Abstract
Sexual isolation is a critical form of reproductive isolation in the early stages of animal speciation, yet little is known about the genetic basis of divergent mate preferences and preference cues in young species. Heliconius butterflies, well known for their diversity of wing color patterns, mate assortatively as a result of divergence in male preference for wing patterns. Here we show that the specific cue used by Heliconius cydno and Heliconius pachinus males to recognize conspecific females is the color of patches on the wings. In addition, male mate preference segregates with forewing color in hybrids, indicating a genetic association between the loci responsible for preference and preference cue. Quantitative trait locus mapping places a preference locus coincident with the locus that determines forewing color, which itself is perfectly linked to the wing patterning candidate gene, wingless. Furthermore, yellow-colored males of the polymorphic race H. cydno alithea prefer to court yellow females, indicating that wing color and color preference are controlled by loci that are located in an inversion or are pleiotropic effects of a single locus. Tight genetic associations between preference and preference cue, although rare, make divergence and speciation particularly likely because the effects of natural and sexual selection on one trait are transferred to the other, leading to the coordinated evolution of mate recognition. This effect of linkage on divergence is especially important in Heliconius because differentiation of wing color patterns in the genus has been driven and maintained by natural selection for Müllerian mimicry.
Keywords: Heliconius, Lepidoptera, sexual isolation, speciation
Empirical research on the genetic basis of reproductive isolation has focused largely on hybrid sterility and inviability, which are the result of epistatic incompatibilities between alleles at different loci (1). Although these genomic incompatibilities serve as a major barrier to interspecific gene flow and are important in maintaining the species boundary between taxa that occasionally hybridize, the fact that a variety of closely related species remain distinct in the absence of hybrid dysfunction suggests that postzygotic isolation may not play a significant role in the initial divergence of many species (2–4). Rather, in the early stages of animal speciation reproductive isolation is often behavioral, with young species remaining distinct simply because they do not interbreed (2–4). In instances of apparent sympatric speciation, sexual isolation is commonly a by-product of ecological shifts, such as those to a new host plant or microhabitat (5, 6). Assortative mating can also result from divergence in mate preference, which may be a consequence of indirect selection, such as selection against hybridization (7, 8), or direct selection on alleles at preference loci (9, 10). Despite their importance in generating species diversity, little is known about the genetic basis of traits that limit mating between species or the evolutionary processes that drive their divergence.
The basic genetic architecture of sexual isolation can profoundly impact the process of speciation but remains largely unexplored in most natural systems. In particular, genetic associations among traits conferring reproductive isolation, in the forms of linkage disequilibrium, physical linkage, and pleiotropy, can facilitate divergence by transferring the effects of natural and sexual selection on some traits to others, resulting in the coordinated evolution of a suite of characteristics that together limit mating between incipient species (11–15). Genetic linkage of sexual isolating traits is particularly widespread in Lepidoptera, where a disproportionately large number of traits distinguishing closely related species are sex-linked (16–18). The neotropical genus Heliconius, a species-rich clade of warningly colored and mimetic butterflies, stands in contrast to other Lepidoptera in that none of the ≈40 wing patterning loci that distinguish various races and species are sex-linked (19, 20). In addition to advertising their unpalatability to predators, these bright wing patterns serve as a cue in male mate choice, with male preference tuned to the appropriate conspecific phenotype (4, 21, 22). Thus, Heliconius offers a rare opportunity to examine the degree to which the two components of mate recognition, mate preference and preference cue, are genetically associated in an active Lepidopteran radiation, independent of sex-linkage.
To explore the relationship between different aspects of mate recognition, we focused on the genetic basis of divergent male mate preference and preference cues in Heliconius cydno galanthus and Heliconius pachinus from Costa Rica. These two species, which diverged within the last 500,000 years, are completely interfertile and occasionally hybridize around Costa Rica's Meseta Central (23), the one location where their adjacent distributions meet. Using a combination of mate choice experiments and genetic mapping, we (i) measured the extent to which males of each species prefer to approach and court conspecific females, (ii) searched for discrete wing pattern elements that mediate male preference, (iii) mapped the locus responsible for the wing pattern preference cue as well as quantitative trait loci (QTL) for male preference, and (iv) measured the association between preference and preference cue in the polymorphic race H. cydno alithea. The results of these analyses point to a tight genetic association between mate preference and preference cue in this rapidly radiating clade.
Results
H. cydno and H. pachinus Males Recognize Conspecific Females Based on Wing Color.
H. cydno and H. pachinus males preferentially approach and court conspecific females. When presented with the mounted bodies of an H. cydno and H. pachinus female, only 1 of 140 total approaches by 45 H. cydno and 25 H. pachinus males (Table 1, which is published as supporting information on the PNAS web site) was directed at the heterospecific female (G1 > 72.08, P < 0.0001, for both comparisons). To isolate the specific cue used by males to recognize conspecific females, we presented males with mounted females displaying four combinations of color and pattern: an unaltered white H. cydno, an H. cydno with the white forewing portion colored yellow, an unaltered yellow striped H. pachinus, and an H. pachinus with the yellow stripes colored white (Fig. 1A). For this experiment we recorded a total of 132 approaches by 65 males (Table 2, which is published as supporting information on the PNAS web site). Males of both species exhibited a pronounced color preference: H. cydno males preferentially approached both white females whereas H. pachinus males approached both yellow females, regardless of female wing pattern per se (Fig. 1A). This male color preference translates into a real mating difference. In an experimental population consisting of H. cydno males, unaltered yellow H. pachinus females, and altered white H. pachinus females (Table 3, which is published as supporting information on the PNAS web site), five of five independent matings were with altered white females (P = 0.0417, exact probability).
Fig. 1.
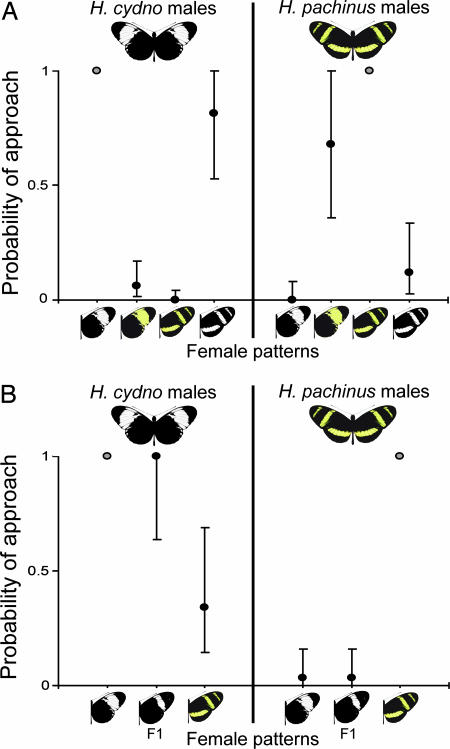
Probability of H. cydno and H. pachinus males approaching female wings displaying various combinations of color and pattern. (A) When given the choice of approaching females displaying four combinations of color and pattern, H. cydno males were as likely to approach the altered white H. pachinus as they were to approach their conspecific female (G1 = 0.93, P = 0.3349). They were significantly less likely to approach either the unaltered H. pachinus or altered yellow H. cydno female (G1 > 51.76, P < 0.0001, for both comparisons). In contrast, the probability of H. pachinus males approaching the yellow H. cydno female was equal to that of their conspecific female (G1 = 1.53, P = 0.2161), whereas the probabilities for both white females were lower (G1 > 20.14, P < 0.0001, for both comparisons). (B) Males of both species responded to F1 female wings (H. pachinus female × H. cydno male) as they did to those of H. cydno. The probability of H. cydno males approaching the hybrid wings was equal to that of approaching the wings of the conspecific female (G1 = 0.00, P = 1.00), whereas H. pachinus males preferred conspecific wings over both H. cydno and hybrid wings (G1 = 34.14, P < 0.0001, for both comparisons). In each experiment, approach probabilities were estimated relative to the probability of approaching the conspecific female (gray point), which was set to 1. Support limits are shown around each estimated probability.
The white/yellow color shift between H. cydno and H. pachinus has a simple genetic basis; a single autosomal locus, the K locus (24, 25), with a dominant white and recessive yellow allele, controls the color of the forewing, and a second, independent locus controls a similar white/yellow switch on the hindwing (23). No area of white is apparent on the hindwing of H. cydno galanthus because an epistatic “shutter” allele at an additional locus replaces these scales with melanic scales, producing a black dorsal hindwing (23). Because the white forewing and hindwing shutter alleles of H. cydno are dominant, first-generation hybrids between H. cydno and H. pachinus display colors similar to H. cydno, but patterns that are intermediate. When we presented males with dissected wings from H. cydno, H. pachinus, and F1 hybrids, we recorded a total of 87 approaches by 53 males (Table 4, which is published as supporting information on the PNAS web site). Consistent with color being the critical mate preference cue, males of both species responded to hybrid female wings in the same way they responded to H. cydno wings; H. cydno males approached hybrid wings as often as they approached H. cydno wings, and H. pachinus males avoided hybrid wings as they did H. cydno wings (Fig. 1B).
Male Color Preference Is Linked to the Forewing Color Locus.
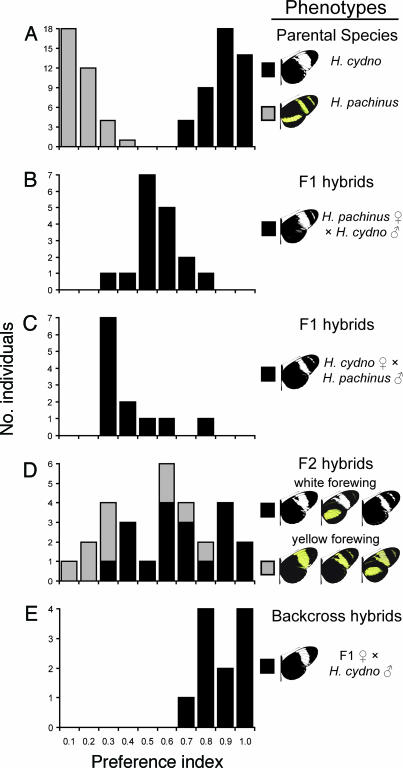
We next examined the genetic basis of male mate preference by testing hybrid males for their preference of H. cydno and H. pachinus, this time by allowing males to court live females. For this experiment we recorded 1,169 courtships and attempted matings by 69 hybrid males as well as 1,054 courtship and attempted matings by 45 H. cydno and 35 H. pachinus males (Table 5, which is published as supporting information on the PNAS web site). The segregation of preference in hybrids suggests that preference is not learned and that it has a simple genetic basis (Fig. 2). For instance, F1 males exhibited preference intermediate to the parental species (Fig. 2 B and C), indicating largely additive genetic variance. Furthermore, the fact that F1 hybrids had color patterns similar to pure H. cydno but both F1 broods differed from pure H. cydno in preference (G1 > 187.63, P < 0.0001, for both comparisons) excludes the possibility that males were “self-matching” (i.e., learning to court or imprinting on their own color pattern). Interestingly, the two F1 broods differed from one another (G1 = 10.50, P = 0.0012), suggesting some directional effect on mate preference. The trimodal distribution in the F2 (Fig. 2D) and the skew in backcross male preference toward H. cydno females (Fig. 2E) indicate that mate preference is largely controlled by few loci. Consistent with these observations, the Castle–Wright estimator of the effective number of loci responsible for male preference was 1.35 ± 0.46. In addition, there was a relationship between forewing color and preference in the F2 brood (Fig. 2D). As a group, males with a yellow forewing (homozygous for the yellow allele) preferred H. pachinus females (G1 = 19.04, P < 0.0001) whereas males with a white forewing (homozygous or heterozygous for the white allele) preferred H. cydno females (G1 = 10.27, P = 0.0014). This relationship indicates physical linkage between a preference locus and the locus that controls forewing color.
Fig. 2.
Segregation of male mate preference in hybrids between H. cydno and H. pachinus. The preference index (x axis) is the proportion of courtship and attempted mating events that were directed toward H. cydno females; a preference index of 1 indicates complete preference for H. cydno (white species), whereas an index of 0 indicates complete preference for H. pachinus (yellow species). (A) Parental populations, H. pachinus (gray bars) and H. cydno (black bars). (B) F1 hybrid males: H. pachinus female × H. cydno male. (C) F1 hybrid males from the reciprocal cross: H. cydno female × H. pachinus male. (D) F2 males produced by interbreeding progeny from the first F1 brood, grouped by yellow forewing (gray bars) and white forewing (black bars). (E) males produced by backcrossing a female from the first F1 brood to a pure H. cydno male.
Color and Color Preference Map to the Genomic Location of the Developmental Gene wingless (wg).
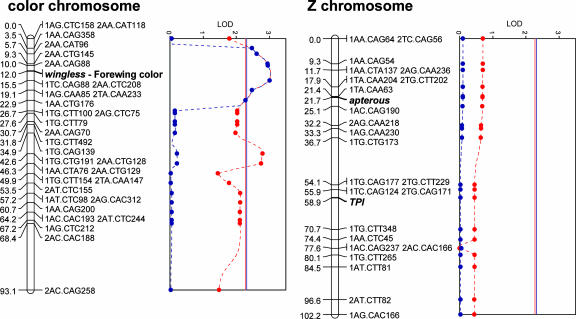
To determine how tightly male mate preference is linked to forewing color, we mapped the chromosome that contains the forewing color gene, and we also mapped the Z chromosome to identify the portion of preference that is sex-linked. As part of the genetic mapping we surveyed allelic variation at two Z-linked single copy nuclear loci, Triose phosphate isomerase (26) and apterous (27), and one wing patterning candidate gene, wg. In another Nymphalid butterfly, Junonia coenia, wg is up-regulated in bands on the developing forewings that eventually contain an orange/red ommochrome pigment, Ommatin-D (28, 29). The yellow pigment on Heliconius wings is also an ommochrome (3-hydroxykynurenine) whereas white is structural (30), making wg a potential candidate for the ommochrome on/off switch that distinguishes H. cydno and H. pachinus. In our sample of 65 F2 hybrids, wg was completely linked to forewing color: all 25 offspring with a yellow forewing were homozygous for the H. pachinus wg allele, and all 40 offspring with a white forewing were either homozygous or heterozygous for the H. cydno wg allele. This association, combined with the spatial pattern of wg expression on the developing wings of J. coenia (28), tentatively implicates wg as the white/yellow switch in Heliconius but further fine-scale genetic mapping and gene expression studies in Heliconius will be required to make a definite link between wing color and wg.
After genotyping the F2 progeny at Tpi, ap, wg, and the mapped amplified fragment length polymorphism (AFLP) loci, we searched for regions of both chromosomes associated with male courtship preference (QTL) using interval mapping (31) and composite interval mapping (32). Although interval mapping detected two potential QTL on the color chromosome, courtship preference was most strongly associated with allelic variation at wg (Fig. 3). Furthermore, there was no detectable association with any of the loci on the Z chromosome (Fig. 3).
Fig. 3.
QTL mapping of male mate preference. Logarithm of odds (LOD) profiles, estimated with both interval mapping (dashed red) and composite interval mapping (dashed blue), are shown alongside genetic linkage maps of the chromosome containing the forewing color locus and the Z chromosome. The 0.05 significance thresholds for interval mapping (solid red) and composite interval mapping (solid blue) are shown. On the chromosome maps, loci are labeled with position in cM (Left) and locus name (Right). Names of AFLP loci begin with a 1 or 2 indicating the phase in the H. cydno male parent.
Wing Color and Color Preference Are Correlated in H. cydno alithea.
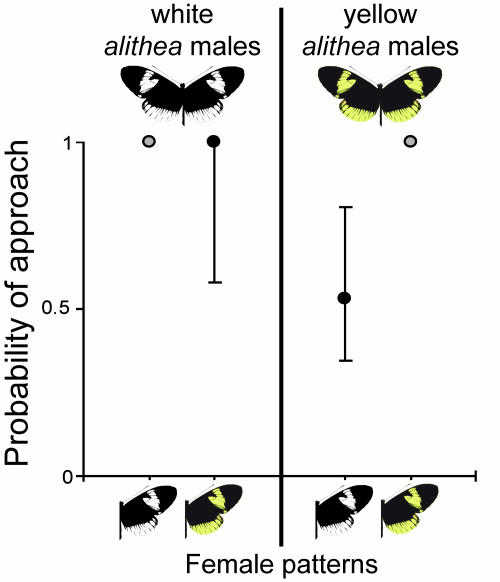
To further characterize the nature of the association between color and preference, we tested H. cydno alithea males for their courtship preference of yellow and white H. cydno alithea females. H. cydno alithea is a naturally polymorphic race from Ecuador that segregates for the same white/yellow switch that distinguishes H. cydno and H. pachinus in Costa Rica (33). These two color morphs mate randomly in nature: field-caught females of either color produce progeny of both colors, and allelic variation at the color locus is in Hardy–Weinberg equilibrium in the wild (34). We recorded a total of 138 courtships and attempted matings by 9 white and 14 yellow H. cydno alithea males (Table 6, which is published as supporting information on the PNAS web site). We expect that if color and preference were controlled by different loci in close proximity on the chromosome, random mating over many generations would erase the correlation between them. In contrast, we found that whereas white males (homozygous or heterozygous for the white allele) had no detectable color preference, yellow males significantly preferred yellow females (Fig. 4). Although we do not know whether white males were homozygous or heterozygous at the color locus, given that the frequency of the yellow allele in the experimental population was ≈0.7, we expect that a majority of them were heterozygotes.
Fig. 4.
Probability of white and yellow H. cydno alithea males courting white and yellow H. cydno alithea females. Although white males (homozygous or heterozygous for the white allele) courted the two female types equally (G1 = 0.00, P = 1.00), yellow males were twice as likely to court yellow females (G1 = 9.33, P = 0.0023). For each male type, probability of between-morph courtship was estimated relative to the probability of within-morph courtship (gray point), which was set to 1. Support limits are shown around each estimated probability.
Discussion
Our experimental results show that wing color plays a critical role in mediating assortative mate preference in Heliconius butterflies and that, as a consequence, the white/yellow color shift that distinguishes H. cydno galanthus from H. pachinus serves as a major barrier to reproduction between the two species. Furthermore, the wing color preference cue is tightly linked to male color preference at the location of the developmental gene wg, and the association between color and preference persists in the polymorphic race H. cydno alithea despite many generations of random mating. The fact that these traits are linked on an autosome is surprising given the disproportionate influence of sex-linked genes in distinguishing other Lepidopteran species (16–18).
A question that arises from these results is, what mechanism causes the linkage between cue and preference? As a whole, the data suggest that the association between color and preference is not simply the result of two tightly linked loci. Rather, the results are most consistent with a genetic structure that prevents recombination between color and preference. Perhaps the loci for color and preference are located in a region of reduced recombination, such as an inversion, which have been shown to play an important role in maintaining genetic isolation between other hybridizing species (35–38). For instance, in some fruit flies (Drosophila) and sunflowers (Helianthus), characters important for reproductive isolation map to inversions that distinguish recently diverged species, and these regions show evidence of reduced interspecific gene flow (35, 39, 40). Although there may be an inversion in the color/preference/wg region that distinguishes H. cydno galanthus from H. pachinus and the white H. cydno alithea morph from the yellow morph, it cannot be very large. Our cydno/pachinus F2 brood showed evidence of recombination between wg and both flanking markers on the H. cydno map, limiting the size of a potential inversion to this 5.5-cM window.
An alternative explanation for the association between color and preference is that these are pleiotropic effects of a single gene. In general there is little reason to expect that the same gene or suite of genes would influence both a morphological trait used in mate recognition and the sensory processes used to recognize that trait (12, 41, 42). In fact, outside of the recent demonstration that the gene desat1 influences both production and discrimination of sex pheromones in Drosophila melanogaster (43), there is no experimental evidence to support such “genetic coupling” in mate recognition. However, in addition to providing the color for Nymphalid butterfly wings, ommochrome pigments are present in the eyes of all insects where, as screening pigments, they influence spectral sensitivity by absorbing and transmitting different wavelengths of light (44, 45). Conceivably, a single gene could influence both the distribution of 3-hydroxykynurenine on the wing and the distribution of 3-hydroxykynurenine or another ommochrome in the eye, thereby influencing sensitivity to color. Although genetic coupling may be highly unlikely generally, the dual role of ommochromes in this system provides a potential link between the processes of signal production and signal reception.
Regardless of the specific nature of the association, the fact that wing color and color preference are very tightly linked genetically has important evolutionary consequences. Wing pattern diversification in the genus Heliconius has been driven by natural selection for Müllerian mimicry (33, 46, 47). Because wing color serves as a critical cue in mate recognition, and male preference for wing color is linked to color, natural selection on wing patterns that is imposed by predators will simultaneously drive the divergence of both mate recognition signal and signal preference. This scenario, which provides a direct link between disruptive natural selection and divergence at loci mediating conspecific recognition and mate choice, facilitates the rapid evolution of sexual isolation. Furthermore, the association between color and preference limits recombination between the two components of mate recognition when species and color pattern races hybridize, a common phenomenon in the genus Heliconius, thus allowing phenotypic differentiation to persist in the face of hybridization. Given the effects of this association on the origin and maintenance of diversity, it has undoubtedly played a significant role in facilitating the explosive adaptive radiation of Heliconius butterflies.
Materials and Methods
Experimental Butterflies.
We established six Heliconius populations in greenhouses at the University of Texas at Austin: three Costa Rican populations of H. cydno galanthus, one from the Organization for Tropical Studies' La Selva Biological Station, one from the town of Guacimo, and one from the town of Vesta; two Costa Rican populations of H. pachinus, one from the town of Santiago de Puriscal and one from Corcovado National Park; and one polymorphic population of H. cydno alithea from western Ecuador. Field-caught population founders were not used in experiments.
Mounted Female Experiments.
To identify the specific wing color pattern characters that males use to identify conspecific females we performed three experiments in which we counted male approaches to mounted females. First, we presented males with one mounted H. cydno and H. pachinus female. Second, we presented males with females representing the four possible combinations of color and pattern. The yellow H. cydno was created by introgressing, from Heliconius melpomene rosina into a population of H. cydno, the allele that colors the forewing yellow. The white H. pachinus was created by washing the wings of an H. pachinus female with acidified methanol, which dissolves the yellow pigment, 3-hydroxykynurenine, leaving the structural white color unaltered. Finally, we presented males with three pairs of female wings: one from H. cydno, one from H. pachinus, and one from a first-generation hybrid (H. pachinus female × H. cydno male). For each experiment, females were placed 20–25 cm apart in full sunlight in each of the population greenhouses, during which time we counted approaches by each male to each female type. Only approaches that were unequivocally directed toward one female were recorded. Males for these three experiments had contact with conspecific females before being tested. Trials, which lasted 30 min, were repeated one to eight times depending on experiment and population.
For analysis, we used likelihood (21, 22, 48) and the approach data for each male to estimate, for each species, the probability of approach to each female type relative to the probability of conspecific approach. The likelihood function was
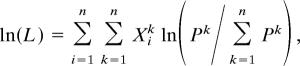 |
where Xik = number of approaches by male i to female type k (k = 1 for the conspecific female) and Pk = relative probability of approach to female type k.
We estimated the relative probability of approach to each experimental female by setting the probability of conspecific approach to one (P1 = 1) and numerically searching for values of Pk that maximized ln(L), using the Solver option in excel (Microsoft). Support limits, which are asymptotically equivalent to 95% confidence intervals, were estimated for each parameter by numerically searching for values that decreased the ln(L) by 2 units (48). For each experiment we tested the hypothesis that the probability of approach to each experimental female was equal to the probability of conspecific approach by comparing the maximized ln(L) to that with P1 = Pk = 1, using a likelihood ratio test with one degree of freedom, assuming a χ2 distributed test statistic.
Color Choice Mating Experiment.
We isolated 7–10 H. cydno males with 1–5 yellow and 1–4 white virgin H. pachinus females on seven nonconsecutive days and monitored all mating (Table 3). Mating couples were removed from the experiment, so all observations were independent. The wings of all females were treated with acidified methanol, but 3-hydroxykynurenine was only dissolved out of those in the white treatment group. This experiment was performed with a second, independent H. cydno population originating from the Organization for Tropical Studies' La Selva Biological Station and a second, independent H. pachinus population originating from Corcovado National Park.
Courtship Experiments.
We preference-tested pure H. cydno and H. pachinus males as well as males from four cydno× pachinus hybrid broods for their courtship preference of live H. cydno and H. pachinus females. All hybrid males and 32 pure males were virgins that had not had contact with females before the start of the experiment. The other 48 pure males were selected from population cages and thus had prior contact with conspecific females. To measure courtship preference, males were placed together in a 2 × 2 × 2-m cage, where they were presented with a live H. cydno and H. pachinus female at the same time. After courting or attempting to mate a female the males were caught, and their identity was recorded. The Castle–Wright estimator and standard error were calculated according to ref. 49 by using angular equivalents of arcsine square root-transformed preference indices, which were computed by using the equation t(p) = (360/2π)(arcsin ) where y = number of courtships/attempted matings directed at the H. cydno female and n = total number courtships/attempted matings.
Using the same experimental design, we tested white and yellow H. cydno alithea males (all had prior contact with conspecific females of both colors) for their courtship preference of live, virgin white and yellow H. cydno alithea females. We analyzed the preference data for yellow and white F2 males and H. cydno alithea males with the likelihood model described previously, but with courtships/attempted matings in place of approaches.
We compared courtship probabilities among H. cydno males and males from the two F1 hybrid broods using the likelihood function
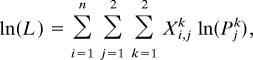 |
where Xi,j1 = number of courtships by male i from population j toward H. cydno females, Xi,j2 = number of courtships by male i from population j toward H. pachinus females, Pj1 = probability of males from population j courting H. cydno females, and Pj2 = probability of males from population j courting H. pachinus females = 1 − Pj1.
For each pairwise comparison among pure H. cydno and the two F1 broods we compared the ln(L) with the observed probabilities to that with the courtship probabilities set equal between the two populations (P11 = P21) using a likelihood ratio test with one degree of freedom, assuming a χ2 distributed test statistic.
Linkage and QTL Mapping.
Using a pseudotestcross design (50) we mapped AFLP loci that were heterozygous in the H. cydno male parent based on segregation data from 33 F1 hybrids. AFLP loci were generated by using the ABI AFLP plant mapping kit (PE Applied Biosystems) and linkage mapping was performed with joinmap 3.0 (51). We typed three single-copy nuclear loci using PCR product length variation (Tpi and ap) or SNPs (wg) to follow male-derived alleles in F1 hybrids. Using allelic variation at the 3 single-copy nuclear loci and 56 AFLP loci, we inferred each F2's genotype at 24 positions along the chromosome that contains the color locus and 20 positions along the Z chromosome. Completely linked AFLP loci in opposite phases were used to score each individual as marker present (homozygous or heterozygous for the H. cydno allele) or marker absent (homozygous for the H. pachinus allele) whereas AFLP loci without a linked marker in the opposite phase were scored as marker present or missing data. We used this scoring scheme because the F2 brood was generated by randomly crossing multiple males and females from the first F1 brood. Although all F2 offspring shared the same H. cydno grandfather and H. pachinus grandmother, we did not assign specific parents to each. Using windows qtl cartographer 2.5 (52) we searched for QTL associated with preference using arcsine square root-transformed preference indices from 29 preference-tested F2 hybrid males. QTL significance thresholds were estimated empirically by 1,000 permutations (53). Linkage maps and logarithm of odds profiles were drawn with mapchart 2.1 (54).
Supplementary Material
Acknowledgments
We thank L. Bae, L. Blume, K. Kronforst, R. Miller, J. Sorrentino, J. Van Reet, and A. Vega for assistance collecting, rearing, and/or testing butterflies. We also thank J. Strassmann and D. Queller for use of laboratory facilities, officials from Costa Rica and Ecuador for collecting and export permits, officials at the U.S. Department of Agriculture Animal and Plant Health Inspection Service for import and breeding permits, and two reviewers for comments on the manuscript. This work was supported by National Science Foundation Grants DEB 0206613 and DEB 0415718, the University of Texas at Austin graduate program in Zoology/Ecology, Evolution, and Behavior, and an Organization for Tropical Studies' graduate research fellowship.
Abbreviations
- QTL
quantitative trait locus/loci
- AFLP
amplified fragment length polymorphism.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Orr H. A., Presgraves D. C. BioEssays. 2000;22:1085–1094. doi: 10.1002/1521-1878(200012)22:12<1085::AID-BIES6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 2.Coyne J. A., Orr H. A. Evolution. 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 3.Coyne J. A., Orr H. A. Evolution. 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- 4.McMillan W. O., Jiggins C. D., Mallet J. Proc. Natl. Acad. Sci. USA. 1997;94:8628–8633. doi: 10.1073/pnas.94.16.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feder J. L., Opp S. B., Wlazlo B., Reynolds K., Go W., Spisak S. Proc. Natl. Acad. Sci. USA. 1994;91:7990–7994. doi: 10.1073/pnas.91.17.7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schluter D. In: Endless Forms: Species and Speciation. Howard D. J., Berlocher S. H., editors. Oxford: Oxford Univ. Press; 1998. pp. 114–129. [Google Scholar]
- 7.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia Univ. Press; 1940. [Google Scholar]
- 8.Howard D. J. In: Hybrid Zones and the Evolutionary Process. Harrison R. G., editor. New York: Oxford Univ. Press; 1993. pp. 46–69. [Google Scholar]
- 9.Servedio M. R. Evolution. 2001;55:1909–1920. doi: 10.1111/j.0014-3820.2001.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 10.Albert A. Y. K., Schluter D. Evolution. 2004;58:1099–1107. doi: 10.1111/j.0014-3820.2004.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoy R. R., Hahn J., Paul R. C. Science. 1977;195:82–84. doi: 10.1126/science.831260. [DOI] [PubMed] [Google Scholar]
- 12.Butlin R. K., Ritchie M. G. Biol. J. Linn. Soc. 1989;37:237–246. [Google Scholar]
- 13.Bakker T. C. M., Pomiankowski A. J. Evol. Biol. 1995;8:129–171. [Google Scholar]
- 14.Hawthorne D. J., Via S. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- 15.Via S., Hawthorne D. J. Genetica. 2005;123:147–156. doi: 10.1007/s10709-004-2731-y. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie M. G., Phillips S. D. F. In: Endless Forms. Howard D. J., Berlocher S. H., editors. New York: Oxford Univ. Press; 1998. pp. 291–308. [Google Scholar]
- 17.Sperling F. A. H. Can. Entomol. 1994;126:807–818. [Google Scholar]
- 18.Prowell D. P. In: Endless Forms. Howard D. J., Berlocher S. H., editors. New York: Oxford Univ. Press; 1998. pp. 309–319. [Google Scholar]
- 19.Sheppard P. M., Turner J. R. G., Brown K. S., Benson W. W., Singer M. C. Philos. Trans. R. Soc. London B. 1985;308:433–613. [Google Scholar]
- 20.Nijhout H. F. The Development and Evolution of Butterfly Wing Patterns. Washington, DC: Smithsonian Institute; 1991. [Google Scholar]
- 21.Jiggins C. D., Naisbit R. E., Coe R. L., Mallet J. Nature. 2001;411:302–305. doi: 10.1038/35077075. [DOI] [PubMed] [Google Scholar]
- 22.Jiggins C. D., Estrada C., Rodrigues A. J. Evol. Biol. 2004;17:680–691. doi: 10.1111/j.1420-9101.2004.00675.x. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert L. E. In: Ecology and Evolution Taking Flight: Butterflies as Model Systems. Boggs C. L., Watt W. B., Ehrlich P. R., editors. Chicago: Univ. of Chicago Press; 2003. pp. 281–318. [Google Scholar]
- 24.Linares M. In: Tropical Biodiversity and Systematics: Proceedings of the International Symposium on Biodiversity and Systematics in Tropical Ecosystems. Ulrich H., editor. Bonn, Germany: Zoologisches Forschungsinstitut und Museum Alexander Koenig; 1997. pp. 93–108. [Google Scholar]
- 25.Naisbit R., Jiggins C. D., Mallet J. Evol. Dev. 2003;5:269–280. doi: 10.1046/j.1525-142x.2003.03034.x. [DOI] [PubMed] [Google Scholar]
- 26.Tobler A., Kapan D., Flanagan N. S., Gonzalez C., Peterson E., Jiggins C. D., Johnston J. S., Heckel D. G., McMillan W. O. Heredity. 2005;94:408–417. doi: 10.1038/sj.hdy.6800619. [DOI] [PubMed] [Google Scholar]
- 27.Jiggins C. D., Mavarez J., Beltrán M., McMillan W. O., Johnston J. S., Bermingham E. Genetics. 2005;171:557–570. doi: 10.1534/genetics.104.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll S. B., Gates J., Keys D. N., Paddock S. W., Panganiban G. E. F., Selegue J. E., Williams J. A. Science. 1994;265:109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- 29.Nijhout H. F. Arch. Insect Biochem. 1997;36:215–222. [Google Scholar]
- 30.Gilbert L. E., Forrest H. S., Schultz T. D., Harvey J. J. Res. Lepidoptera. 1988;26:141–160. [Google Scholar]
- 31.Lander E. S., Botstein D. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng Z. Proc. Natl. Acad. Sci. USA. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapan D. D. Nature. 2001;409:338–340. doi: 10.1038/35053066. [DOI] [PubMed] [Google Scholar]
- 34.Kapan D. D. Ph.D. thesis. Vancouver, BC, Canada: Univ. of British Columbia; 1998. [Google Scholar]
- 35.Noor M. A. F., Grams K. L., Bertucci L. A., Reiland J. Proc. Natl. Acad. Sci. USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieseberg L. Trends Ecol. Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- 37.Ortiz-Barrientos D., Reiland J., Hey J., Noor M. A. F. Genetica. 2002;116:167–178. [PubMed] [Google Scholar]
- 38.Butlin R. K. Mol. Ecol. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 39.Rieseberg L. H., Whitton J., Gardner K. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machado C. A., Kliman R. M., Markert J. A., Hey J. Mol. Biol. Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- 41.Coyne J. A., Orr H. A. Philos. Trans. R. Soc. London B. 1998;353:287–305. doi: 10.1098/rstb.1998.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moehring A. J., Li J., Schug M. D., Smith S. G., deAngelis M., Mackay T. F. C., Coyne J. A. Genetics. 2004;167:1265–1274. doi: 10.1534/genetics.103.024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcillac F., Grosjean Y., Ferveur J. Proc. Biol. Sci.; 2005. pp. 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linzen B. Adv. Insect Physiol. 1974;10:117–246. [Google Scholar]
- 45.Briscoe A. D., Chittka L. Annu. Rev. Entomol. 2001;46:471–510. doi: 10.1146/annurev.ento.46.1.471. [DOI] [PubMed] [Google Scholar]
- 46.Benson W. W. Science. 1972;176:936–939. doi: 10.1126/science.176.4037.936. [DOI] [PubMed] [Google Scholar]
- 47.Mallet J., Barton N. H. Evolution. 1989;43:421–431. doi: 10.1111/j.1558-5646.1989.tb04237.x. [DOI] [PubMed] [Google Scholar]
- 48.Edwards A. W. F. Likelihood. Cambridge, U.K.: Cambridge Univ. Press; 1972. [Google Scholar]
- 49.Lande R. Genetics. 1981;99:541–553. doi: 10.1093/genetics/99.3-4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grattapaglia D., Sederoff R. Genetics. 1994;137:1121–1137. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Ooijen J. W., Voorrips R. E. joinmap 3.0, Software for the Calculation of Genetic Linkage Maps. Wageningen, The Netherlands: Plant Res. Int.; 2001. [Google Scholar]
- 52.Wang S., Basten C. J., Zeng Z. B. windows qtl cartographer 2.5. Raleigh: North Carolina State Univ.; 2005. [Google Scholar]
- 53.Doerge R. W., Churchill G. A. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voorrips R. E. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.