from the closest CCB site; Mina el Rosario is 43 km from CCB, 59 km from the farthest Calaveras site, and 41 km from the farthest El Hundido site. El Hundido is 39 km from CCB, 55 km from Calaveras farthest site, and 34 km from el Rosario.
Cultivated Bacteria and Archaea.
Marine agar and H medium with different salt concentrations (0–250 g·liter−1) as well as Archaeal media and Luria broth were used to cultivate organisms from water samples. The compositions of the media used can be found at www.atcc.org. Colonies with different morphologies were selected from each medium, and axenic cultures were obtained. (The complete list of the morphotypes is in Table 5, which is published as supporting information on the PNAS web site) Genomic DNA was extracted from colonies of each strain by using the PureGene DNA extraction Kit (Gentra Systems).
Molecular Analysis.
Genomic DNA was extracted from water and sediment samples in the field by using the UltraClean Water DNA kit (MoBio Laboratories, Carlsbad, CA) and the Ultra Clean Soil DNA kit (MoBio Laboratories), respectively. DNA was stored at −20°C.
For T-RFLP analysis, clone library construction from genomic DNA, and the cloning of 16S rRNA genes from cultivated Bacteria and Archaea, a region of the 16S rRNA genes in each sample was PCR amplified by using domain-specific primers (35). The forward primer was F515 (5′-GCGGATCCTCTAGACTGCAGTGCCAGCAGCCGCGGTAA-3′), and the reverse primer was R1492 (5′-GGCTCGAGCGGCCGCCCGGGTTACCTTGTTACGACTT-3′). In the case of T-RFLP, the F515 and R1492 primers were fluorescently labeled with VIC and FAM, respectively. Each PCR reaction contained 1× PCR buffer, 1.65 mM MgCl2, 0.2 mM dNTP mixture, 0.06 mM of each primer, 1 unit of Taq polymerase (Applied Biosystems), and 5% DMSO. All reactions were carried out in a thermocycler (MJ Research, Watertown, MA) with the following program: 94°C for 4 min then 35 cycles of 92°C for 1.5 min, 50°C for 1.5 min, 72°C for 2 min, and completing with 72°C for 10 min. To have a better representation of each sample, three independent PCRs were performed per sample. All were mixed and purified from a 2% agarose gel by using the protocol provided for the QIAquick gel extraction kit (Qiagen).
Clone Libraries.
Amplified 16S rRNA genes were pooled from three reaction mixtures from each site and cloned into the pCR2.1 vector according to the manufacturer’s instructions (Invitrogen). Plasmid DNAs containing inserts were isolated for sequencing with the SNAP Miniprep kit (Invitrogen) and were sequenced by using vector-based primers M13F and M13R by the DNA Laboratory at Arizona State University.
Sequence Analysis.
The partial sequences of 16S rRNA genes from 98 clones from total community DNA and the 350 cultivated strains were initially compared with reference sequences by using blast (45) (www.ncbi.nlm.nih.gov/BLAST) to determine their phylogenetic affiliations and orientation of the cloned inserts. The sequences were manually checked and aligned to 16S rRNA gene sequence data by using the program sequence aligner from the Ribosomal Database Project (RDP) (46). Chimeric sequences were identified by using the check_chimera and bellerephon programs (47), and discrepancies in the phylogenetic trees were identified by comparing the branching order obtained with trees constructed by using two regions of the gene sequences (533–873 and 874-1215, Escherichia coli numbering). The unique partial sequences of 16S rRNA genes (hereafter referred to as phylotypes) were submitted to blast and sequence match at RDP, and the most closely related phylotypes were identified. Neighbor-joining and maximum-parsimony analyses were performed by using mega2 (48) to determine the phylogeny of these populations.
T-RFLP Analysis of 16S rRNA Genes.
The T-RFLP profiles of 16S rRNA genes in samples used to construct clone libraries (described above) were determined. The 16S rRNA genes were amplified from community DNA by using fluorescently labeled PCR primers, as described above, and restricted by using AluI (Promega) in 20-μl reactions for 3 h. Each reaction contained 10 units of AluI and 50 ng of the PCR product. The reactions were incubated in an MJ Research thermocycler for 3 h at 37°C, followed by 65°C for 30 min. The sizes and abundances of fluorescently labeled terminal restriction fragments (T-RFs) were determined by using an ABI 3100 PRISM DNA analyzer (Applied Biosystems). Each T-RF was considered to be an operational taxonomic unit (OTU), and only peaks with heights ≥50 fluorescence units were considered to be true OTUs.
Affiliation Comparisons and Statistical Analysis.
We wished to determine whether the known habitat of closely related organisms could be used to accurately assess the origin of an “undescribed population.” To do so, we used data from eight published studies that had characterized the composition of microbial communities in different saline, freshwater, and soil environments (29, 35 –41) and compare them with our study. The selected studies were the only ones that complied with the following characteristics: The methodology is similar, they present GenBank accession numbers, and they represent different habitats. A nearest-neighbor analysis was performed with each cloned sequence wherein the sequences from these genes were compared with those reported for the most closely related reference sequences in databases. The habitat of origin of the reference sequence was assigned to the undescribed population. The accuracy of the method was quantified by comparing the predicted habitat with the actual habitat of origin. A marine affiliation score was assigned to the five most closely related clones by assigning a value of 1 if a sequence was from a marine environment or a value of 0 if it was not and then averaging across the five taxa. If the sequence similarity was <90%, the data were not used in the calculation. ANOVA was then used to determine whether there was a significant effect of sample site on the mean marine affiliation score for the various studies considered; differences between a particular pair of comparison sites (e.g., CCB vs. Mono Lake) were assessed by a t test. These parametric tests were applied after corroboration of the assumptions of normality and variance homogeneity.
Supplementary Material
Acknowledgments
We dedicate this work to W. J. Minckley. We thank A. Valera, A. Cruz, C. Granados, and R. Cerritos for work on the cultivated strains; J. Domínguez and A. Castillo for work in bioinformatics; M. Kyle, J. Schampel, J. Watts, and all of the personnel of the Area de Protección de Flora y Fauna of Cuatro Cienegas (in particular, Arturo Contreras and Susana Moncada) for logistical and technical support; the La Venta Exploration Team for providing samples from subsurface springs and mines; and M. Travisano, B. Bohanan, and S. Peacock for comments on an early draft. This work was supported by grants from the National Aeronautics and Space Administration Astrobiology Institute, Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO) Grant AE015, and Consejo Nacional Ciencia y Tecnologia, Fondo Sectorial de la Secretaria del Medio Ambiente y Recursos Naturales (CONACyT/SEMARNAT) Grant CO1–0237. CONABIO provided the map that forms the basis of Fig. 3.
Abbreviations
- CCB
Cuatro Cienegas basin
- T-RFLP
terminal restriction site polymorphism.
Footnotes
References
- 1.Postel S. L., Daily G. C., Ehrlich P. R. Science. 1996;271:785–788. [Google Scholar]
- 2.Gleick P. H. Annu. Rev. Environ. Resour. 2003;28:275–314. [Google Scholar]
- 3.Jackson R. B., Carpenter S. R., Dahm C. N., McKnight D. M., Naiman R. J., Postel S. L., Running S. W. Ecol. Appl. 2001;11:1027–1045. [Google Scholar]
- 4.Naiman R. J., Turner M. G. Ecol. Appl. 2000;10:958–970. [Google Scholar]
- 5.Contreras S., Lozano M. L. Conserv. Biol. 1994;8:379–387. [Google Scholar]
- 6.Shepard W. D. Aquat. Conserv. Mar. Freshwater Ecosyst. 1993;3:351–359. [Google Scholar]
- 7.McNeeley J. A. J. Arid Environ. 2003;54:914–928. [Google Scholar]
- 8.Minckley W. L. Proc. Az.–Nv. Acad. Sci.; 1992. pp. 89–119. [Google Scholar]
- 9.Winsborough B. M., Seeler J.-S., Golubic S., Folk R. L., Maguire B., Jr . In: Phanerozoic Stromatolites II. Bertrand-Sarfati J., Monty C., editors. Amsterdam: Kluwer; 1994. pp. 71–100. [Google Scholar]
- 10.Minckley W. L. Univ. Tx. El Paso Sci. Ser. 1969;2:1–65. [Google Scholar]
- 11.Garcia-Pichel F., Al-Horani F. A., Farmer J. D., Ludwig R., Wade B. D. Geobiology. 2004;2:49–57. [Google Scholar]
- 12.Gomez-Pompa A., Dirzo R. Reservas de la Biosfera y Otras Áreas Naturales Protegidas de México. Mexico City: SEMARNAT-CONABIO; 1995. [Google Scholar]
- 13.McKee J. W., Jones N. W., Long L. E. Geol. Soc. Am. Bull. 1990;102:593–614. [Google Scholar]
- 14.Elser J. J., Schampel J. H., Kyle M., Watts J., Carson E. W., Dowling T. E., Tang C., Roopnarine P. D. Freshwater Biol. 2005;50:1808–1825. [Google Scholar]
- 15.Meyer E. R. Ecology. 1973;54:982–995. [Google Scholar]
- 16.Johannesson K. H., Cortes A., Kilroy K. C. J. S. Am. Earth Sci. 2004;17:171–180. [Google Scholar]
- 17.Kuske C. R., Ticknor L. O., Miller M. E., Dunbar J. M., Davis J. A., Barns S. M., Belnap J. Appl. Environ. Microbiol. 2002;68:1854–1863. doi: 10.1128/AEM.68.4.1854-1863.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukow T., Dunfield P. F., Liesak W. FEMS Microbiol. Ecol. 2000;32:1854–1863. doi: 10.1111/j.1574-6941.2000.tb00717.x. [DOI] [PubMed] [Google Scholar]
- 19.Dilly O., Bloem J., Vos A., Munch J. C. Appl. Environ. Microbiol. 2004;70:468–474. doi: 10.1128/AEM.70.1.468-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nee S. TREE. 2003;18:62–63. [Google Scholar]
- 21.Torsvik V., #x00D8;vreåas L., Thingstad T. F. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 22.Konstantinidis K. T., Isaacs N., Fett J., Simpson S., Long D. T., Marsh T. L. Microb. Ecol. 2003;45:191–202. doi: 10.1007/s00248-002-1035-y. [DOI] [PubMed] [Google Scholar]
- 23.Casamayor E. O., Massana R., Benllonch S., Ovreas L., Diez B., Goddard V. J., Gasol J. M., Joint I., Rodriguez-Valera F., Pedros-Alio C. Environ. Microbiol. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. [DOI] [PubMed] [Google Scholar]
- 24.Ricklefs R. E., Miller G. L. Ecology. New York: Freeman; 2000. [Google Scholar]
- 25.Zhou J., Xia B., Treves D. S., Wu L.-Y., Marsh T. L., O’Neill R. V., Palumbo A. V., Tiedje J. M. Appl. Environ. Microbiol. 2002;68:326–334. doi: 10.1128/AEM.68.1.326-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vreeland R. H., Piselli A. F., McDonnough S., Meyers S. S. Extremophyles. 1998;2:321–331. doi: 10.1007/s007920050075. [DOI] [PubMed] [Google Scholar]
- 27.Powers D. W., Vreeland R. H., Rosenzweig W. D. Nature. 2001;411:155–156. [Google Scholar]
- 28.Inagaki F., Okada H., Tsapin A. I., Nealson K. H. Astrobiology. 2005;5:141–153. doi: 10.1089/ast.2005.5.141. [DOI] [PubMed] [Google Scholar]
- 29.Demergasso C, Casamayor E. O., Chong G, Galleguillos P, Escudero L, Pedrós-Alió C. FEMS Microbiol. Ecol. 2004;48:57–69. doi: 10.1016/j.femsec.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Cho J., Tieje J. M. Appl. Environ. Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badino G., Bernabei T., De Vivo A., Giulivo I., Savino G. Under the Desert: The Mysterious Waters of Cuatro Cienegas. Treviso, Italy: Associazione Geografica La Venta; 2004. [Google Scholar]
- 32.Danielpol D. L., Griebler C., Gunatilaka A., Notenboom J. Environ. Conserv. 2003;30:104–130. [Google Scholar]
- 33.Lemly A. D., Kingsford R. T., Thompson J. R. Environ. Manage. 2000;25:485–512. doi: 10.1007/s002679910039. [DOI] [PubMed] [Google Scholar]
- 34.Pringle D. M. Ecol. Appl. 2001;11:981–998. [Google Scholar]
- 35.Angert E. R., Northup D. E., Reysenbach A. L., Peek A. S., Goebel B. M., Pace N. R. Am. Mineral. 1998;83:1583–1592. [Google Scholar]
- 36.Kuske C. R., Barns S. M., Busch J. D. Appl. Environ. Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madrid V. M., Taylor G. T., Scranton M. I., Chistoserdov A. Y. Appl. Environ. Microbiol. 2001;67:1663–1674. doi: 10.1128/AEM.67.4.1663-1674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humayoun S. B., Bano N., Hollibaugh J. T. Appl. Environ. Microbiol. 2003;69:1030–1042. doi: 10.1128/AEM.69.2.1030-1042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekigushi H., Watanabe M., Nakahara T., Xu B., Uchiyama H. Appl. Environ. Microbiol. 2002;68:5142–5150. doi: 10.1128/AEM.68.10.5142-5150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casamayor E. O., Schafer H., Baneras L., Pedros-Alio C., Muyzer G. Appl. Environ. Microbiol. 2000;67:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams M. M., Domingo J. W., Meckes M. C., Kelty C. A., Rochon H. S. J. Appl. Microbiol. 2004;96:954–964. doi: 10.1111/j.1365-2672.2004.02229.x. [DOI] [PubMed] [Google Scholar]
- 42.Bowman J. P., McCuaig R. D. Appl. Environ. Microbiol. 2003;69:2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W. T., Marsh T., Cheng H., Forney L. J. Appl. Environ. Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blackwood C. B., Marsh T., Kim S. H., Paul E. A. Appl. Environ. Microbiol. 2003;69:926–932. doi: 10.1128/AEM.69.2.926-932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul D. F., Gish W., Miller W., Myers E., Lipman D. J. J. Mol. Biol. 1990;215:522–552. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Cole J. R., Chai B., Marsh T. L., Farris R. J., Wang Q., Kulam S. A., Chandra S., McGarrell D. M., Schmidt T. M., Garrity G. M., Tiedje J. M. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber T., Falukner G., Hugenholtz P. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S., Tamura K., Jakobsen I. B., Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.